Cell-Free Tumor DNA (ctDNA) Utility in Detection of Original Sensitizing and Resistant EGFR Mutations in Non-Small Cell Lung Cancer (NSCLC)
Abstract
:1. Introduction
2. Materials and Methods
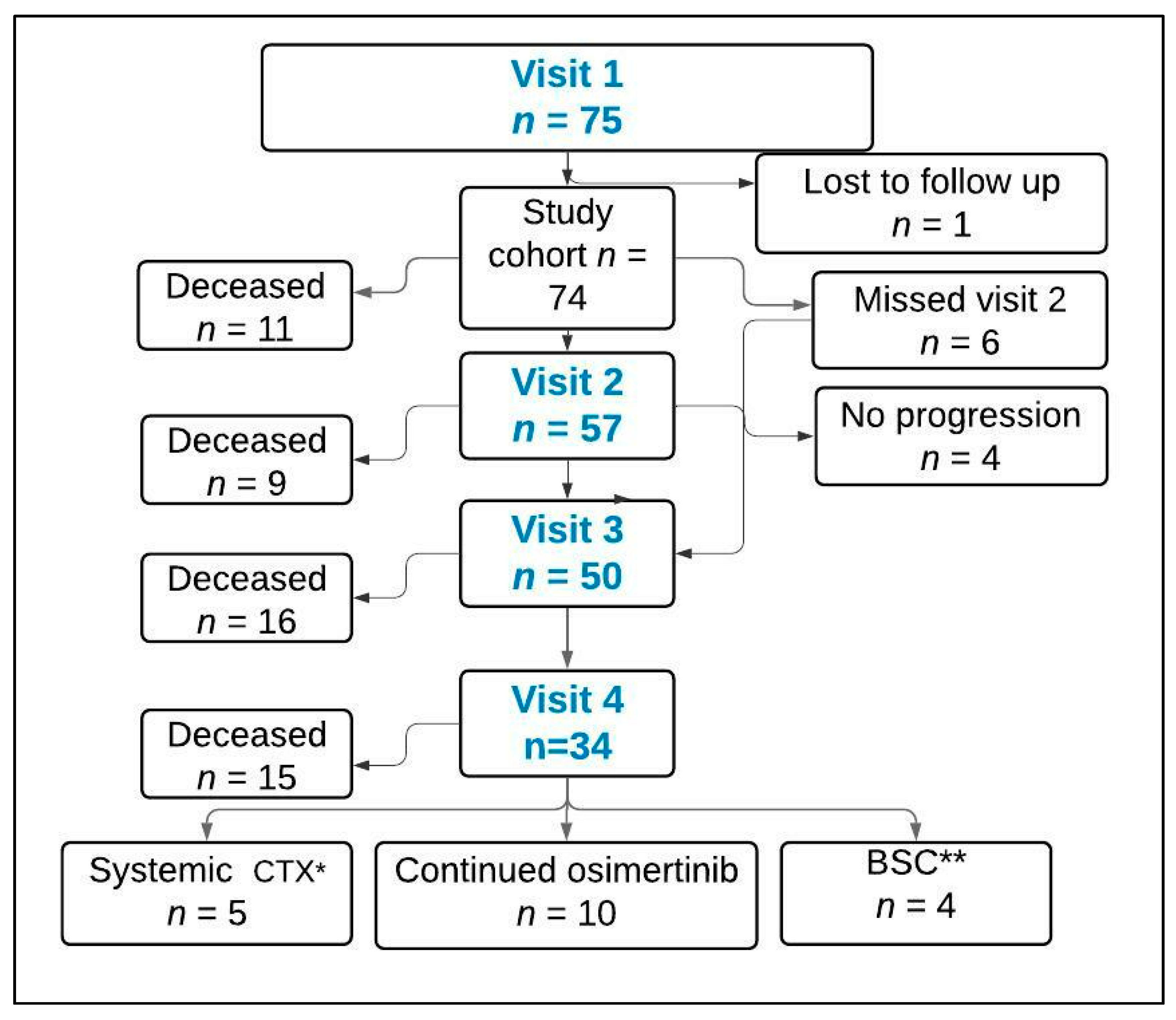
- The 1st cohort participants were TKI-treated patients. These patients were currently on EGFR-TKI therapy for EGFRm metastatic NSCLC and had not yet experienced disease progression on first-line treatment. The mean time of enrollment from the start of EGFR-TKI in this cohort was 10.5 (range 3–44) months. Four longitudinal blood samples were obtained throughout the course of treatment: at the time of enrollment (baseline), at the time of 1st follow up CT scan, at the time of progression, and 1–3 months after starting second-line therapy (Table 1).
