A Case Series of Metastatic Malignant Gastrointestinal Neuroectodermal Tumors and Comprehensive Genomic Profiling Analysis of 20 Cases
Abstract
:1. Introduction
2. Clinical Cases Presentation
2.1. Case 1
2.2. Case 2
2.3. Case 3
3. Clinicopathological Characterization and Comprehensive Genomic Profiling (CGP) of 20 Additional GNET Cases from Foundation Medicine
3.1. Pathology
3.1.1. Methodology
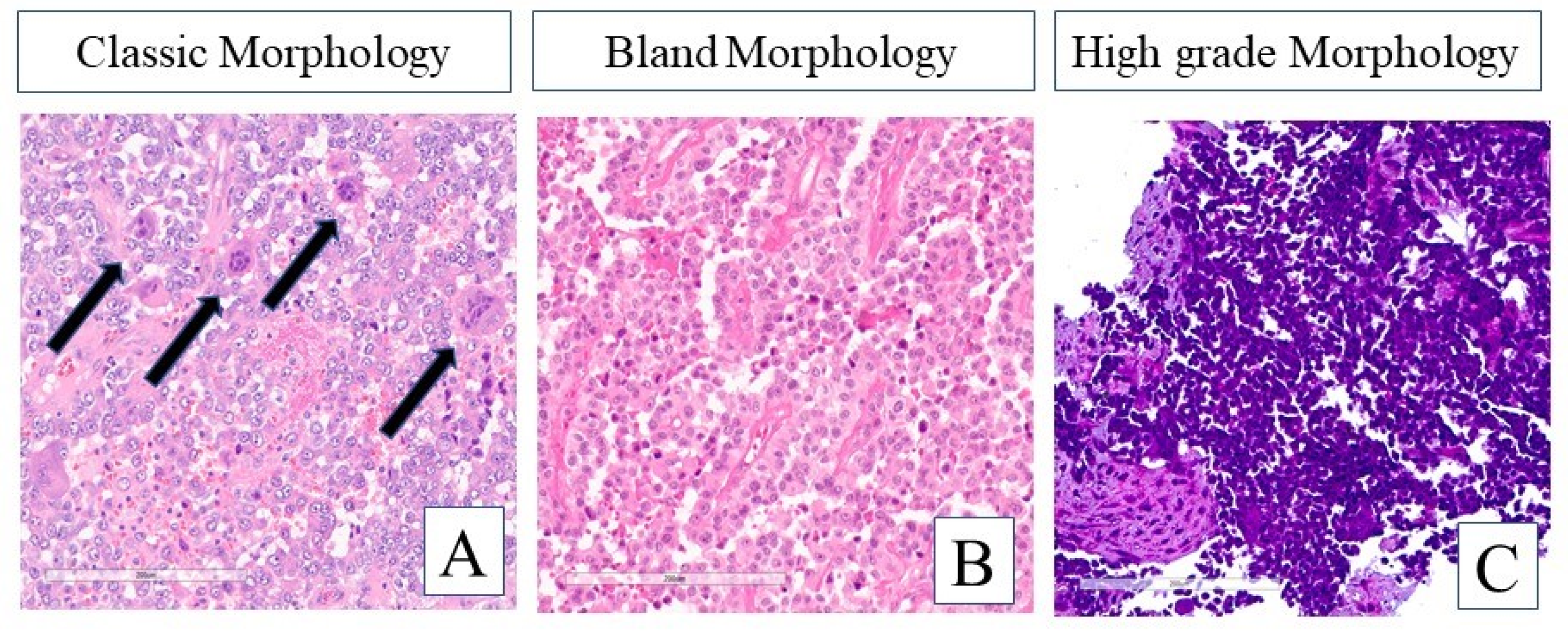
3.1.2. Histomorphologic Features
3.1.3. Immunohistochemistry and EWSR1 FISH
3.2. Genomic Profiling Analysis
3.2.1. Methodology
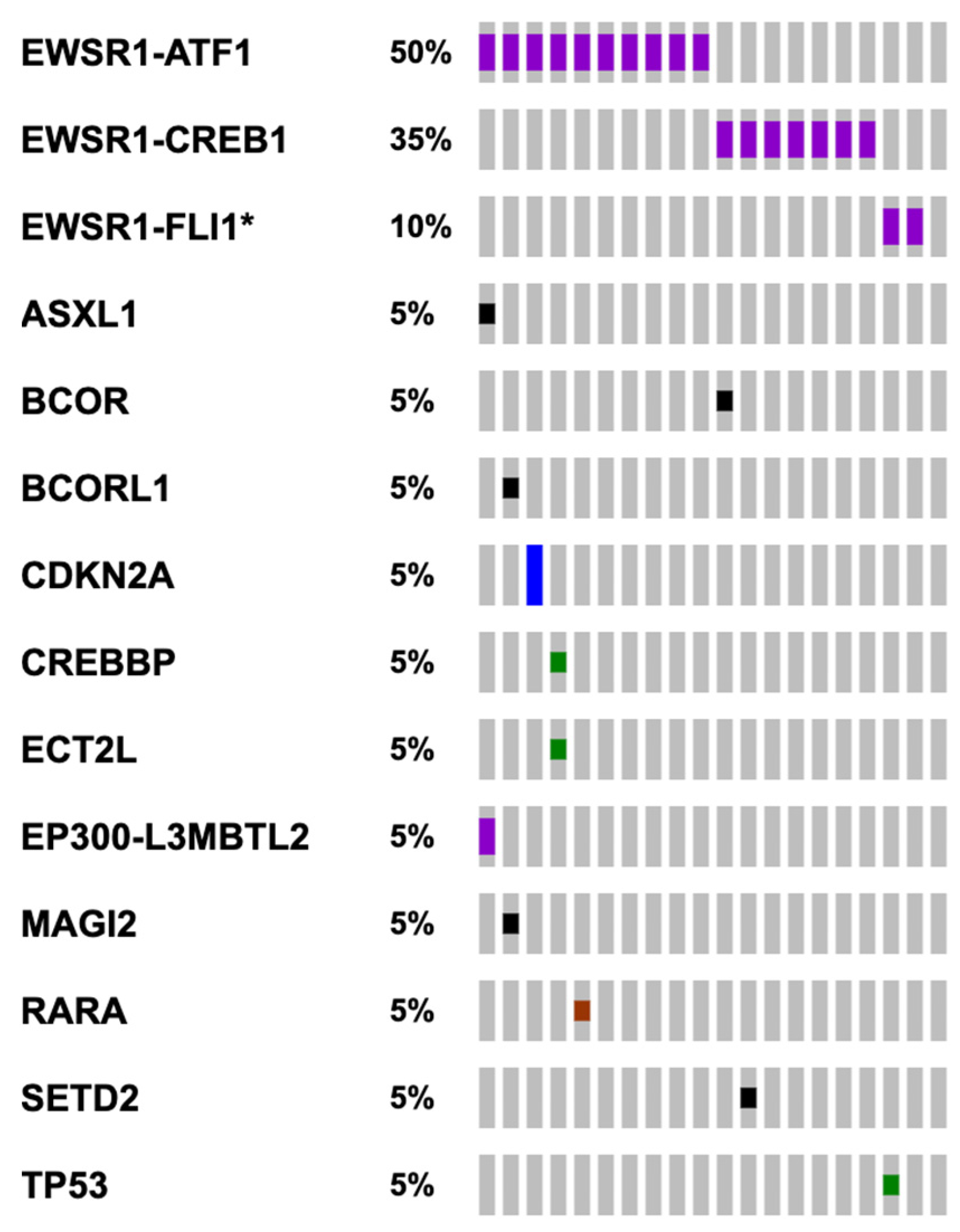
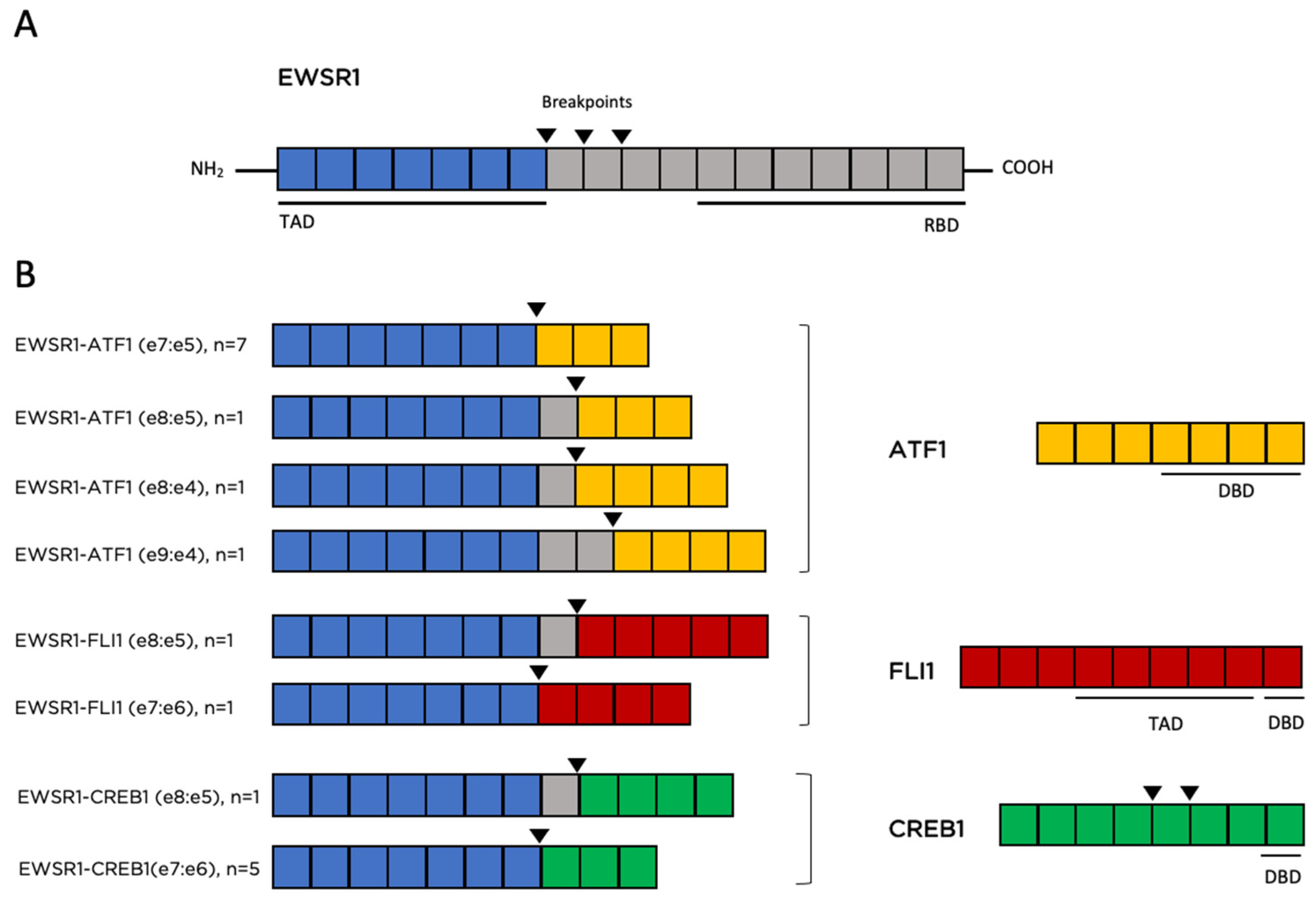
3.2.2. Genomic Findings
4. Literature Review and Discussion
4.1. Clinical Presentation Is Variable and Nonspecific
4.2. Accurate Pathological Diagnosis Remains a Significant Challenge
4.3. Initially Thought to Be a Highly Aggressive Tumor, Prognosis of GNET Is Variable
4.4. Surgery Remains a Main Treatment Modality in Both Localized and Metastatic Settings
4.5. Efficacy of Chemotherapy and Targeted Therapies Remains Largely Unknown and Appears to Be Variable
4.5.1. Adjuvant Chemotherapy for Localized Disease
4.5.2. Systemic Treatments for Metastatic Disease
4.6. Radiotherapy Potentially Beneficial in the Metastatic Setting
4.7. EWSR1 Translocations Are the Most Recurrent Genomic Alteration and Other Potentially Targetable Genomic Alterations Are Rarely Identified
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, B.; Yu, L.; Guo, W.; Sheng, W.; Wang, L.; Lao, I.; Huang, D.; Bai, Q.; Wang, J. Malignant Gastrointestinal Neuroectodermal Tumor: Clinicopathologic, Immunohistochemical, and Molecular Analysis of 19 Cases. Am. J. Surg. Pathol. 2020, 44, 456–466. Available online: https://journals.lww.com/ajsp/Abstract/2020/04000/Malignant_Gastrointestinal_Neuroectodermal_Tumor_.3.aspx (accessed on 27 September 2021). [CrossRef] [PubMed]
- Zambrano, E.; Reyes-Mugica, M.; Franchi, A.; Rosai, J. An osteoclast-rich tumor of the gastrointestinal tract with features resembling clear cell sarcoma of soft parts: Reports of 6 cases of a GIST simulator. Int. J. Surg. Pathol. 2003, 11, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Stockman, D.L.; Miettinen, M.; Suster, S.; Spagnolo, D.; Dominguez-Malagon, H.; Hornick, J.L.; Adsay, V.; Chou, P.M.; Amanuel, B.; Vantuinen, P.; et al. Malignant gastrointestinal neuroectodermal tumor: Clinicopathologic, immunohistochemical, ultrastructural, and molecular analysis of 16 cases with a reappraisal of clear cell sarcoma-like tumors of the gastrointestinal tract. Am. J. Surg. Pathol. 2012, 36, 857–868. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Abdel-Wahab, O.; Nahas, M.K.; Wang, K.; Rampal, R.K.; Intlekofer, A.M.; Patel, J.; Krivstov, A.; Frampton, G.M.; Young, L.E.; et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016, 127, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer. Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Damle, A.; Sreenija, Y.; Mathews, N.R.; Nair, K.; Philp, A.; Pavithran, K.; Paulose, R.R. Malignant Gastrointestinal Neuroectodermal Tumour-Case Report with Review of Literature. J. Gastrointest. Cancer 2021, 52, 1125–1130. [Google Scholar] [CrossRef]
- Alyousef, M.J.; Alratroot, J.A.; ElSharkawy, T.; Shawarby, M.A.; Al Hamad, M.A.; Hashem, T.M.; Alsayyah, A. Malignant gastrointestinal neuroectodermal tumor: A case report and review of the literature. Diagn. Pathol. 2017, 12, 29. [Google Scholar] [CrossRef] [Green Version]
