The Clinicopathological Significance of BiP/GRP-78 in Breast Cancer: A Meta-Analysis of Public Datasets and Immunohistochemical Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Public Datasets
2.2. Meta-Analysis of BiP Immunohistochemistry in Breast Cancer Samples
2.2.1. Search Strategy
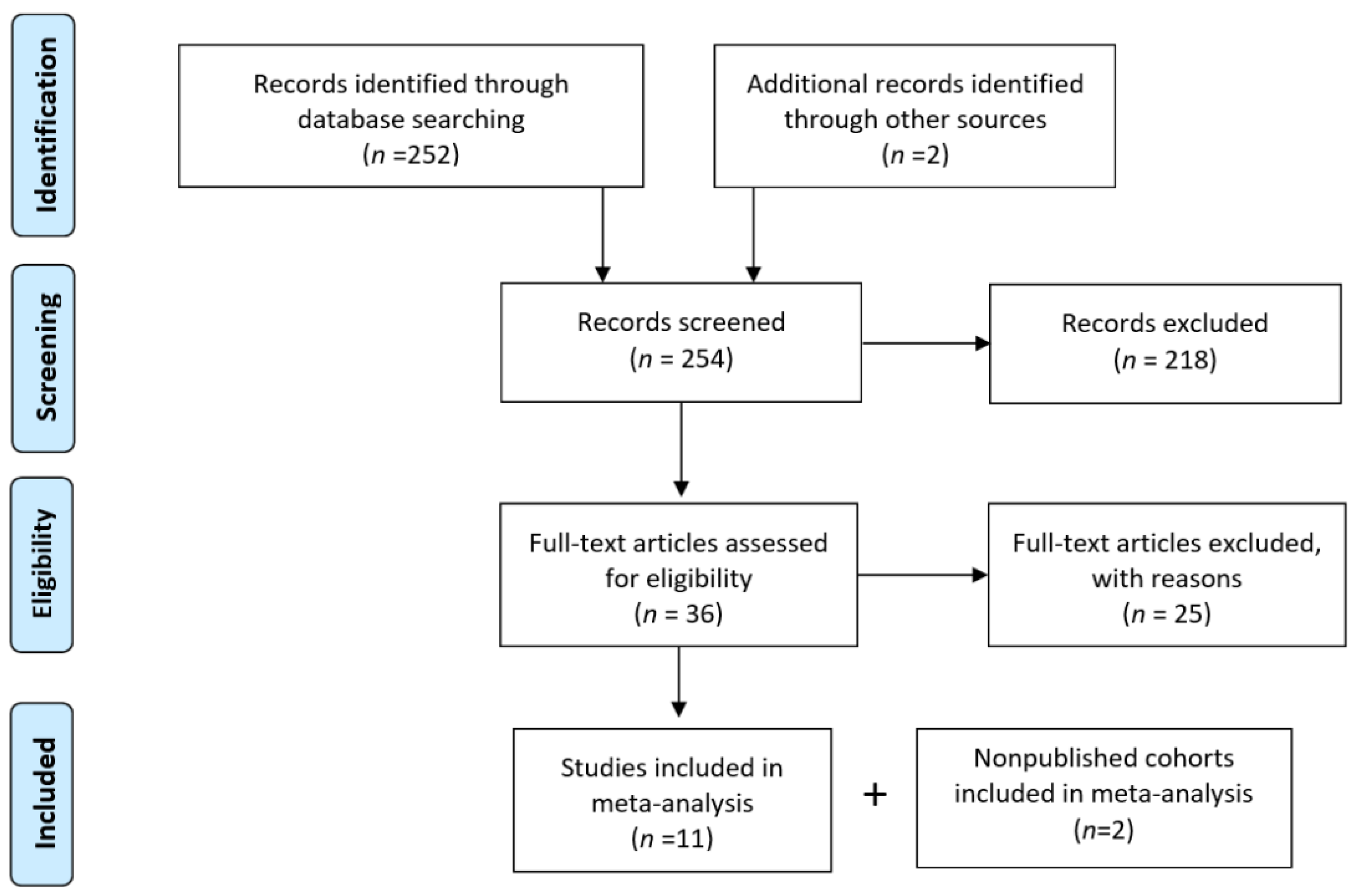
2.2.2. Eligibility and Data Collection
2.2.3. Data Analysis
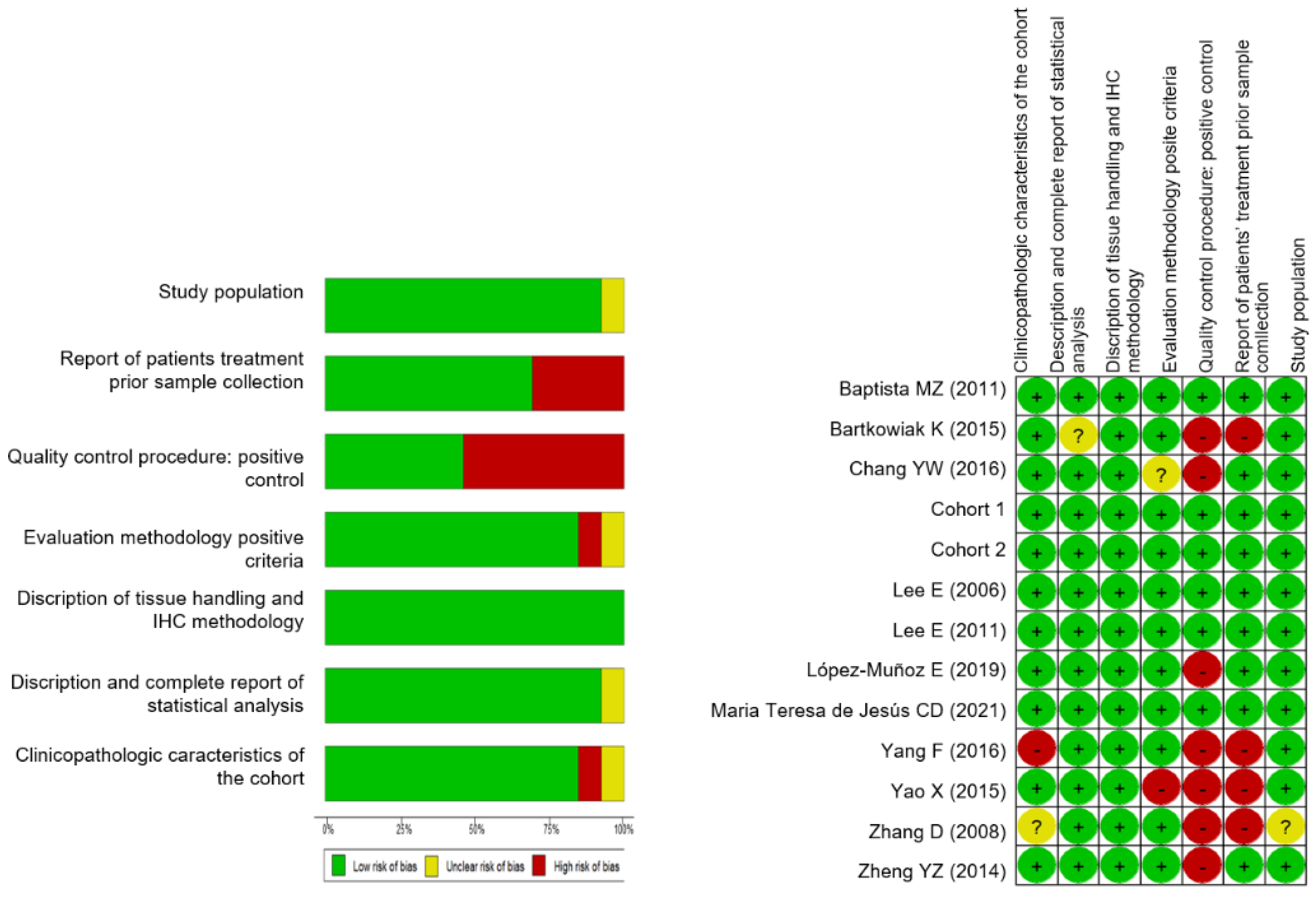
2.3. Immunohistochemical Detection of BiP
2.3.1. Patient Sample Collection and Characterization
2.3.2. Immunohistochemistry
2.3.3. Statistical Analysis
3. Results
3.1. Stratification of Public Datasets by BiP Differential Expression Correlates with Breast Cancer Molecular Subtype and Immune Score
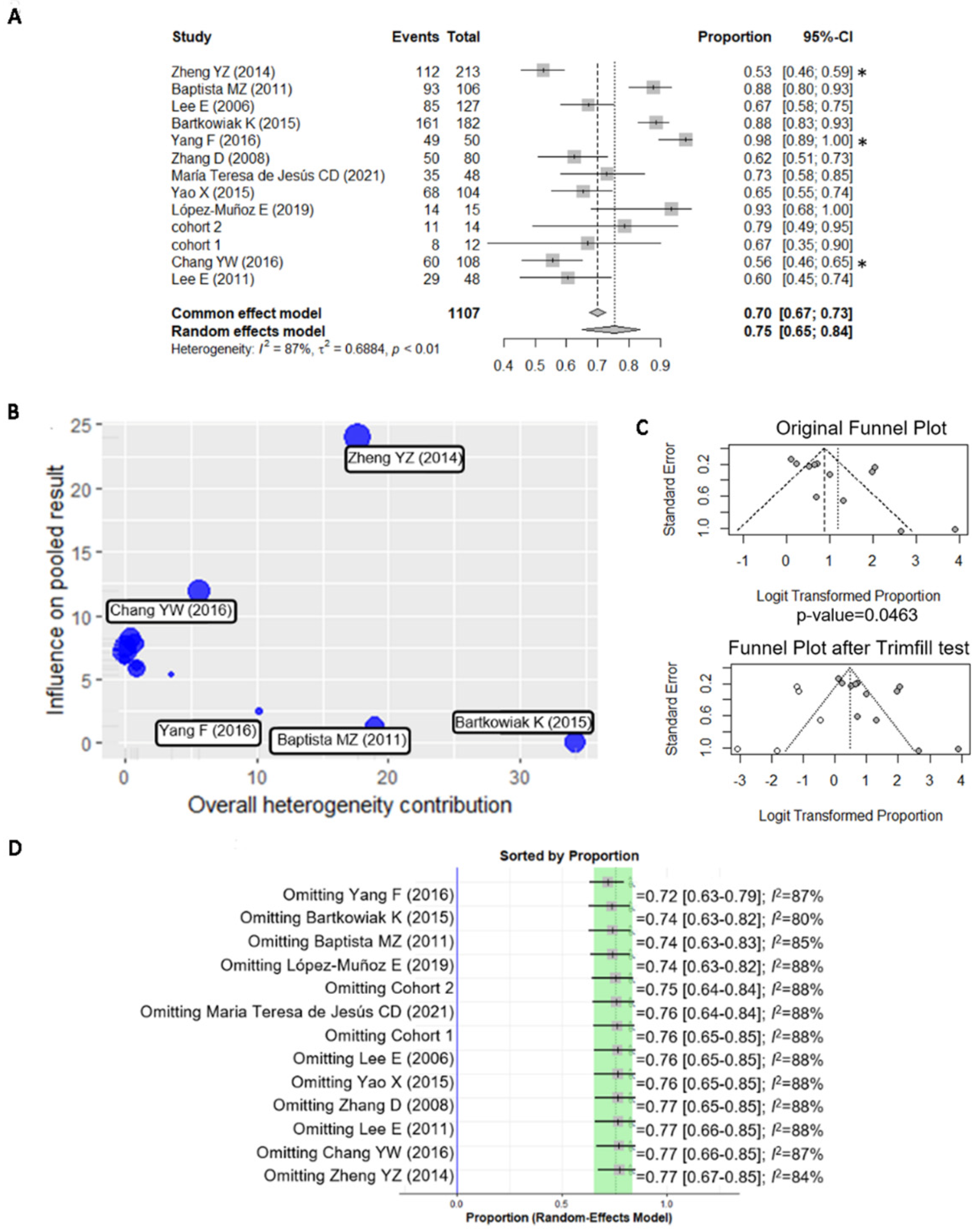
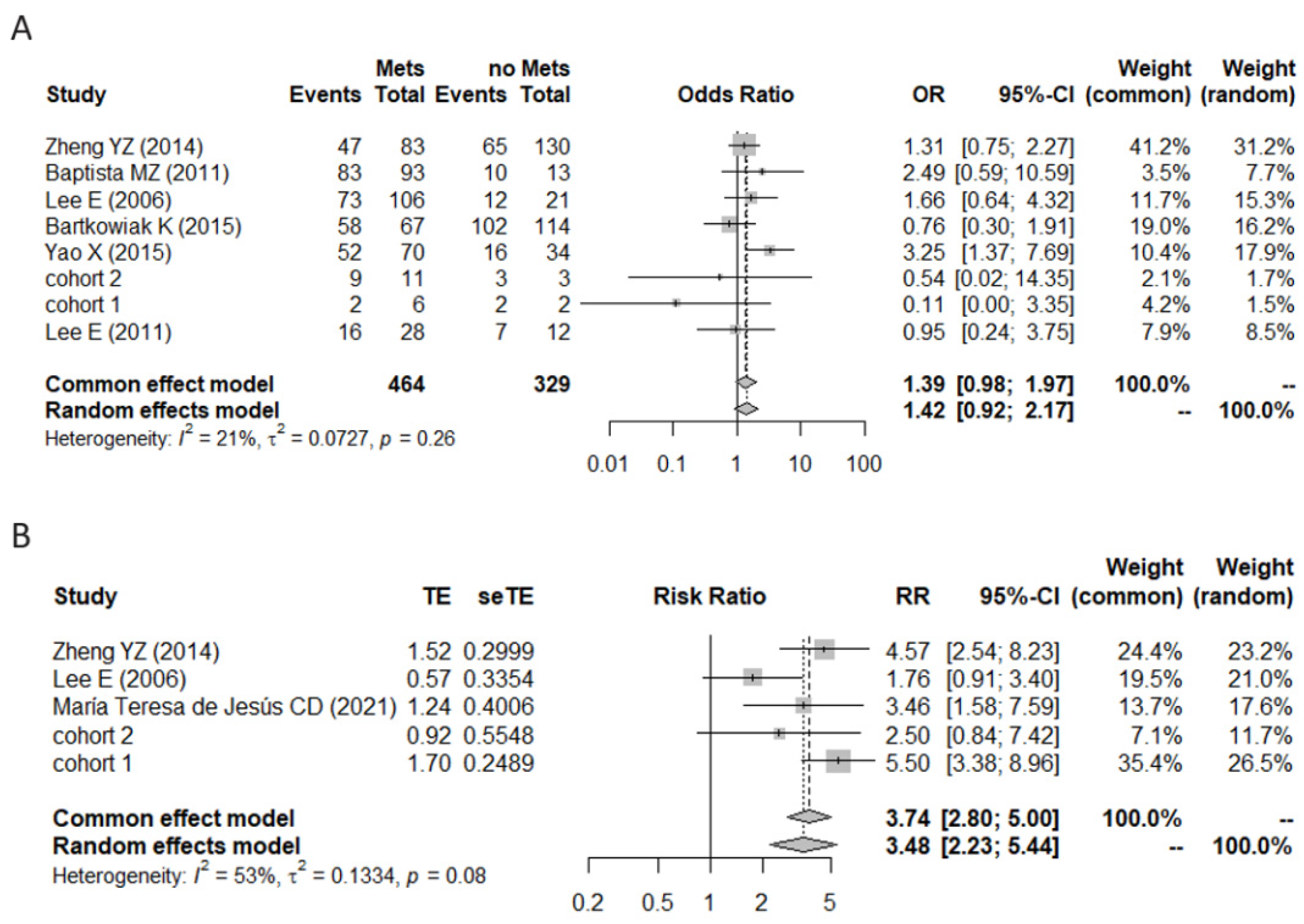
3.2. A Meta-Analysis of BiP Immunohistochemistry Identifies An Association with A Higher Risk of Recurrence
| Study | N | Positive Cases | Antibody | Histology | Sample | Cutoff | Pre/Post Menopause | Low/High Stage | Low/ High Grade | Lymph Nodes +/ Lymph Nodes − | LVC Invasion/ No LVC Invasion | ER+/ ER− | HER2+/ HER2− |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baptista, M.Z. (2011) [6] | 106 | 93 | C50B12 | N/A | TMA | score | 68/38 | 25/81 | 20/86 | 93/13 | 33/73 | 76/30 | 27/79 |
| Zheng, Y.Z. (2014) [10] | 213 | 112 | 11587-1-AP | IDC | TMA | score | 98/115 | N/A | 123/55 | 83/130 | N/A | 90/123 | 83/130 |
| Lee, E. (2006) [41] | 127 | 85 | sc-13968 | IDC + ILC + other | Tissue | score | 66/61 | 116/11 | 52/52 | 106/21 | 50/77 | 97/27 | 23/76 |
| Bartkowiak, K. (2015) [42] | 182 | 161 | C50B12 | IDC + ILC + other | TMA | score | N/A | 167/14 | 102/73 | 67/114 | 15/131 | 124/37 | 6/157 |
| Chang, Y.W. (2016) [43] | 108 | 60 | sc-13968 | IDC | Tissue | score | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Yao, X. (2015) [5] | 104 | 68 | sc-1051 | IDC + other | Tissue | score | 66/38 | 70/34 | 44/60 | 70/34 | N/A | 69/35 | 31/73 |
