Overexpression of SCYL1 Is Associated with Progression of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagengts
2.2. Gene Expression Analysis
2.3. The Prognostic Value of SCYL1 Analysis
2.4. Human Tissue Specimens and Patient Information
2.5. Immunohistochemistry (IHC) Staining
2.6. Knockdown of SCYL1 by the shRNA
2.7. Cell Lysates Preparation and Immunoblotting
2.8. Cell Proliferation Assay
2.9. Cell Migration Assays
2.10. Statistical Analysis
3. Results
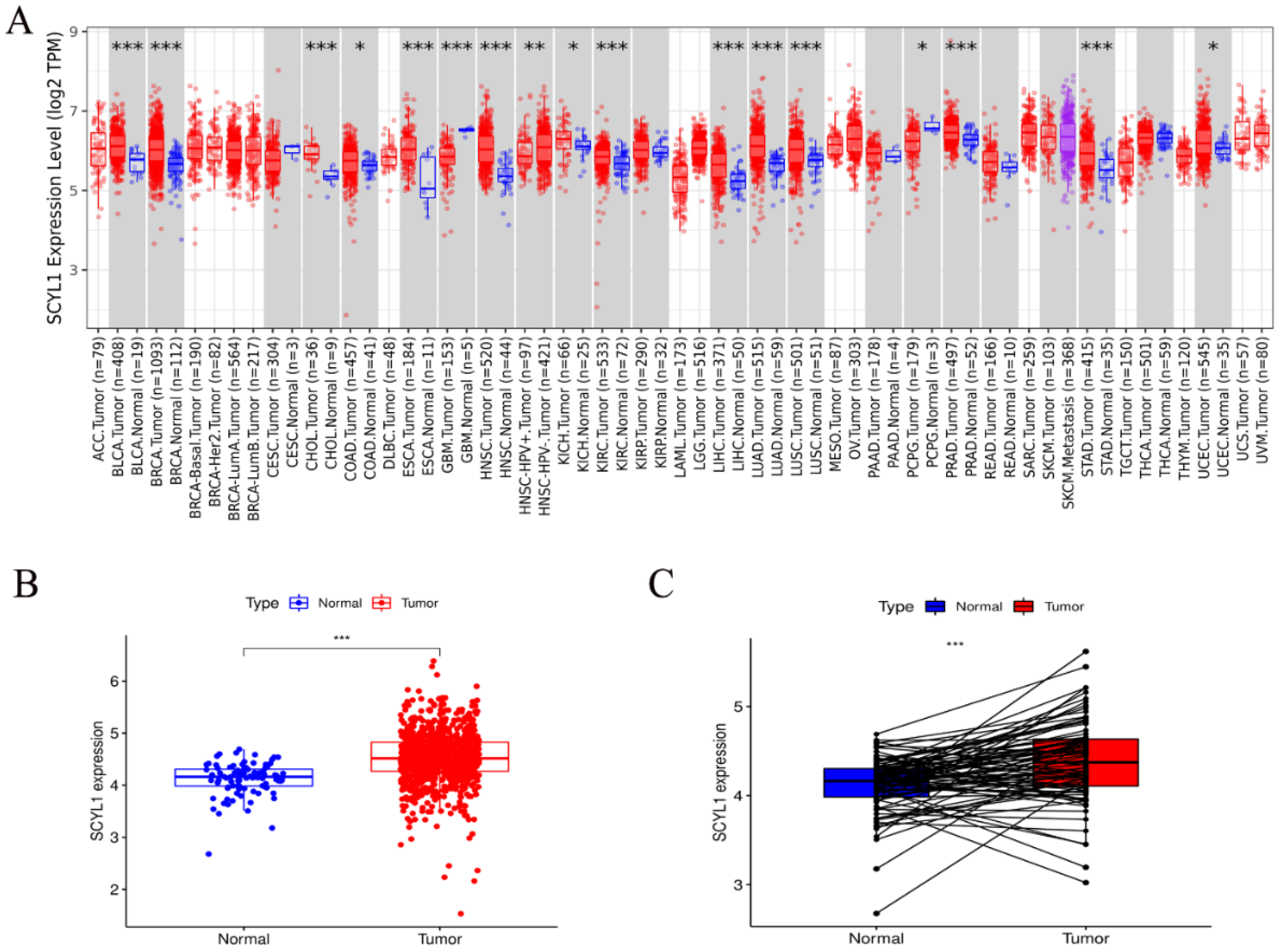
3.1. Expression of SCYL1 mRNA Is Increased in Breast Cancer Patients Than Normal Controls