- Antonescu, C.R.; Nafa, K.; Segal, N.H.; Dal Cin, P.; Ladanyi, M. EWS-CREB1: A recurrent variant fusion in clear cell sarcoma—Association with gastrointestinal location and absence of melanocytic differentiation. Clin. Cancer Res. 2006, 12, 5356–5362. [Google Scholar] [CrossRef] [Green Version]
- Askan, G.; Kombak, F.E.; Seven, I.E.; Basturk, O. Clear Cell Sarcoma-Like Tumor of the Gastrointestinal Tract. J. Gastrointest. Cancer 2019, 50, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yin, W.; Wang, X.; Li, P.; Chen, Y.; Jin, X.; Yang, P.; Wu, H. Synchronous Malignant Gastrointestinal Neuroectodermal Tumor and SMARCA4-Deficient Undifferentiated Carcinoma with Independent Origins in the Small Intestine: A Case Report. Front. Oncol. 2021, 11, 665056. [Google Scholar] [CrossRef] [PubMed]
- Gadde, R.; Linos, K.; Lisovsky, M.; Kerrigan, T.; Loehrer, A.P.; Kasumova, G.; Kerr, D.A.; Liu, X. Fine Needle Aspiration Cytology of Malignant Digestive System Gastrointestinal Neuroectodermal Tumor in a Lymph Node Metastasis from a Previously Diagnosed Liver Primary: A Case Report and Review of Literature. Diagn. Cytopathol. 2021, 49, E130–E136. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/dc.24624 (accessed on 29 September 2021). [CrossRef] [PubMed]
- Harshavardhini, S.; Saishalini, C.N.; Pavithra, V.; Shah, N.M.; Sankar, S. Malignant gastrointestinal neuroectodermal tumor—A case report. Indian J. Pathol. Microbiol. 2021, 64, 373. Available online: https://www.ijpmonline.org/article.asp?issn=0377-4929;year=2021;volume=64;issue=2;spage=373;epage=375;aulast=Harshavardhini;type=0 (accessed on 27 September 2021). [CrossRef] [PubMed]
- Huang, G.X.; Chen, Q.Y.; Zhong, L.L.; Chen, H.; Zhang, H.P.; Liu, X.F.; Tang, F. Primary malignant gastrointestinal neuroectodermal tumor occurring in the ileum with intra-abdominal granulomatous nodules: A case report and review of the literature. Oncol. Lett. 2019, 17, 3899–3909. [Google Scholar] [CrossRef]
- Huang, H.J.; He, Y.H.; Fan, D.G.; Chen, X.Y. Malignant gastrointestinal neuroectodermal tumor: Clinicopathological analyses of four cases. Zhonghua Bing Li Xue Za Zhi 2020, 49, 821–826. [Google Scholar] [CrossRef]
- Kansal, S.; Rao, S. Malignant Gastrointestinal Neuroectodermal Tumor: A Unique Rare Neoplasm. Indian J. Surg. Oncol. 2017, 8, 630–633. [Google Scholar] [CrossRef]
- Keditsu, K.K.; Patkar, S.; Bal, M.; Shrikhande, S.V.; Goel, M. Gastrointestinal Neuroectodermal Tumor: A Diagnostic Dilemma. Indian J. Surg. 2017, 79, 166–168. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28442847 (accessed on 27 September 2021). [CrossRef] [Green Version]
- Kong, J.; Li, N.; Wu, S.; Guo, X.; Gu, C.; Feng, Z. Malignant gastrointestinal neuroectodermal tumor: A case report and review of the literature. Oncol. Lett. 2014, 8, 2687–2690. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4214465/ (accessed on 27 September 2021). [CrossRef]
- Li, R.; Cao, J.; Chen, L.; Cui, F.; Chen, S.; Feng, Z.; Li, N. Malignant Gastrointestinal Neuroectodermal Tumors: Clinicopathological and Prognostic Features of 96 Patients. Onco Targets Ther. 2020, 13, 9731–9740. [Google Scholar] [CrossRef]
- Sbaraglia, M.; Zanatta, L.; Toffolatti, L.; Spallanzani, A.; Bertolini, F.; Mattioli, F.; Lami, F.; Presutti, L.; Dei Tos, A.P. Clear cell sarcoma-like/malignant gastrointestinal neuroectodermal tumor of the tongue: A clinicopathologic and molecular case report. Virchows Arch. 2021, 478, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Atieh, M.K.; Russell, M.A.; Kittaneh, M. Malignant Gastrointestinal Neuroectodermal Tumor (GNET) with Prolonged Disease-Free Survival after Platinum-Based Chemotherapy. Case Rep. Oncol. Med. 2020, 2020, e8880202. Available online: https://www.hindawi.com/journals/crionm/2020/8880202/ (accessed on 27 September 2021). [CrossRef] [PubMed]