| Yang, F. (2016) [44] | 50 | 49 | sc-13968 | N/A | Tissue | score | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Zhang, D. (2008) [45] | 80 | 50 | 610979 | N/A | TMA | score | N/A | N/A | 6/8 | 8/6 | N/A | 6/8 | 32/48 |
| María Teresa de Jesús, C.D. (2021) [46] | 48 | 35 | ab21685 | IDC + other | TMA | score | 24/31 | N/A | N/A | N/A | N/A | 27/26 | 13/40 |
| López-Muñoz, E. (2019) [47] | 15 | 14 | ab21685 | IDC | Tissue | score | N/A | 11/4 | N/A | 11/4 | N/A | 11/4 | 1/13 |
| Lee, E. (2011) [48] | 48 | 29 | sc-13968 | IDC + other | Tissue | score | 30/18 | N/A | 17/30 | 28/12 | N/A | N/A | 21/25 |
| Study | N | Positive Cases | Antibody | Histology | Sample | Cutoff Criteria | Low Stage/ High Stage | Low Grade/ High Grade | Lnodes/ No Lnodes | ER+/ ER− | HER2+/ HER2− |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cohort 1 | 14 | 11 | HPA038845 | IDC + ILC + other | Tissue | score | 9/5 | 11/3 | 1/13 | 14/0 | 2/12 |
| cohort 2 | 12 | 9 | HPA038845 | IDC + ILC + other | Tissue | score | 7/5 | 5/7 | 12/0 | 12/0 | 3/9 |
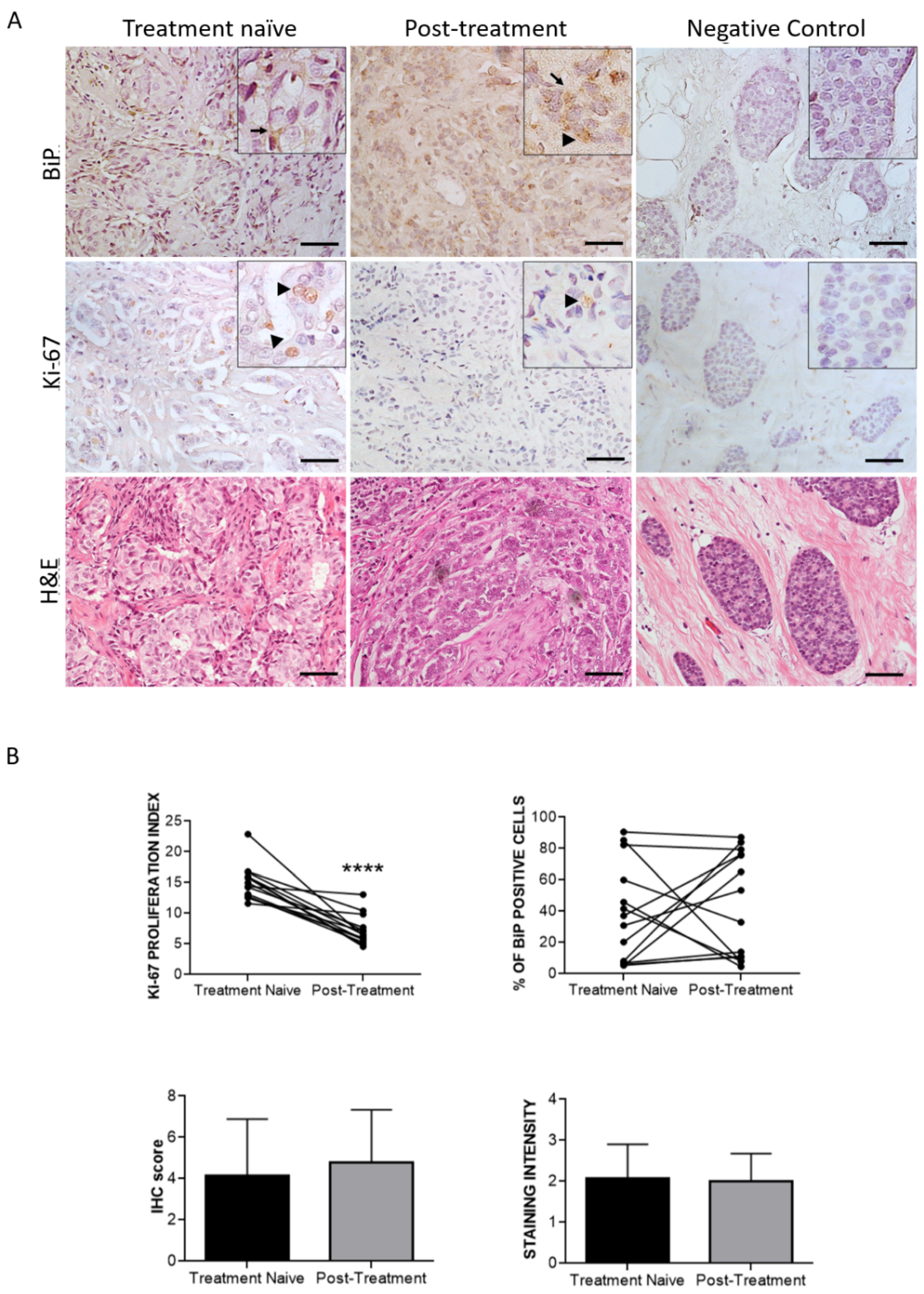
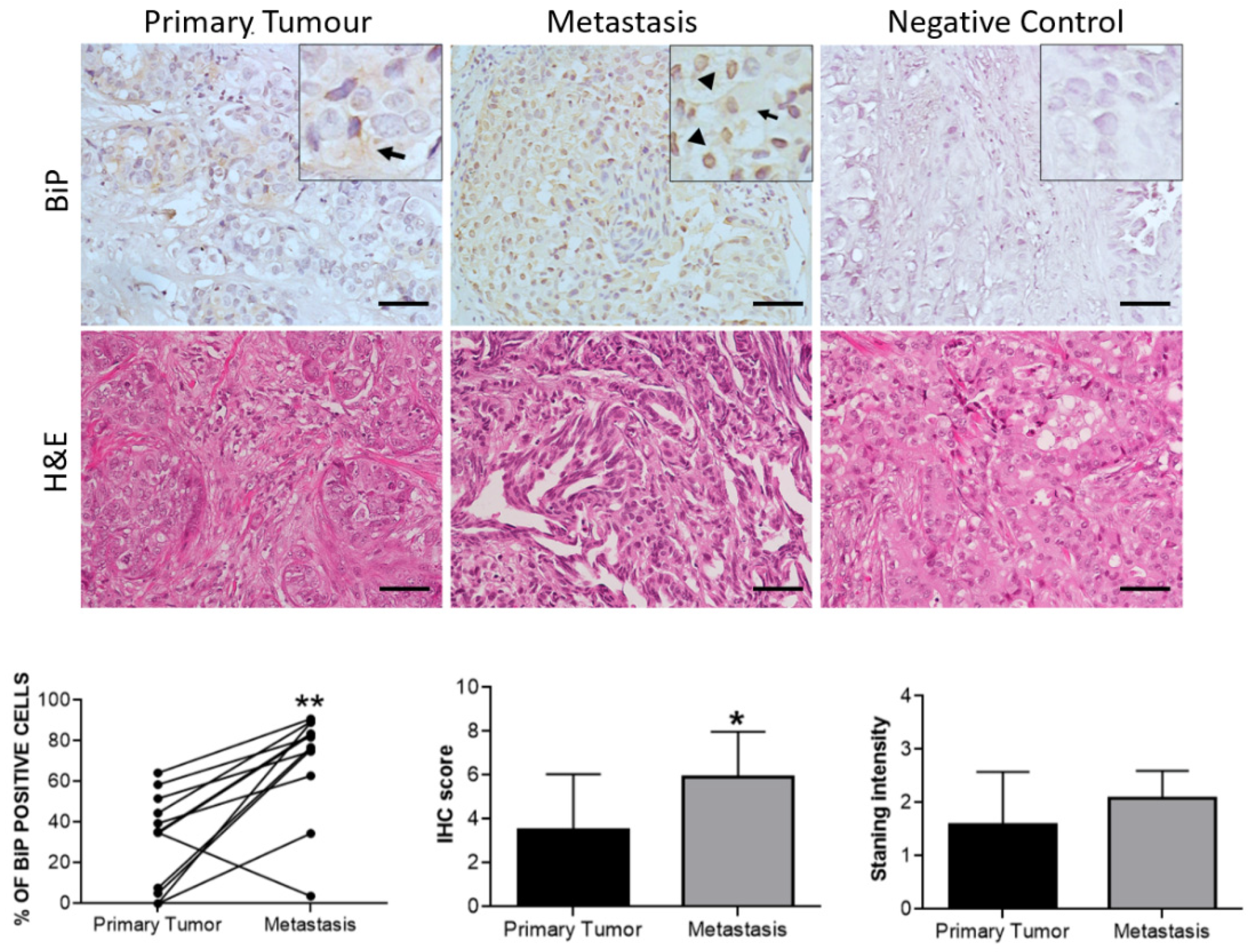
3.3. Effect of Therapy and Metastasis on BiP Expression in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Fact Sheets: Breast Cancer. Available online: http://gco.iarc.fr/today (accessed on 27 December 2021).
- Direito, I.; Fardilha, M.; Helguero, L.A. Contribution of the Unfolded Protein Response to Breast and Prostate Tissue Homeostasis and Its Significance to Cancer Endocrine Response. Carcinogenesis 2019, 40, 203–215. [Google Scholar] [CrossRef]
- Wang, J.; Lee, J.; Liem, D.; Ping, P. HSPA5 Gene Encoding Hsp70 Chaperone BiP in the Endoplasmic Reticulum. Gene 2017, 618, 14. [Google Scholar] [CrossRef]
- Carvalho, H.H.; Silva, P.A.; Mendes, G.C.; Brustolini, O.J.B.; Pimenta, M.R.; Gouveia, B.C.; Valente, M.A.S.; Ramos, H.J.O.; Soares-Ramos, J.R.L.; Fontes, E.P.B. The Endoplasmic Reticulum Binding Protein BiP Displays Dual Function in Modulating Cell Death Events. Plant Physiol. 2014, 164, 654. [Google Scholar] [CrossRef]