- The 2nd cohort participants were TKI-naive newly diagnosed EGFRm metastatic NSCLC. The same 4 longitudinal blood samples were obtained throughout the course of treatment, with the exception of the 1st sample, which was drawn prior to TKI treatment (baseline) (Table 1).
3. Results
3.1. Patient Characteristics
3.2. Original Mutation (OM) Detection
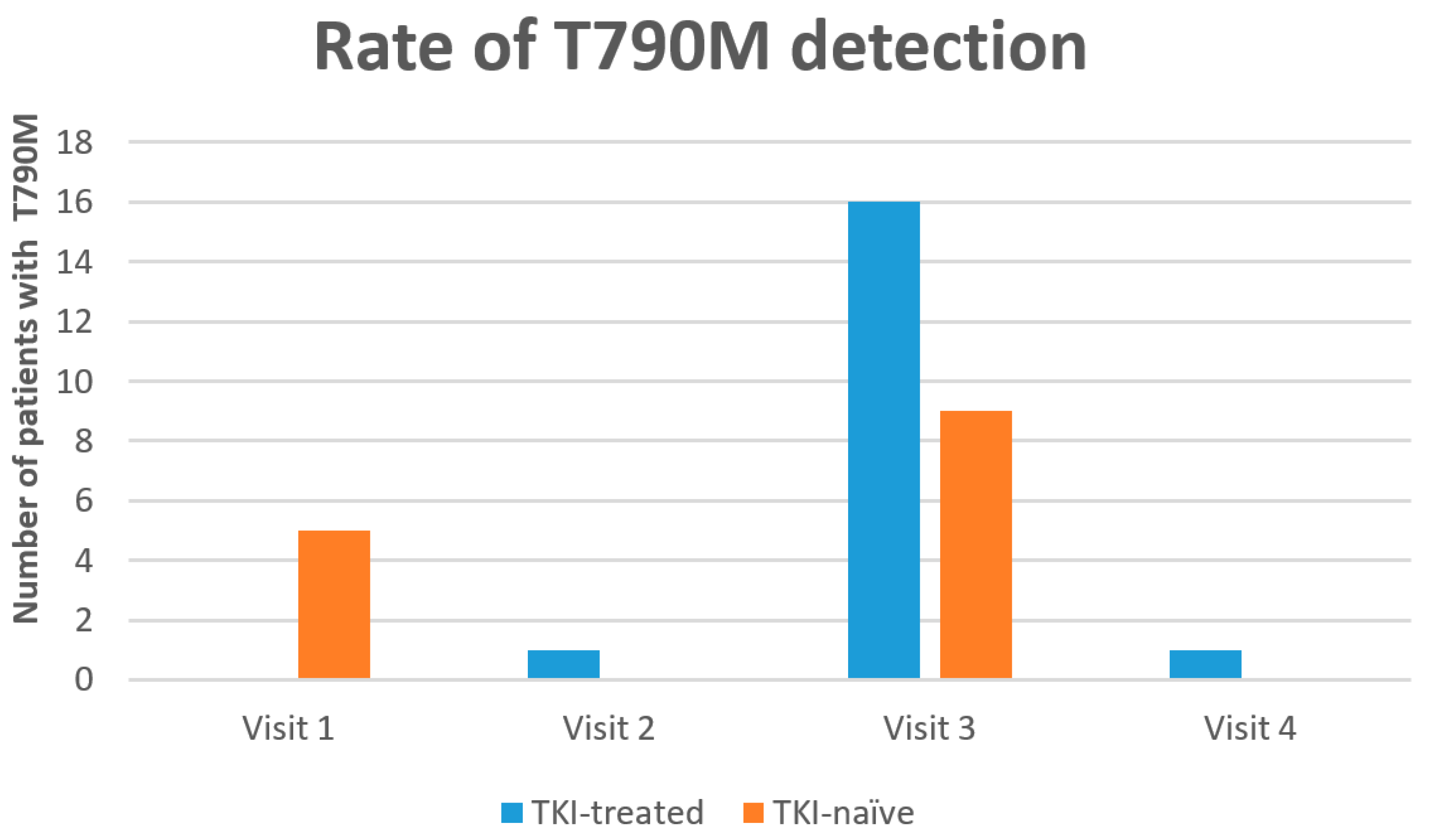
3.3. Resistant Mutation (T790M) Detection
3.4. Treatment Outcomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mouliere, F.; Messaoudi, S.E.; Pang, D.; Dritschilo, A.; Thierry, A.R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 2014, 8, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Li, H.; Hu, Z.; Wu, H.; Liu, C.; Li, Y.; Zhang, X.; Lin, P.; Hou, Q.; Ding, G.; et al. Noninvasive diagnosis and monitoring of mutations by deep sequencing of circulating tumor DNA in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2016, 471, 596–602. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26876573 (accessed on 10 January 2021). [CrossRef] [PubMed]
- Messaoudi, S.E.; Mouliere, F.; Manoir, S.D.; Bascoul-Mollevi, C.; Gillet, B.; Nouaille, M.; Fiess, C.; Crapez, E.; Bibeau, F.; Theillet, C.; et al. Circulating DNA as a strong multi-marker prognostic tool for metastatic colorectal cancer patient management care. Clin. Cancer Res. 2016, 22, 3067–3077. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26847055 (accessed on 12 February 2021). [CrossRef] [PubMed] [Green Version]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. Available online: http://www.ncbi.nlm.nih.gov/pubmed/19692680 (accessed on 11 March 2021). [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.-L.; Chen, G.; Feng, J.; Liu, X.-Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21783417 (accessed on 15 March 2021). [CrossRef]