- Thway, K.; Fisher, C. Mesenchymal Tumors with EWSR1 Gene Rearrangements. Surg. Pathol. Clin. 2019, 12, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhao, T.W.; Ma, J.; Wu, C.Y.; Chen, L.; Ru, G.Q.; He, X.L. Clinicopathologic and molecular characteristics of malignant gastrointestinal neuroectodermal tumors. Zhonghua Bing Li Xue Za Zhi 2017, 46, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, D.; Li, W.; Zhang, L.; Li, Z.; Zhou, J. Primary malignant neuroectodermal tumor of the ileum with predominantly uncommon pseudopapillary architecture. Int. J. Clin. Exp. Pathol. 2014, 7, 8967–8971. [Google Scholar]
- Konstantinidis, A.; Cheesman, E.; O’Sullivan, J.; Pavaine, J.; Avula, S.; Pizer, B.; Kilday, J.P. Intracranial Angiomatoid Fibrous Histiocytoma with EWSR1-CREB Family Fusions: A Report of 2 Pediatric Cases. World Neurosurg. 2019, 126, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Green, C.; Spagnolo, D.V.; Robbins, P.D.; Fermoyle, S.; Wong, D.D. Clear cell sarcoma of the gastrointestinal tract and malignant gastrointestinal neuroectodermal tumour: Distinct or related entities? A review. Pathology 2018, 50, 490–498. [Google Scholar] [CrossRef]
- Shah, A.A.; Grosh, W.W.; Frierson, H.F. Malignant gastrointestinal neuroectodermal tumour of the oesophagus with pulmonary metastasis and protracted survival. Histopathology 2015, 67, 927–930. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/his.12740 (accessed on 1 October 2021). [CrossRef]
- Okada, T.; Hirano, Y.; Ishikawa, S.; Kondo, H.; Ishii, T.; Yamaguchi, S. A long-term survivor of clear cell sarcoma-like tumor of the gastrointestinal tract with liver metastasis: A case report. Surg. Case Rep. 2020, 6, 260. [Google Scholar] [CrossRef]
- Suárez-Vilela, D.; Izquierdo, F.M.; Tojo-Ramallo, S.; R. Riera-Velasco, J.; Escobar-Stein, J. Malignant Gastrointestinal Neuroectodermal Tumor Showing Overlapped Immunophenotype with Synovial Sarcoma: CD99 and SOX10 Antibodies Are Useful in Differential Diagnosis. Am. J. Surg. Pathol. 2012, 36, 1905–1908. Available online: https://journals.lww.com/ajsp/Citation/2012/12000/Malignant_Gastrointestinal_Neuroectodermal_Tumor.24.aspx (accessed on 28 September 2021). [CrossRef]
- Song, S.H.; Shin, J.H.; Ryu, H.J.; Kim, D.J.; Park, S.Y. Successful Surgical Treatment of a Recurrent Esophageal Malignant Gastrointestinal Neuroectodermal Tumor. Korean J. Thorac. Cardiovasc. Surg. 2018, 51, 142–145. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5894580/ (accessed on 27 September 2021). [CrossRef] [Green Version]
- Sivasubramaniam, P.; Tiegs-Heiden, C.A.; Sturgis, C.D.; Hagen, C.E.; Hartley, C.P.; Thangaiah, J.J. Malignant gastrointestinal neuroectodermal tumor: Cytologic, histologic, immunohistochemical, and molecular pitfalls. Ann. Diagn. Pathol. 2021, 55, 151813. Available online: https://www.sciencedirect.com/science/article/pii/S1092913421001131 (accessed on 29 September 2021). [CrossRef] [PubMed]
- Wolak, P.; Wincewicz, A.; Czauderna, P.; Spałek, M.; Kruczak, A.; Urbaniak-Wąsik, S.; Ryś, J.; Michalak, E.; Woltanowska, M.; Sulkowski, S. Malignant gastrointestinal neuroectodermal tumor (clear cell sarcoma-like tumor of the gastrointestinal tract) of the small intestine in a 12-year-old boy. Dev. Period. Med. 2018, 22, 358–363. [Google Scholar] [PubMed]