- Yao, X.; Liu, H.; Zhang, X.; Zhang, L.; Li, X.; Wang, C.; Sun, S. Cell Surface GRP78 Accelerated Breast Cancer Cell Proliferation and Migration by Activating STAT3. PLoS ONE 2015, 10, e0125634. [Google Scholar] [CrossRef]
- Baptista, M.Z.; Sarian, L.; Vassallo, J.; Pinto, G.A.; Soares, F.A.; de Souza, G.A. Prognostic Significance of GRP78 Expression Patterns in Breast Cancer Patients Receiving Adjuvant Chemotherapy. Int. J. Biol. Markers 2011, 26, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, P.M.; Tabbara, S.O.; Jacobs, L.K.; Manning, F.C.R.; Tsangaris, T.N.; Schwartz, A.M.; Kennedy, K.A.; Patierno, S.R. Overexpression of the Glucose-Regulated Stress Gene GRP78 in Malignant but Not Benign Human Breast Lesions. Breast Cancer Res. Treat. 2000, 59, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, I.; Woods, A.G.; Gawinowicz, M.A.; Roy, U.; Darie, C.C. Identification of Potential Tumor Differentiation Factor (TDF) Receptor from Steroid-Responsive and Steroid-Resistant Breast Cancer Cells. J. Biol. Chem. 2012, 287, 1719. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Shajahan, A.N.; Wärri, A.; Jin, L.; Hilakivi-Clarke, L.A.; Clarke, R. Glucose-Regulated Protein 78 Controls Cross-Talk between Apoptosis and Autophagy to Determine Antiestrogen Responsiveness. Cancer Res. 2012, 72, 3337. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cao, Z.; Hu, X.; Shao, Z. The Endoplasmic Reticulum Stress Markers GRP78 and CHOP Predict Disease-Free Survival and Responsiveness to Chemotherapy in Breast Cancer. Breast Cancer Res. Treat. 2014, 145, 349–358. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, Z.; Zou, Y.; Gao, G.; Liu, L.; Xu, H.; Liu, F. Expression of Glucose-Regulated Protein 78 as Prognostic Biomarker for Triple-Negative Breast Cancer. Histol. Histopathol. 2020, 35, 559–568. [Google Scholar] [CrossRef]