- Connolly, I.D.; Li, Y.; Gephart, M.H.; Nagpal, S. The “Liquid Biopsy”: The Role of Circulating DNA and RNA in Central Nervous System Tumors. Curr. Neurol. Neurosci. Rep. 2016, 16, 25. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26838352 (accessed on 10 March 2021). [CrossRef]
- Diaz, L.A.J.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24449238 (accessed on 6 June 2021). [CrossRef]
- Zhang, Z.; Ramnath, N.; Nagrath, S. Current Status of CTCs as Liquid Biopsy in Lung Cancer and Future Directions. Front. Oncol. 2015, 5, 209. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26484313 (accessed on 10 June 2021). [CrossRef]
- Lin, C.-C.; Huang, W.-L.; Wei, F.; Su, W.-C.; Wong, D.T. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev. Mol. Diagn. 2015, 15, 1427–1440. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26420338 (accessed on 10 June 2021). [CrossRef] [Green Version]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. Available online: http://www.ncbi.nlm.nih.gov/pubmed/18596266 (accessed on 10 June 2021). [CrossRef] [Green Version]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Jänne, P.A.; Oxnard, G.R. Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016, 2, 1014–1022. Available online: http://www.ncbi.nlm.nih.gov/pubmed/27055085 (accessed on 1 September 2021). [CrossRef] [PubMed] [Green Version]
- Agulnik, J.; Law, J.H.; Juergens, R.; Laskin, J.; Laurie, S.; Hao, D.; Ezeife, D.A.; Le, L.W.; Kiedrowski, L.A.; Lanman, R.B.; et al. Defining VALUE: Routine liquid biopsy in NSCLC diagnosis—A Canadian trial in progress in AACR. Philadelphia 2020, 26, 1557–3265. Available online: https://clincancerres.aacrjournals.org/content/26/11_Supplement/A26 (accessed on 1 September 2021).