- Libertini, M.; Thway, K.; Noujaim, J.; Puls, F.; Messiou, C.; Fisher, C.; Jones, R.L. Clear Cell Sarcoma-like Tumor of the Gastrointestinal Tract: Clinical Outcome and Pathologic Features of a Molecularly Characterized Tertiary Center Case Series. Anticancer Res. 2018, 38, 1479–1483. [Google Scholar] [PubMed] [Green Version]
- Narasimhan, V.; Tan, S.; Kong, J.; Pham, T.; Michael, M.; Ramsay, R.; Warrier, S.; Heriot, A. Prognostic factors influencing survival in patients undergoing cytoreductive surgery with hyperthermic intraperitoneal chemotherapy for isolated colorectal peritoneal metastases: A systematic review and meta-analysis. Colorectal Dis. 2020, 22, 1482–1495. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/codi.15003 (accessed on 1 October 2021). [CrossRef]
- Hübner, M.; Kusamura, S.; Villeneuve, L.; Al-Niaimi, A.; Alyami, M.; Balonov, K.; Bell, J.; Bristow, R.; Guiral, D.C.; Fagotti, A.; et al. Guidelines for Perioperative Care in Cytoreductive Surgery (CRS) with or without hyperthermic IntraPEritoneal chemotherapy (HIPEC): Enhanced recovery after surgery (ERAS®) Society Recommendations—Part I: Preoperative and intraoperative management. Eur. J. Surg. Oncol. 2020, 46, 2292–2310. Available online: https://www.ejso.com/article/S0748-7983(20)30665-X/abstract (accessed on 1 October 2021). [CrossRef]
- Klempner, S.J.; Ryan, D.P. HIPEC for colorectal peritoneal metastases. Lancet Oncol. 2021, 22, 162–164. Available online: https://www.thelancet.com/journals/lanonc/article/PIIS1470-2045(20)30693-8/abstract (accessed on 1 October 2021). [CrossRef]
- Matushansky, I.; Taub, R.N. Adjuvant chemotherapy in 2011 for patients with soft-tissue sarcoma. Nat. Rev. Clin. Oncol. 2011, 8, 434–438. [Google Scholar] [CrossRef]
- Friedrichs, N.; Testi, M.A.; Moiraghi, L.; Modena, P.; Paggen, E.; Plötner, A.; Wiechmann, V.; Mantovani-Löffler, L.; Merkelbach-Bruse, S.; Buettner, R.; et al. Clear Cell Sarcoma-like Tumor with Osteoclast-like Giant Cells in the Small Bowel: Further Evidence for a New Tumor Entity. Int. J. Surg. Pathol. 2005, 13, 313–318. [Google Scholar] [CrossRef]
- Washimi, K.; Takagi, M.; Hisaoka, M.; Kawachi, K.; Takeyama, M.; Hiruma, T.; Narimatsu, H.; Yokose, T. Clear cell sarcoma-like tumor of the gastrointestinal tract: A clinicopathological review. Pathol. Int. 2017, 67, 534–536. [Google Scholar] [CrossRef]
- Yegen, G.; Gulluoglu, M.; Mete, O.; Onder, S.; Kapran, Y. Clear cell sarcoma-like tumor of the gastrointestinal tract: A case report and review of the literature. Int. J. Surg. Pathol. 2015, 23, 61–67. [Google Scholar] [CrossRef]
- Lyle, P.L.; Amato, C.M.; Fitzpatrick, J.E.; Robinson, W.A. Gastrointestinal Melanoma or Clear Cell Sarcoma? Molecular Evaluation of 7 Cases Previously Diagnosed as Malignant Melanoma. Am. J. Surg. Pathol. 2008, 32, 858–866. Available online: https://journals.lww.com/ajsp/Abstract/2008/06000/Gastrointestinal_Melanoma_or_Clear_Cell_Sarcoma_.7.aspx (accessed on 27 September 2021). [CrossRef] [PubMed]
- Yagi, T.; Nagata, S.; Yamamoto, T.; Wakamatsu, T.; Imura, Y.; Tamiya, H.; Sabe, H.; Yamashita, K.; Takenaka, S. Malignant gastrointestinal neuroectodermal tumor with BRAF mutation and a history of malignant melanoma: A case report. Mol. Clin. Oncol. 2021, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Boland, J.M.; Folpe, A.L. Oncocytic variant of malignant gastrointestinal neuroectodermal tumor: A potential diagnostic pitfall. Hum. Pathol. 2016, 57, 13–16. [Google Scholar] [CrossRef]