- Yerushalmi, R.; Raiter, A.; Nalbandyan, K.; Hardy, B. Cell Surface GRP78: A Potential Marker of Good Prognosis and Response to Chemotherapy in Breast Cancer. Oncol. Lett. 2015, 10, 2149. [Google Scholar] [CrossRef]
- Raiter, A.; Yerushalmi, R.; Hardy, B. Pharmacological Induction of Cell Surface GRP78 Contributes to Apoptosis in Triple Negative Breast Cancer Cells. Oncotarget 2014, 5, 11452. [Google Scholar] [CrossRef]
- Dhamad, A.E.; Zhou, Z.; Zhou, J.; Du, Y. Systematic Proteomic Identification of the Heat Shock Proteins (Hsp) That Interact with Estrogen Receptor Alpha (ERα) and Biochemical Characterization of the ERα-Hsp70 Interaction. PLoS ONE 2016, 11, 160312. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lee, A. Stress Induction of GRP78/BiP and Its Role in Cancer. Curr. Mol. Med. 2006, 6, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wey, S.; Zhang, Y.; Ye, R.; Lee, A.S. Role of the Unfolded Protein Response Regulator GRP78/BiP in Development, Cancer, and Neurological Disorders. Antioxid Redox Signal. 2009, 11, 2307. [Google Scholar] [CrossRef]
- Fu, Y.; Lee, A.S. Glucose Regulated Proteins in Cancer Progression, Drug Resistance and Immunotherapy. Cancer Biol. 2006, 5, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Gazit, G.; Lu, J.; Lee, A.S. De-Regulation of GRP Stress Protein Expression in Human Breast Cancer Cell Lines. Breast Cancer Res. Treat. 1999, 54, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z. Glucose Regulated Protein 78: A Critical Link between Tumor Microenvironment and Cancer Hallmarks. Biochim. Biophys. Acta 2012, 1826, 13–22. [Google Scholar] [CrossRef] [PubMed]
- La, X.; Zhang, L.; Li, H.; Li, Z.; Song, G.; Yang, P.; Yang, Y. Ajuba Receptor Mediates the Internalization of Tumor-Secreted GRP78 into Macrophages through Different Endocytosis Pathways. Oncotarget 2018, 9, 15464–15479. [Google Scholar] [CrossRef][Green Version]
- PanCanAtlas Publications|NCI Genomic Data Commons. Available online: https://gdc.cancer.gov/about-data/publications/pancanatlas (accessed on 26 July 2022).