- Alegre, E.; Fusco, J.P.; Restituto, P.; Salas-Benito, D.; Rodríguez-Ruiz, M.E.; Andueza, M.P.; Pajares, M.J.; Patiño-García, A.; Pio, R.; Lozano, M.D.; et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour. Biol. 2016, 37, 13687–13694. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27473086 (accessed on 1 September 2021). [CrossRef] [PubMed]
- Mayo-de-Las-Casas, C.; Garzón Ibáñez, M.; Jordana-Ariza, N.; García-Peláez, B.; Balada-Bel, A.; Villatoro, S.; Malapelle, U.; Karachaliou, N.; Troncone, G.; Rosell, R.; et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev. Mol. Diagn. 2018, 18, 35–45. Available online: http://www.ncbi.nlm.nih.gov/pubmed/29172773 (accessed on 10 September 2021). [CrossRef] [PubMed]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.-M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and Prognostic Significance of Circulating Tumor Cells in Patients With Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2011, 29, 1556–1563. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21422424 (accessed on 10 September 2021). [CrossRef]
- Bauernhofer, T.; Zenahlik, S.; Hofmann, G.; Balic, M.; Resel, M.; Pirchmoser, R.; Regitnig, P.; Ambros, P.; Dandachi, N.; Samonigg, H. Association of disease progression and poor overall survival with detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer. Oncol. Rep. 2005, 13, 179–184. Available online: http://www.ncbi.nlm.nih.gov/pubmed/15643496 (accessed on 10 September 2021).
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br. J. Cancer 2014, 111, 1482–1489. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25157833 (accessed on 15 September 2021). [CrossRef] [Green Version]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non–Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30325992 (accessed on 15 September 2021). [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30988079 (accessed on 20 September 2021). [CrossRef] [Green Version]
- Wei, Z.; Wang, W.; Shu, Z.; Zhou, X.; Zhang, Y. Correlation Between Circulating Tumor DNA Levels and Response to Tyrosine Kinase Inhibitors (TKI) Treatment in Non-Small Cell Lung Cancer. Med Sci. Monit. 2017, 23, 3627–3634. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28742791 (accessed on 20 September 2021). [CrossRef] [Green Version]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25829397 (accessed on 20 September 2021). [CrossRef] [PubMed] [Green Version]
- Jovelet, C.; Madic, J.; Remon, J.; Honoré, A.; Girard, R.; Rouleau, E.; André, B.; Besse, B.; Droniou, M.; Lacroix, L. Crystal digital droplet PCR for detection and quantification of circulating EGFR sensitizing and resistance mutations in advanced non-small cell lung cancer. PLoS ONE 2017, 12, e0183319. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5567481/pdf/pone.0183319.pdf (accessed on 20 September 2021). [CrossRef] [PubMed] [Green Version]
- Rosell, R.; Karachaliou, N. Lung cancer: Using ctDNA to track EGFR and KRAS mutations in advanced-stage disease. Nat. Rev. Clin. Oncol. 2016, 13, 401–402. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27245284 (accessed on 20 September 2021). [CrossRef] [PubMed]