- Çomunoğlu, N.; Dervişoğlu, S.; Elçin, B.B.; Tekant, G.A.; Apak, H. Malignant Extragastrointestinal Neuroectodermal Tumor Located at Right Cervical Region. Open J. Pathol. 2015, 5, 125–128. Available online: http://www.scirp.org/Journal/Paperabs.aspx?paperid=60331 (accessed on 27 October 2021). [CrossRef] [Green Version]
- Insabato, L.; Guadagno, E.; Natella, V.; Somma, A.; Bihl, M.; Pizzolorusso, A.; Mainenti, P.P.; Apice, G.; Tornillo, L. An unusual association of malignant gastrointestinal neuroectodermal tumor (clear cell sarcoma-like) and Ewing sarcoma. Pathol. Res. Pract. 2015, 211, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Lee, S.H.; Gu, M.J. Esophageal subepithelial lesion diagnosed as malignant gastrointestinal neuroectodermal tumor. World J. Gastroenterol. 2015, 21, 5739–5743. Available online: http://lib.cqvip.com/qk/84123X/201518/90888889504849534956485155.html (accessed on 29 September 2021). [CrossRef]
- Li, Z.; Pu, X.; He, L.; Fu, Y.; Li, L.; Xu, Y.; Guan, W.; Fan, X. Malignant Gastrointestinal Neuroectodermal Tumor in the Right Heart: A Report of an Extremely Rare Case Presenting with a Cardiac Mass. Front. Cardiovasc. Med. 2021, 8, 702215. [Google Scholar] [CrossRef]
- Subbiah, V.; Holmes, O.; Gowen, K.; Spritz, D.; Amini, B.; Wang, W.L.; Schrock, A.B.; Meric-Bernstam, F.; Zinner, R.; Piha-Paul, S.; et al. Activity of c-Met/ALK Inhibitor Crizotinib and Multi-Kinase VEGF Inhibitor Pazopanib in Metastatic Gastrointestinal Neuroectodermal Tumor Harboring EWSR1-CREB1 Fusion. Oncology 2016, 91, 348–353. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, T.; Lu, X.; Zhang, B. Malignant gastrointestinal neuroectodermal tumor in the small intestine with liver metastasis: First case report worldwide. Asian J. Surg. 2020, 43, 769–772. Available online: https://www.sciencedirect.com/science/article/pii/S1015958420300488 (accessed on 27 September 2021). [CrossRef]
- Zhan, M.N.; Yu, J.; Luo, R.K.; Hou, Y.Y. Malignant gastrointestinal neuroectodermal tumor, presenting as a second malignancy after gastric adenocarcinoma: A case report and literature review. J. Gastrointest. Oncol. 2019, 10, 1144–1150. [Google Scholar] [CrossRef]
- Ferrari, A.; Casanova, M.; Bisogno, G.; Mattke, A.; Meazza, C.; Gandola, L.; Sotti, G.; Cecchetto, G.; Harms, D.; Koscielniak, E.; et al. Clear cell sarcoma of tendons and aponeuroses in pediatric patients. Cancer 2002, 94, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- Schöffski, P.; Wozniak, A.; Stacchiotti, S.; Rutkowski, P.; Blay, J.-Y.; Lindner, L.H.; Strauss, S.J.; Anthoney, A.; Duffaud, F.; Richter, S.; et al. Activity and safety of crizotinib in patients with advanced clear-cell sarcoma with MET alterations: European Organization for Research and Treatment of Cancer phase II trial 90101 ‘CREATE’. Ann. Oncol. 2017, 28, 3000–3008. Available online: https://www.sciencedirect.com/science/article/pii/S0923753419353906 (accessed on 27 September 2021). [CrossRef] [PubMed]
- Youssef, B.; Asberry, D.; Mohamed, R. Malignant Gastrointestinal Neuroectodermal Tumor: A case report and a review of the literature. Am. J. Clin. Pathol. 2021, 156, S66–S67. [Google Scholar] [CrossRef]
- Cantile, M.; Marra, L.; Franco, R.; Ascierto, P.; Liguori, G.; De Chiara, A.; Botti, G. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med. Oncol. 2013, 30, 412. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3586390/ (accessed on 1 December 2021). [CrossRef] [PubMed] [Green Version]
| Characteristic | Result |
|---|---|
| Median age (range), years | 37.5 (15–64) |
| Sex | 13 female, 7 male |
| Specimen site | |