- CPTAC|Office of Cancer Clinical Proteomics Research. Available online: https://proteomics.cancer.gov/programs/cptac (accessed on 26 July 2022).
- CBioPortal for Cancer Genomics. Available online: https://www.cbioportal.org/ (accessed on 26 July 2022).
- Bartha, Á.; Győrffy, B. TNMplot.Com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef] [PubMed]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 26 July 2022).
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New Functionalities in the TCGAbiolinks Package for the Study and Integration of Cancer Data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef] [PubMed]
- Bioconductor—MetagenomeSeq. Available online: https://bioconductor.org/packages/release/bioc/html/metagenomeSeq.html (accessed on 16 August 2022).
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the EdgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive Evaluation of Transcriptome-Based Cell-Type Quantification Methods for Immuno-Oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef]
- Balduzzi, S.; Rücker, G.; Schwarzer, G. How to Perform a Meta-Analysis with R: A Practical Tutorial. Evid. Based Ment. Health 2019, 22, 153–160. [Google Scholar] [CrossRef]
- Package “netmeta” Title Network Meta-Analysis Using Frequentist Methods. Available online: https://cran.r-project.org/web/packages/netmeta/netmeta.pdf (accessed on 22 June 2021).
- Higgins, J.P.T.; Thompson, S.G. Quantifying Heterogeneity in a Meta-Analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- HSPA5—Antibodies—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000044574-HSPA5/antibody (accessed on 26 July 2022).
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the Population Abundance of Tissue-Infiltrating Immune and Stromal Cell Populations Using Gene Expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting Recommendations for Tumour MARKer Prognostic Studies (REMARK). Br. J. Cancer 2005, 93, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Nichols, P.; Spicer, D.; Groshen, S.; Yu, M.C.; Lee, A.S. GRP78 as a Novel Predictor of Responsiveness to Chemotherapy in Breast Cancer. Cancer Res. 2006, 66, 7849–7853. [Google Scholar] [CrossRef]
- Bartkowiak, K.; Kwiatkowski, M.; Buck, F.; Gorges, T.M.; Nilse, L.; Assmann, V.; Andreas, A.; Müller, V.; Wikman, H.; Riethdorf, S.; et al. Disseminated Tumor Cells Persist in the Bone Marrow of Breast Cancer Patients through Sustained Activation of the Unfolded Protein Response. Cancer Res. 2015, 75, 5367–5377. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Tseng, C.; Wang, M.; Chang, W.; Lee, C.; Chen, L.; Hung, M.; Su, J. Deacetylation of HSPA5 by HDAC6 Leads to GP78-Mediated HSPA5 Ubiquitination at K447 and Suppresses Metastasis of Breast Cancer. Oncogene 2016, 35, 1517–1528. [Google Scholar] [CrossRef]
- Yang, F.; Tang, X.Y.; Liu, H.; Jiang, Z.W. Inhibition of Mitogen-Activated Protein Kinase Signaling Pathway Sensitizes Breast Cancer Cells to Endoplasmic Reticulum Stress-Induced Apoptosis. Oncol. Rep. 2016, 35, 2113–2120. [Google Scholar] [CrossRef]
- Zhang, D.; Tai, L.K.; Wong, L.L.; Putti, T.C.; Sethi, S.K.; Teh, M.; Koay, E.S. Proteomic characterization of differentially expressed proteins in breast cancer: Expression of hnRNP H1, RKIP and GRP78 is strongly associated with HER-2/neu status. Proteom. Clin. Appl. 2008, 2, 99–107. [Google Scholar] [CrossRef]
- María Teresa de Jesús, C.D.; Agni Jaim, M.G.; Cindy Karina, V.V.; Víctor Alberto, O.C.; Nicolás, R.T.; Verónica, G.O.; Fabio Abdel, S.G.; Patricia, P.S.; Sergio, F.H.; Eunice, L.M. BIK and GRP78 Protein Expression as Possible Markers of Response to Preoperative Chemotherapy and Survival in Breast Cancer. Taiwan J. Obs. Gynecol. 2021, 60, 245–252. [Google Scholar] [CrossRef]
- López-Muñoz, E.; Corres-Molina, M.; García-Hernández, N. Correlation of the protein expression of GRP78 and BIK/NBK with prognostic markers in patients with breast cancer and neoadjuvant chemotherapy. J. Obstet. Gynaecol. 2019, 40, 419–426. [Google Scholar] [CrossRef]
- Lee, E.; Nichols, P.; Groshen, S.; Spicer, D.; Lee, A.S. GRP78 as Potential Predictor for Breast Cancer Response to Adjuvant Taxane Therapy. Int. J. Cancer 2011, 128, 726. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elfiky, A.A. GRP78: A Cell’s Response to Stress. Life Sci. 2019, 226, 156. [Google Scholar] [CrossRef]
- Scriven, P.; Coulson, S.; Haines, R.; Balasubramanian, S.; Cross, S.; Wyld, L. Activation and Clinical Significance of the Unfolded Protein Response in Breast Cancer. Br. J. Cancer 2009, 101, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Conner, C.; Lager, T.W.; Guldner, I.H.; Wu, M.; Hishida, Y.; Hishida, T.; Ruiz, S.; Yamasaki, A.E.; Gilson, R.C.; Belmonte, J.; et al. Cell Surface GRP78 Promotes Stemness in Normal and Neoplastic Cells. Sci. Rep. 2020, 10, 3474. [Google Scholar] [CrossRef]