- Usui, K.; Yokoyama, T.; Naka, G.; Ishida, H.; Kishi, K.; Uemura, K.; Ohashi, Y.; Kunitoh, H. Plasma ctDNA monitoring during epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor treatment in patients with EGFR-mutant non-small cell lung cancer (JP-CLEAR trial). Jpn. J. Clin. Oncol. 2019, 49, 554–558. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30809659 (accessed on 20 September 2021). [CrossRef]
- Takahama, T.; Azuma, K.; Shimokawa, M.; Takeda, M.; Ishii, H.; Kato, T.; Saito, H.; Daga, H.; Tsuboguchi, Y.; Okamoto, I.; et al. Plasma screening for the T790M mutation of EGFR and phase 2 study of osimertinib efficacy in plasma T790M–positive non–small cell lung cancer: West Japan Oncology Group 8815L/LPS study. Cancer 2020, 126, 1940–1948. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32022929 (accessed on 11 October 2021). [CrossRef]
- Jenkins, S.; Yang, J.C.-H.; Jänne, P.A.; Thress, K.S.; Yu, K.; Hodge, R.; Weston, S.; Dearden, S.; Patel, S.; Cantarini, M.; et al. EGFR Mutation Analysis for Prospective Patient Selection in Two Phase II Registration Studies of Osimertinib. J. Thorac. Oncol. 2017, 12, 1247–1256. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28527899 (accessed on 1 September 2021). [CrossRef]
- Wang, X.; Li, X.; Guo, H.; Zhu, L.; Peng, Z.; Wang, J.; Yang, F.; Guo, Y. Highly Sensitive Droplet Digital PCR Method for Detection of de novo EGFR T790M Mutation in Patients with Non-Small Cell Lung Cancer. OncoTargets Ther. 2020, 13, 10621–10630. Available online: https://www.ncbi.nlm.nih.gov/pubmed/33116639 (accessed on 11 October 2021).
- Wang, X.; Zhong, D. Advanced Research on Non-small Cell Lung Cancer with De Novo T790M Mutation. Zhongguo Fei Ai Za Zhi 2019, 22, 324–328. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31109443 (accessed on 11 October 2021).
- Laurie, S.; Agulnik, J.; Hao, D.; Juergens, R.; Ezeife, D.; Law, J.; Le, L.; Kiedrowski, L.; Shepherd, F.; Cohen, V.; et al. 1195P The value of detecting resistance through liquid biopsy. Ann. Oncol. 2020, 31, S786–S787. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.-H.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28428148 (accessed on 10 October 2021). [CrossRef] [PubMed] [Green Version]
- Cho, M.-S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS ONE 2020, 15, e0230622. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32196518 (accessed on 10 September 2021). [CrossRef] [PubMed] [Green Version]


| Visits | Cohort 1 | Cohort 2 |
|---|---|---|
| Visit 1 | Time of enrollment 1 | Prior to any TKI treatment |
| Visit 2 | Time of 1st follow up CT scan 2 | Time of 1st follow up CT scan 2 |
| Visit 3 | Time of progression | Time of progression |
| Visit 4 | 1–3 months after starting second line therapy | 1–3 months after starting second line therapy |
| Characteristics | TKI-Treated n (%) n = 52 | TKI-Naïve n (%) n = 23 | p Value | Total n (%) n = 75 |
|---|---|---|---|---|
| Gender: | ||||
| Male | 17 (33) | 8 (35) | 0.86 | 26 (35) |
| Female | 35 (67) | 15 (65) | 49 (65) | |
| Ethnicity: | ||||
| Caucasian | 34 (65) | 13 (56) | 0.68 | 47 (63) |
| Asian | 19 (35) | 9 (44) | 28 (37) | |
| Smoking history: | ||||
| Ex/current smokers | 14 (27) | 6 (26) | 0.94 | 20 (27) |