| Small intestine | 9 |
| Liver | 5 |
| Soft tissue | 4 |
| Appendix | 1 |
| Stomach | 1 |
| Immunohistochemistry | |
| S100 | 14/14 positive |
| SOX-10 | 7/8 positive |
| HMB-45/Melan-A/melanoma cocktail | 0/13 positive |
| EWSR1 FISH | 11/11 positive for translocation |
| Reference | Case No. | Age/Gender | Local/Recurrent/Metastatic Disease | Adjuvant/Metastatic Treatment (All Patients Underwent Primary Tumor Resection) | Clinical Outcome |
|---|---|---|---|---|---|
| Alyousef et al., 2017 [9] | 1 | 18 M | Local recurrent | No | DFS 36 mos, DD 12 mos after local recurrence |
| Antonescu et al., 2006 [10] | 3 | 81 F | De novo metastatic | Metastatic: Liver metastectomy and peritoneal implants removal (NA) | NA |
| 42 F | Local | NA | NA | ||
| 42 F | De novo metastatic | NA | NA | ||
| Boland et al., 2016 [44] | 1 | 46 F | Local | NA | NA |
| Chang et al., 2020 [1] | 19 | 29 F | De novo metastatic | NA | NA |
| 44 M | De novo metastatic | No | OS 61 mos | ||
| 41 M | Recurrent metastatic | Adjuvant: no Metastatic: radiofrequency ablation of liver metastasis (CR), no systemic treatment | DFS 8 mos NED F/U 63 mos | ||
| 42 M | Local | Adjuvant: dacarbazine + cisplatin | NED F/U 46 mos | ||
| 51 F | Recurrent metastatic | NA | DFS 11 mos OS 36 mos | ||
| 30 F | Recurrent metastatic | Adjuvant: no Metastatic: 1st line: sunitinib (stopped due to SE), 2nd line: anlotinib (PR for 6 mos) | DFS 14 mos AD F/U 43 mos | ||
| 48 F | NA | NA | NA | ||
| 42 F | Local | No | NED F/U 3 mos | ||
| 41 F | Recurrent metastatic | NA | DFS 4 mos AD F/U NA | ||
| 56 F | De novo metastatic | Metastatic: Liver metastectomy, no systemic treatment | NED F/U 33 mos | ||
| 30 F | NA | NA | NA | ||
| 59 M | Local | No | NED F/U 29 mos | ||
| 27 F | Recurrent Metastatic | Adjuvant: no Metastatic: apatinib (PR for 4 mos stopped due to SE) | DFS 13 mos AD F/U 26 mos | ||
| 47 M | Recurrent metastatic | Adjuvant: no Metastatic: 1st line: pazopanib + pembrolizumab (stopped due to SE), 2nd line: lenvatinib + pembrolizumab (PD), 3rd line: epirubicin + ifosfamide (SD for 3 mos) | DFS 27 mos AD F/U 48 mos | ||
| 36 M | Local | Adjuvant: Vincristine amide + Adriamycin + cyclophosphamide | NED F/U 3 mos | ||
| 40 F | Local | Adjuvant: Adriamycin + dacarbazine | NED F/U 8 mos | ||
| 41 F | Recurrent Metastatic | Adjuvant: no Metastatic: 1st line: oxaliplatin (PD), 2nd line: irinotecan (PD), 3rd line: paclitaxel (PD), apatinib (SD for 5 mos) | DFS 17 mos AD F/U 29 mos | ||
| 64 M | Local | Adjuvant: adriamycin + ifosfomaide | NED F/U 14 mos | ||
| 55 M | NA | NA | NA | ||
| Comunoglu et al. 2015 [45] | 1 | 9 M | Local | Adjuvant: chemotherapy (NA) | NED F/U 12 mos |
| Damle et al., 2021 [8] | 1 | 56 M | De Novo Metastatic | Metastatic: VAC/IE (PR after 3 cycles then PR/SD additional 11 cycles) | AD F/U 3 mos |
| Friedrichs et al., 2005 [39] | 1 | 41 M | Recurrent Metastatic | Adjuvant: no Metastatic: ifosfamide, vincristine, actinomycin D, followed by ifosfamide and epirubicin (NA) | DFS: 6 mos AD F/U NA |
| Gadde et al., 2021 [13] | 1 | 36 F | De novo metastatic | Metastatic: liver metastectomy, dacarbazine + gemcitabine (PD) | OS 4 mos |
| Harshavardhini et al., 2021 [14] | 1 | 33 M | Local | Adjuvant: vincristine, etoposide, adriamycin, cyclophosphamide, mesna and ifosfamide | NED F/U 8 mos |
| Huang et al., 2019 [15] | 1 | 30 F | Local | Adjuvant: ifosfamide + epirubicin | NED F/U 6 mos |
| Huang et al. 2020 [16] | 4 | 45 F | Local | No | NED F/U 41 mos |
| 34 F | Local | No | NED F/U 17 mos | ||