- Chang, Y.; Chen, H.; Tseng, C.; Hong, C.; Ma, J.; Hung, M.; Wu, C.; Huang, M.; Su, J. De-Acetylation and Degradation of HSPA5 Is Critical for E1A Metastasis Suppression in Breast Cancer Cells. Oncotarget 2014, 5, 10558. [Google Scholar] [CrossRef] [PubMed]
- Ethier, J.; Desautels, D.; Templeton, A.; Shah, P.S.; Amir, E. Prognostic Role of Neutrophil-to-Lymphocyte Ratio in Breast Cancer: A Systematic Review and Meta-Analysis. Breast Cancer Res. 2017, 19, 2. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Murthy, V.; Tian, W.; Yan, L.; Angarita, F.; Nagahashi, M.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; et al. Low Intratumoral Genetic Neutrophil-to-Lymphocyte Ratio (NLR) Is Associated with Favorable Tumor Immune Microenvironment and with Survival in Triple Negative Breast Cancer (TNBC). Am. J. Cancer Res. 2021, 11, 5743. [Google Scholar]
- Xu, J.; Ni, C.; Ma, C.; Zhang, L.; Jing, X.; Li, C.; Liu, Y.; Qu, X. Association of Neutrophil/Lymphocyte Ratio and Platelet/Lymphocyte Ratio with ER and PR in Breast Cancer Patients and Their Changes after Neoadjuvant Chemotherapy. Clin. Transl. Oncol. 2017, 19, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Slimane, K.; Bachelot, T.; Dunant, A.; Namer, M.; Barrelier, A.; Kabbaj, O.; Spano, J.; Marsiglia, H.; Rouzier, R.; et al. Breast Cancer With Synchronous Metastases: Trends in Survival During a 14-Year Period. J. Clin. Oncol. 2004, 22, 3302–3308. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, S.; Ding, N.; Li, N.; Huang, J.; Xiao, Z. Platelet/Lymphocyte Ratio Is Superior to Neutrophil/Lymphocyte Ratio as a Predictor of Chemotherapy Response and Disease-Free Survival in Luminal B-like (HER2−) Breast Cancer. Clin. Breast Cancer 2020, 20, e403–e409. [Google Scholar] [CrossRef]
- Jeong, J.; Suh, Y.; Jung, K. Context Drives Diversification of Monocytes and Neutrophils in Orchestrating the Tumor Microenvironment. Front. Immunol. 2019, 10, 1817. [Google Scholar] [CrossRef]
- Direito, I.; Monteiro, L.; Melo, T.; Figueira, D.; Lobo, J.; Enes, V.; Moura, G.; Henrique, R.; Santos, M.A.S.; Jerónimo, C.; et al. Protein Aggregation Patterns Inform about Breast Cancer Response to Antiestrogens and Reveal the RNA Ligase RTCB as Mediator of Acquired Tamoxifen Resistance. Cancers 2021, 13, 3195. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, J.; Lee, A.S. GRP78/BiP Inhibits Endoplasmic Reticulum BIK and Protects Human Breast Cancer Cells against Estrogen Starvation-Induced Apoptosis. Cancer Res. 2007, 67, 3734–3740. [Google Scholar] [CrossRef]
- Ariazi, E.A.; Cunliffe, H.E.; Lewis-Wambi, J.S.; Slifker, M.J.; Willis, A.L.; Ramos, P.; Tapia, C.; Kim, H.R.; Yerrum, S.; Sharma, C.G.N.; et al. Estrogen Induces Apoptosis in Estrogen Deprivation-Resistant Breast Cancer through Stress Responses as Identified by Global Gene Expression across Time. Proc. Natl. Acad. Sci. USA 2011, 108, 18879–18886. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.K.; Mao, C.; Baumeister, P.; Austin, R.C.; Kaufman, R.J.; Lee, A.S. Endoplasmic Reticulum Chaperone Protein GRP78 Protects Cells from Apoptosis Induced by Topoisomerase Inhibitors: Role of ATP Binding Site in Suppression of Caspase-7 Activation. J. Biol. Chem. 2003, 278, 20915–20924. [Google Scholar] [CrossRef]
- Barker, S.; Weinfeld, M.; Zheng, J.; Li, L.; Murray, D. Identification of Mammalian Proteins Cross-Linked to DNA by Ionizing Radiation. J. Biol. Chem. 2005, 280, 33826–33838. [Google Scholar] [CrossRef]
- Zhai, L.; Kita, K.; Wano, C.; Wu, Y.; Sugaya, S.; Suzuki, N. Decreased Cell Survival and DNA Repair Capacity after UVC Irradiation in Association with Down-Regulation of GRP78/BiP in Human RSa Cells. Exp. Cell Res. 2005, 305, 244–252. [Google Scholar] [CrossRef]
- François-Moutal, L.; Scott, D.; Ambrose, A.J.; Zerio, C.J.; Rodriguez-Sanchez, M.; Dissanayake, K.; May, D.G.; Carlson, J.M.; Barbieri, E.; Moutal, A.; et al. Heat Shock Protein Grp78/BiP/HspA5 Binds Directly to TDP-43 and Mitigates Toxicity Associated with Disease Pathology. Sci. Rep. 2022, 12, 8140. [Google Scholar] [CrossRef]
- Raiter, A.; Lipovetsky, J.; Hyman, L.; Mugami, S.; Ben-Zur, T.; Yerushalmi, R. Chemotherapy Controls Metastasis Through Stimulatory Effects on GRP78 and Its Transcription Factor CREB3L1. Front. Oncol. 2020, 10, 1500. [Google Scholar] [CrossRef]
- Jakobsen, C.G.; Rasmussen, N.; Laenkholm, A.; Ditzel, H.J. Phage Display Derived Human Monoclonal Antibodies Isolated by Binding to the Surface of Live Primary Breast Cancer Cells Recognize GRP78. Cancer Res. 2007, 67, 9507–9517. [Google Scholar] [CrossRef]