| Non-smokers | 38 (73) | 17 (74) | 55 (73) | |
| EGFR alterations: | ||||
| Exon 19 deletion | 31 (60) | 15 (65) | 46 (62) | |
| Exon 21 (L858R) | 19 (36) | 8 (35) | 0.64 | 27 (36) |
| Exon 21 (L681Q) | 1 (2) | 0 (0) | 1 (1) | |
| Exon 18 | 1 (2) | 0 (0) | 1 (1) | |
| First line TKI: | ||||
| gefitinib | 43 (82) | 16 (70) | 59 (80) | |
| afatinib | 6 (12) | 3 (13) | 0.78 | 9 (12) |
| erlotinib | 3 (6) | 0 (0) | 3 (4) | |
| osimertinib | 0 | 3 (13) | 3 (4) | |
| unknown (lost to follow up) | 0 | 1 (4) | 1 | |
| Best response to 1st line treatment: | ||||
| Complete response (CR) | 4 (8) | 2 (9) | 6 (8) | |
| Partial response (PR) | 29 (56) | 13 (57) | 42 (56) | |
| Stable disease (SD) | 12 (23) | 6 (26) | 0.75 | 18 (24) |
| Mixed response | 1 (2) | 1 (4) | 2 (3) | |
| Progressive disease (PD) | 6 (11) | 0 (0) | 6 (8) | |
| Unknown (lost to follow up) | 0 | 1 (4) | 1 (1) | |
| Adequacy of DNA in plasma | ||||
| Adequate quantity | 51 (98) | 22 (96) | 0.99 | 73 (97) |
| Undetectable quantity: | 1 (2) | 1 (4) | 2 (3) |
| Variable | TKI-Treated n = 50 * | TKI-Naïve n = 23 | Total n = 73 | Pearson Chi-Square |
|---|---|---|---|---|
| OM ** detected n(%) | 19 (38) | 16 (70) | 35 (48) | 0.012 |
| OM not detected n(%) | 31 (62) | 7 (30) | 38 (52) | |
| Total | 50 (100) | 23 (100) | 73 (100) |
| Time | OM Detection | T790M Detection | Total | p Value | |
|---|---|---|---|---|---|
| Detected | Not Detected | ||||
| Visit1 a | Detected | 5 | 30 | 73 | n/a |
| Not detected | 0 | 38 | |||
| Visit 2 | Detected | 1 | 23 | 57 | n/a |
| Not detected | 0 | 33 | |||
| Visit 3 | Detected | 24 | 12 | 50 | 0.001 |
| Not detected | 1 | 13 | |||
| Visit 4 | Detected | 8 | 7 | 31 | n/a |
| Not detected | 0 | 16 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agulnik, J.S.; Papadakis, A.I.; Pepe, C.; Sakr, L.; Small, D.; Wang, H.; Kasymjanova, G.; Spatz, A.; Cohen, V. Cell-Free Tumor DNA (ctDNA) Utility in Detection of Original Sensitizing and Resistant EGFR Mutations in Non-Small Cell Lung Cancer (NSCLC). Curr. Oncol. 2022, 29, 1107-1116. https://doi.org/10.3390/curroncol29020094
Agulnik JS, Papadakis AI, Pepe C, Sakr L, Small D, Wang H, Kasymjanova G, Spatz A, Cohen V. Cell-Free Tumor DNA (ctDNA) Utility in Detection of Original Sensitizing and Resistant EGFR Mutations in Non-Small Cell Lung Cancer (NSCLC). Current Oncology. 2022; 29(2):1107-1116. https://doi.org/10.3390/curroncol29020094
Chicago/Turabian StyleAgulnik, Jason S., Andreas I. Papadakis, Carmela Pepe, Lama Sakr, David Small, Hangjun Wang, Goulnar Kasymjanova, Alan Spatz, and Victor Cohen. 2022. "Cell-Free Tumor DNA (ctDNA) Utility in Detection of Original Sensitizing and Resistant EGFR Mutations in Non-Small Cell Lung Cancer (NSCLC)" Current Oncology 29, no. 2: 1107-1116. https://doi.org/10.3390/curroncol29020094
APA StyleAgulnik, J. S., Papadakis, A. I., Pepe, C., Sakr, L., Small, D., Wang, H., Kasymjanova, G., Spatz, A., & Cohen, V. (2022). Cell-Free Tumor DNA (ctDNA) Utility in Detection of Original Sensitizing and Resistant EGFR Mutations in Non-Small Cell Lung Cancer (NSCLC). Current Oncology, 29(2), 1107-1116. https://doi.org/10.3390/curroncol29020094