| 81 M | Local with residual disease | No | AD F/U 8 mos | ||
| 68 M | De novo metastatic | Metastatic: chemotherapy (PD) | OS 5 mos | ||
| Insabato et al., 2015 [46] | 1 | 29 M | Recurrent metastatic | Adjuvant: no Metastatic: 1st line: IE (PR/SD for 8 mos) 2nd line: ifosfamide alone (SD) | DFS 36 mos AD F/U 39 mos |
| Kansal & Rao, 2017 [17] | 1 | 55 F | NA | NA | NA |
| Keditsu et al., 2017 [18] | 1 | 37 F | De novo metastatic | Metastatic: liver metastectomy followed by pseudoadjuvant VAC/IE | NED F/U 36 mos |
| Kim et al. [47] | 1 | 21 M | Local | Adjuvant: RT | NED F/U 5 mos |
| Kong et al., 2014 [19] | 1 | 17 M | NA | NA | NA |
| Li et al., 2020 [48] | 2 | 17 M | Local | No | NED F/U 10 mos |
| 62 M | De novo metastatic | No | AD F/U 6 mos | ||
| Libertini et al., 2018 [34] | 6 | 59 F | Recurrent metastatic | Adjuvant: no Metastatic: dacarbazine (PD) | DFS 11 mos OS 18 mos |
| 28 F | Local recurrent | Resection for local recurrence, no systemic treatment | DFS 109 mos NED F/U 161 mos | ||
| 27 F | Recurrent metastatic | NA | DFS 2 mos OS 4 mos | ||
| 33 M | Recurrent metastatic | Adjuvant: no Metastatic: no | DFS 2 mos OS 8 mos | ||
| 48 M | Recurrent local/metastatic | NA | DD F/U NA | ||
| 27 M | De novo metastatic | Metastatic: metastectomy (peritoneal resection), no systemic treatment | NED F/U 2 mos | ||
| Lyle et al., 2008 [42] | 7 | 46 M | Local | Adjuvant: chemotherapy (NA) | NED F/U 7 mos |
| 62 M | De novo metastatic | Metastatic: chemotherapy (NA) | OS 12 mos | ||
| 49 M | De novo metastatic | No | OS 2 mos | ||
| 60 F | De novo metastatic | NA | NA | ||
| 29 F | De novo metastatic | NA | NA | ||
| 60 M | De novo metastatic | Metastatic: chemotherapy (NA) | OS 28 mos | ||
| 55 F | NA | NA | NA | ||
| Okada et al., 2020 [29] | 1 | 38 F | De novo metastatic | Metastatic: liver Metastectomy, no systemic treatment | NED F/U 36 mos |
| Shah et al., 2015 [28] | 1 | 28 F | Recurrent metastatic | Adjuvant: RT, no systemic treatment Metastatic: IL-2 (PD), anti-CTLA4 (SD for 10 mos), anti-PD-L1 (SD for 7 mos), IL-15 (PD) | DFS 48 mos AD F/U 72 mos |
| Singh et al., 2020 [22] | 1 | 61 M | Recurrent metastatic | Adjuvant: cisplatin+etoposide Metastatic: 1st line: capecitabine+temozolomide (PD), 2nd line: everolimus (SD for 5 mos), 3rd line: pazopanib (PD), 4th line: sunitinib (SD for 3 mos) | DFS 84 mos OS 13–15 mos |
| Sivasubramaniam et al., 2021 [32] | 1 | 46 F | Recurrent metastatic | Adjuvant: no Metastatic: metastectomy (intraabdominal lymph nodes) followed by pseudoadjuvant pazopanib | DFS 17 mos NED F/U NA |
| Stockman et al., 2012 [3] | 16 | 30 F | NA | NA | AD F/U 21 mos |
| 35 M | NA | NA | OS 18 mos | ||
| 33 M | NA | NA | AD F/U 1.5 mos | ||
| 50 F | NA | NA | AD F/U 24 mos | ||
| 20 F | NA | NA | NED F/U 20 mos | ||
| 52 M | NA | NA | OS 22 mos | ||
| 46 M | NA | NA | NA | ||
| 34 F | NA | NA | OS 19 mos | ||
| 37 F | NA | NA | NA | ||
| 77 F | NA | NA | OS 106 mos | ||
| 31 M | NA | NA | OS 3 mos | ||
| 17 M | NA | NA | NA | ||
| 30 M | NA | NA | AD F/U 36 mos | ||
| 60 F | NA | NA | NED F/U 41 mos | ||
| 56 M | NA | NA | NA | ||
| 28 F | NA | NA | OS 23 mos | ||
| Song et al., 2018 [31] | 1 | 23 M | Local recurrence | Adjuvant: RT Local recurrence: surgical resection, no systemic treatment | DFS 12 mos NED F/U 24 mos |
| Subbiah et al., 2016 [49] | 1 | 27 F | De novo metastatic | Metastatic: metastectomy (liver and others), cryotherapy, palliative RT, crizotinib + pazopanib (PR for 1.5 yrs) | AD F/U 2.8yrs |
| Wang et al., 2020 [50] | 1 | 30 F | De novo metastatic | Metastatic: chemotherapy (NA, PR) | AD F/U 6 mos |