- Hernandez, I.; Cohen, M. Linking Cell-Surface GRP78 to Cancer: From Basic Research to Clinical Value of GRP78 Antibodies. Cancer Lett. 2022, 524, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, R.; Ni, M.; Gill, P.; Lee, A.S. Cell Surface Relocalization of the Endoplasmic Reticulum Chaperone and Unfolded Protein Response Regulator GRP78/BiP. J. Biol. Chem. 2010, 285, 15065–15075. [Google Scholar] [CrossRef] [PubMed]

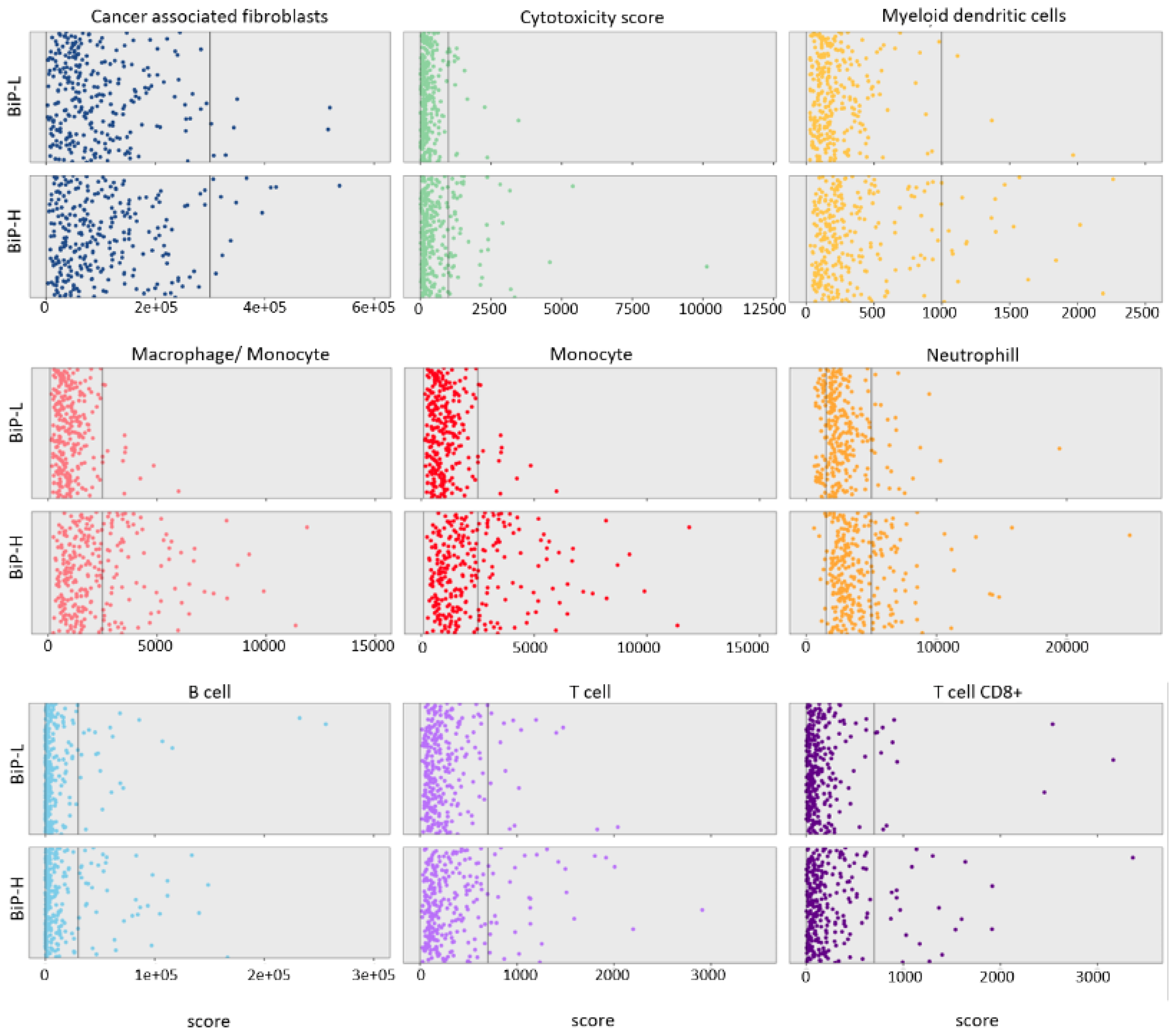
| Clinical Attribute | Attribute Type | Statistical Test | p-Value | q-Value | Higher in |
|---|---|---|---|---|---|
| TOP2A Proteogenomic Status | Patient | Chi-squared Test | 5.67 × 10−14 | 1.82 × 10−12 | BiP-H |
| ERBB2 Proteogenomic Status | Patient | Chi-squared Test | 8.17 × 10−13 | 1.31 × 10−11 | BiP-L |
| TNBREAST CANCER Updated Clinical Status | Patient | Chi-squared Test | 1.38 × 10−10 | 1.47 × 10−9 | BiP-H |
| xCell Immune Score | Patient | Wilcoxon Test | 8.45 × 10−5 | 5.41 × 10−4 | BiP-H |
| ESTIMATE Immune Score | Patient | Wilcoxon Test | 3.10 × 10−4 | 1.66 × 10−3 | BiP-H |
| PAM50 | Sample | Chi-squared Test | 9.61 × 10−4 | 4.39 × 10−3 | |
| CIBERSORT Absolute Score | Patient | Wilcoxon Test | 3.10 × 10−3 | 0.0123 | BiP-H |
| ER Updated Clinical Status | Patient | Chi-squared Test | 3.46 × 10−3 | 0.0123 | BiP-L |
| ESTIMATE TumorPurity | Patient | Wilcoxon Test | 7.61 × 10−3 | 0.0244 | BiP-L |
| PR Clinical Status | Patient | Chi-squared Test | 0.012 | 0.035 | BiP-L |
| #term ID | Term Description | Genes Mapped | Enrichment Score | Direction | FDR | Method |
|---|---|---|---|---|---|---|
| KEGG PATHWAYS | ||||||
| hsa04141 | Protein processing in the endoplasmic reticulum | 43 | 399.369 | BiP-H | 3.47 × 10−12 | ks |
| hsa05169 | Epstein–Barr virus infection | 40 | 363.869 | BiP-H | 1.94 × 10−7 | ks |
| hsa04612 | Antigen processing and presentation | 21 | 493.643 | BiP-H | 1.97 × 10−7 | ks |
| hsa04110 | Cell cycle | 29 | 409.514 | BiP-H | 8.80 × 10−7 | ks |
| hsa04650 | Natural killer cell-mediated cytotoxicity | 26 | 390.086 | BiP-H | 1.91 × 10−6 | ks |
| hsa05164 | Influenza A | 30 | 351.733 | BiP-H | 4.67 × 10−6 | ks |
| hsa05332 | Graft-versus-host disease | 15 | 461.606 | BiP-H | 1.40 × 10−5 | afc |
| hsa04940 | Type I diabetes mellitus | 13 | 479.743 | BiP-H | 2.75 × 10−5 | afc |
| hsa04145 | Phagosome | 38 | 26.468 | BiP-H | 6.58 × 10−5 | ks |
| hsa05152 | Tuberculosis | 28 | 285.612 | BiP-H | 6.58 × 10−5 | ks |
| hsa03050 | Proteasome | 14 | 420.107 | BiP-H | 0.00023 | afc |
| hsa04142 | Lysosome | 22 | 26.693 | BiP-H | 0.0031 | ks |
| hsa05020 | Prion disease | 42 | 125.948 | BiP-H | 0.0087 | ks |
| REACTOME PATHWAYS | ||||||
| HSA-168249 | Innate immune system | 178 | 269.185 | BiP-H | 2.16 × 10−24 | ks |
| HSA-6798695 | Neutrophil degranulation | 90 | 348.227 | BiP-H | 1.06 × 10−19 | ks |
| HSA-1280218 | Adaptive immune system | 132 | 248.407 | BiP-H | 2.22 × 10−14 | ks |
| HSA-1280215 | Cytokine Signaling in the immune system | 149 | 234.508 | BiP-H | 5.12 × 10−12 | ks |
| HSA-913531 | Interferon signaling | 47 | 415.029 | BiP-H | 6.46 × 10−12 | ks |
| HSA-1236975 | Antigen processing cross-presentation | 31 | 421.988 | BiP-H | 1.09 × 10−9 | ks |
| HSA-909733 | Interferon alpha/beta signaling | 27 | 501.242 | BiP-H | 2.01 × 10−9 | ks |
| HSA-72766 | Translation | 23 | 42.642 | BiP-H | 4.59 × 10−9 | ks |
| HSA-1236974 | ER–phagosome pathway | 28 | 420.465 | BiP-H | 1.62 × 10−8 | ks |
| HSA-983169 | Class I MHC-mediated antigen processing and presentation | 54 | 311.864 | BiP-H | 1.68 × 10−8 | ks |
| HSA-5688426 | Deubiquitination | 45 | 299.335 | BiP-H | 3.27 × 10−8 | ks |
| HSA-381119 | Unfolded protein response (UPR) | 20 | 437.966 | BiP-H | 8.16 × 10−7 | afc |
| HSA-2132295 | MHC class II antigen presentation | 31 | 321.922 | BiP-H | 1.24 × 10−5 | ks |
| HSA-381070 | IRE1alpha activates chaperones | 14 | 459.057 | BiP-H | 1.24 × 10−5 | afc |
| HSA-449147 | Signaling by Interleukins | 97 | 186.579 | BiP-H | 1.79 × 10−5 | ks |
| HSA-381038 | XBP1(S) activates chaperone genes | 13 | 428.195 | BiP-H | 0.00017 | afc |
| HSA-977225 | Amyloid fiber formation | 13 | 330.029 | BiP-H | 0.0055 | afc |
| HSA-983168 | Antigen processing: ubiquitination and proteasome degradation | 32 | 217.782 | BiP-H | 0.0060 | ks |
| #term ID | Term Description | Genes Mapped | Enrichment Score | Direction | FDR | Method |
|---|---|---|---|---|---|---|
| KEGG PATHWAYS | ||||||
| hsa04141 | Protein processing in the endoplasmic reticulum | 40 | 183.735 | BiP-H | 9.15 × 10−6 | ks |
| hsa04657 | IL-17 signaling pathway | 15 | 325.286 | BiP-H | 0.00014 | afc |
| hsa01100 | Metabolic pathways | 151 | 0.503067 | Both | 0.0049 | ks |
| REACTOME PATHWAYS | ||||||