| Washimi et al., 2017 [40] | 1 | 32 F | Recurrent metastatic | Adjuvant: ifosfamide + adriamycin Metastatic: NA | DFS 38 mos AD F/U NA |
| Wolak et al., 2018 [33] | 1 | 12 M | Recurrent metastatic | Adjuvant: vincristine + adriamycin + ifosfamide + dactinomycin, followed by carboplatin + epirubicin + vincristine + actinomycin D + ifosfamide + etoposide Metastatic: thermal ablation (PD), liver metastectomy (PD), 1st line: carboplatin, epirubicin, vincristine, actinomycin D, ifosfamide and etoposide (PD), 2nd line: pazopanib (PD) | DFS 8 weeks OS 18 mos |
| Yagi et al., 2020 [43] | 1 | 66 F | De novo metastatic | Metastatic: 1st line: dabrafenib + trametinib (PR for 3 mos), 2nd line: nivolumab + ipilimumab (PD) | OS 21 mos |
| Yegen et al., 2015 [41] | 1 | 25 F | De novo metastatic | Metastatic: Liver Metastectomy followed by chemotherapy (NA, PD) | AD F/U 47 mos |
| Zambrano et al., 2003 [2] | 6 | 15 F | De novo metastatic | Metastatic: chemotherapy (NA, PD) | OS 16 mos |
| 21 F | De novo metastatic | Metastatic chemotherapy (NA, PD) | OS 12 mos | ||
| 35 F | Recurrent metastatic | Adjuvant: No Metastatic: NA | DFS 12 mos AD F/U NA | ||
| 37 F | Local | NA | NA | ||
| 13 M | Recurrent local/metastatic | Adjuvant: chemotherapy (NA) Recurrence: total gastrectomy for local recurrence followed by pseudoadjuvant chemotherapy (NA) | DFS 7 mos AD F/U 5 mos | ||
| 32 M | Local | NA | NA | ||
| Zhan et al., 2019 [51] | 1 | 33 F | Recurrent metastatic | Adjuvant: chemotherapy (NA) Metastatic: metastectomy (mesentery), sunitinib (SD for 12 mos) | DFS 14 mos AD F/U 12 mos |
| Zhao et al., 2014 [25] | 1 | 33 F | Local | Adjuvant: ifosfamide + epirubicin | NED F/U NA |
| Zhao et al., 2017 [24] | 2 | 57 M | local | No | NED F/U 16 mos |
| 24 M | De novo metastatic | Metastatic: 1st line: paclitaxel + gemcitabine (PD), 2nd line: vinorelbine + gemcitabine (PD), 3rd line: apatinib (SD for 2 mos), 4th line: apatinib+temozolomide (SD for 3–4 mos) | AD F/U 55 mos |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandler, T.; Cortez, E.; Clinton, L.; Hemmerich, A.; Ahmed, O.; Wong, R.; Forns, T.; MacNeill, A.J.; Hamilton, T.D.; Khorasani, M.; et al. A Case Series of Metastatic Malignant Gastrointestinal Neuroectodermal Tumors and Comprehensive Genomic Profiling Analysis of 20 Cases. Curr. Oncol. 2022, 29, 1279-1297. https://doi.org/10.3390/curroncol29020109
Kandler T, Cortez E, Clinton L, Hemmerich A, Ahmed O, Wong R, Forns T, MacNeill AJ, Hamilton TD, Khorasani M, et al. A Case Series of Metastatic Malignant Gastrointestinal Neuroectodermal Tumors and Comprehensive Genomic Profiling Analysis of 20 Cases. Current Oncology. 2022; 29(2):1279-1297. https://doi.org/10.3390/curroncol29020109
Chicago/Turabian StyleKandler, Taylor, Eliane Cortez, Lani Clinton, Amanda Hemmerich, Osama Ahmed, Ralph Wong, Taylor Forns, Andrea J. MacNeill, Trevor D. Hamilton, Mohammadali Khorasani, and et al. 2022. "A Case Series of Metastatic Malignant Gastrointestinal Neuroectodermal Tumors and Comprehensive Genomic Profiling Analysis of 20 Cases" Current Oncology 29, no. 2: 1279-1297. https://doi.org/10.3390/curroncol29020109
APA StyleKandler, T., Cortez, E., Clinton, L., Hemmerich, A., Ahmed, O., Wong, R., Forns, T., MacNeill, A. J., Hamilton, T. D., Khorasani, M., & Feng, X. (2022). A Case Series of Metastatic Malignant Gastrointestinal Neuroectodermal Tumors and Comprehensive Genomic Profiling Analysis of 20 Cases. Current Oncology, 29(2), 1279-1297. https://doi.org/10.3390/curroncol29020109