| HSA-6798695 | Neutrophil degranulation | 94 | 23.212 | BiP-H | 1.65 × 10−9 | ks |
| HSA-168249 | Innate immune system | 170 | 156.502 | BiP-H | 3.49 × 10−8 | ks |
| HSA-1474244 | Extracellular matrix organization | 37 | 249.065 | BiP-H | 1.52 × 10−7 | ks |
| HSA-6799990 | Metal sequestration by antimicrobial proteins | 5 | 737.368 | BiP-H | 2.17 × 10−6 | afc |
| HSA-6803157 | Antimicrobial peptides | 12 | 5.985 | BiP-H | 2.17 × 10−6 | afc |
| HSA-72766 | Translation | 58 | 113.467 | BiP-H | 1.46 × 10−5 | ks |
| HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 46 | 119.151 | BiP-H | 0.00011 | ks |
| HSA-1474228 | Degradation of the extracellular matrix | 16 | 280.973 | BiP-H | 0.00041 | afc |
| HSA-71291 | Metabolism of amino acids and derivatives | 75 | 102.951 | BiP-H | 0.00041 | ks |
| HSA-1442490 | Collagen degradation | 10 | 321.766 | BiP-H | 0.0020 | afc |
| HSA-877300 | Interferon gamma signaling | 18 | 222.754 | BiP-H | 0.0063 | afc |
| HSA-381119 | Unfolded protein response (UPR) | 18 | 21.652 | BiP-H | 0.0077 | afc |
| HSA-1280215 | Cytokine signaling in the immune system | 80 | 115.149 | BiP-H | 0.0089 | ks |
| HSA-198933 | Immunoregulatory interactions between a lymphoid and a non-lymphoid cell | 13 | 248.601 | BiP-H | 0.0089 | afc |
| HSA-1236975 | Antigen processing cross-presentation | 30 | 159.939 | BiP-H | 0.0095 | ks |
| HSA-1280218 | Adaptive immune system | 103 | 0.869687 | BiP-H | 0.0098 | ks |
| HSA-5617833 | Cilium assembly | 32 | 251172 | BiP-L | 1.85 × 10−5 | ks |
| HSA-1852241 | Organelle biogenesis and maintenance | 38 | 211028 | BiP-L | 0.00019 | ks |
| HSA-73894 | DNA repair | 31 | 198671 | BiP-L | 0.00099 | ks |
| HSA-9018519 | Estrogen-dependent gene expression | 15 | 328834 | BiP-L | 0.0020 | afc |
| HSA-74160 | Gene expression (Transcription) | 111 | 0.735739 | BiP-L | 0.0050 | ks |
| HSA-5620924 | Intraflagellar transport | 16 | 296691 | BiP-L | 0.0060 | afc |
| Stratification | No. of Studies | p-Value | I2 (%) |
|---|---|---|---|
| Menopause status | 6 | 0.1092 | 70.00 |
| Tumor stage | 7 | 0.7248 | 75.84 |
| Tumor grade | 9 | 0.0498 * | 74.77 |
| Lymph node metastasis | 10 | 0.9594 | 84.40 |
| ER expression | 10 | 0.0371 * | 68.72 |
| HER2 expression | 11 | <0.0001 * | 47.08 |
| Age | 5 | 0.0018 * | 0.00 |
| Clinical Factor | Cases, No. | p-Value |
|---|---|---|
| Histology | 14 | 0.449 |
| Age | 14 | 1.000 |
| Grade | 14 | 0.014 * |
| HER2+ | 14 | 0.046 * |
| Stage | 14 | 0.946 |
| Lymph node metastasis | 14 | 0.308 |
| Vital status | 14 | 0.353 |
| Response to HT | 14 | 0.120 |
| Ki-67 index | 14 | 0.373 # |
| Clinical Factor | Cases, No. | p-Value |
|---|---|---|
| HER2+ | 14 | 1.000 |
| Grade | 14 | 0.209 |
| Stage | 14 | 0.038 * |
| Lymph node metastasis | 14 | 0.209 |
| Vital status | 14 | 0.043 * |
| Response to HT | 14 | 0.326 |
| Clinical Factor | Cases, No. | p-Value |
|---|---|---|
| Age | 12 | 1.000 |
| Histology | 12 | 0.510 |
| Grade | 12 | 0.567 |
| HER2+ | 12 | 1.000 |
| Stage | 11 | 1.000 |
| Lymph node metastasis | 8 | 1.000 |
| Time to metastasis | 12 | 0.834 |
| Clinical Factor | Cases, No. | p-Value |
|---|---|---|
| HER2+ | 12 | 1.000 |
| Grade | 12 | 1.000 |
| Stage | 11 | 0.900 |
| Lymph node metastasis | 8 | 0.604 |
| Time to metastasis | 12 | 0.682 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Direito, I.; Gomes, D.; Monteiro, F.L.; Carneiro, I.; Lobo, J.; Henrique, R.; Jerónimo, C.; Helguero, L.A. The Clinicopathological Significance of BiP/GRP-78 in Breast Cancer: A Meta-Analysis of Public Datasets and Immunohistochemical Detection. Curr. Oncol. 2022, 29, 9066-9087. https://doi.org/10.3390/curroncol29120710
Direito I, Gomes D, Monteiro FL, Carneiro I, Lobo J, Henrique R, Jerónimo C, Helguero LA. The Clinicopathological Significance of BiP/GRP-78 in Breast Cancer: A Meta-Analysis of Public Datasets and Immunohistochemical Detection. Current Oncology. 2022; 29(12):9066-9087. https://doi.org/10.3390/curroncol29120710
Chicago/Turabian StyleDireito, Inês, Daniela Gomes, Fátima Liliana Monteiro, Isa Carneiro, João Lobo, Rui Henrique, Carmen Jerónimo, and Luisa Alejandra Helguero. 2022. "The Clinicopathological Significance of BiP/GRP-78 in Breast Cancer: A Meta-Analysis of Public Datasets and Immunohistochemical Detection" Current Oncology 29, no. 12: 9066-9087. https://doi.org/10.3390/curroncol29120710
APA StyleDireito, I., Gomes, D., Monteiro, F. L., Carneiro, I., Lobo, J., Henrique, R., Jerónimo, C., & Helguero, L. A. (2022). The Clinicopathological Significance of BiP/GRP-78 in Breast Cancer: A Meta-Analysis of Public Datasets and Immunohistochemical Detection. Current Oncology, 29(12), 9066-9087. https://doi.org/10.3390/curroncol29120710