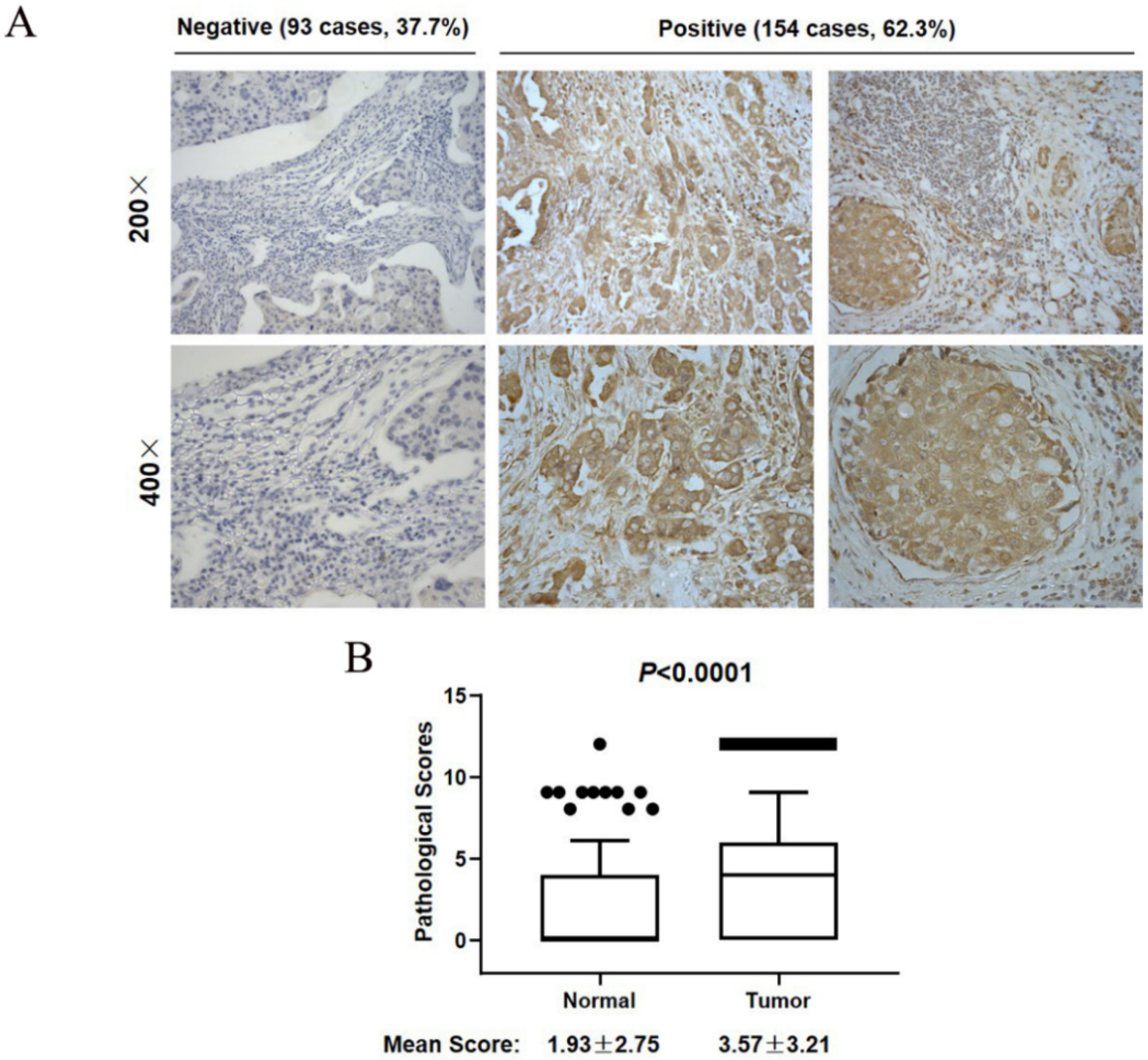
3.2. SCYL1 Is Abundantly Expressed in Breast Cancer Tumor Tissues
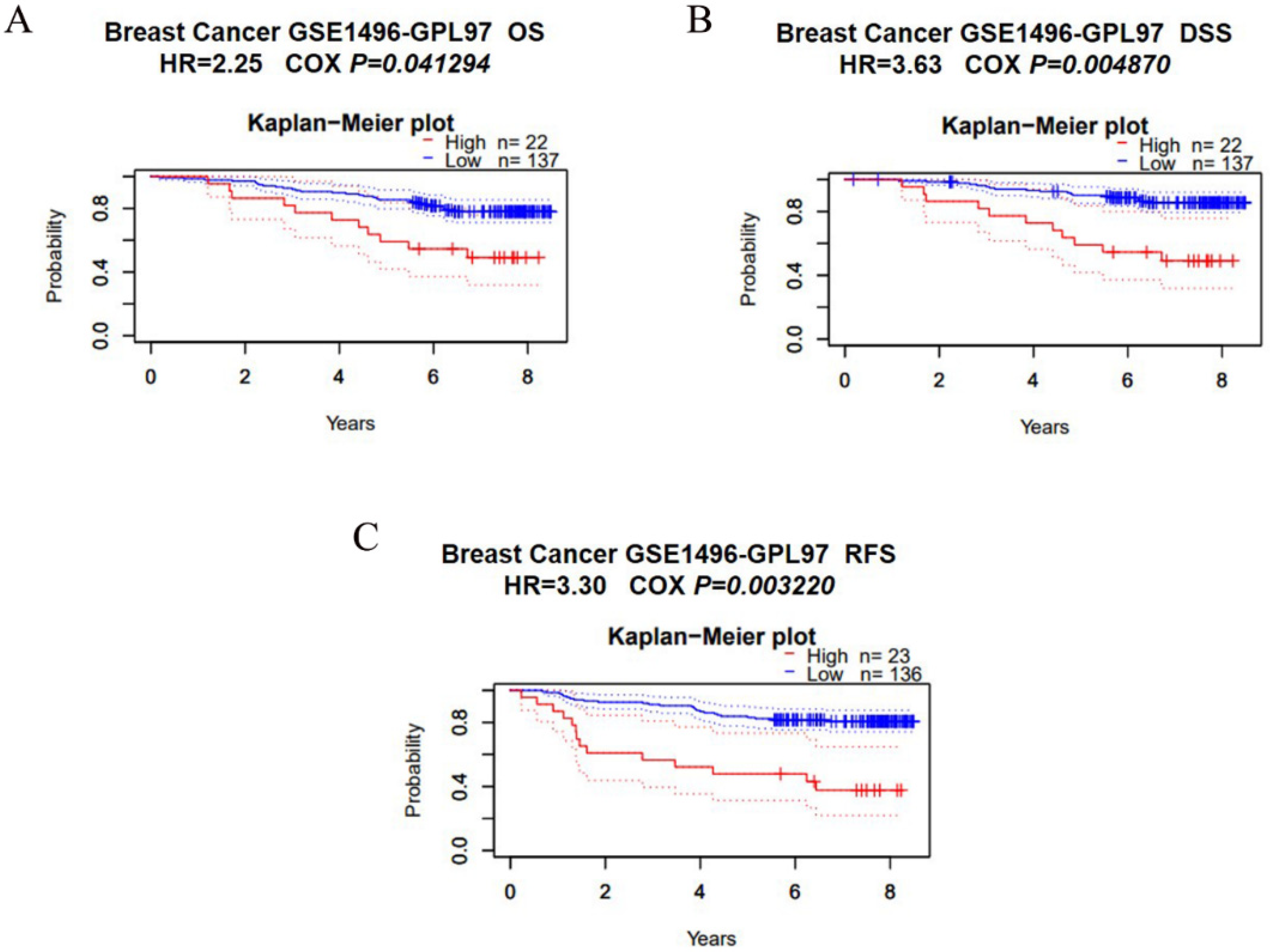
3.3. Overexpression of SCYL1 Is Associated with a Poorer Prognosis in Breast Cancer
3.4. Downregulation of SCYL1 Inhibits Breast Cancer Cell Proliferation
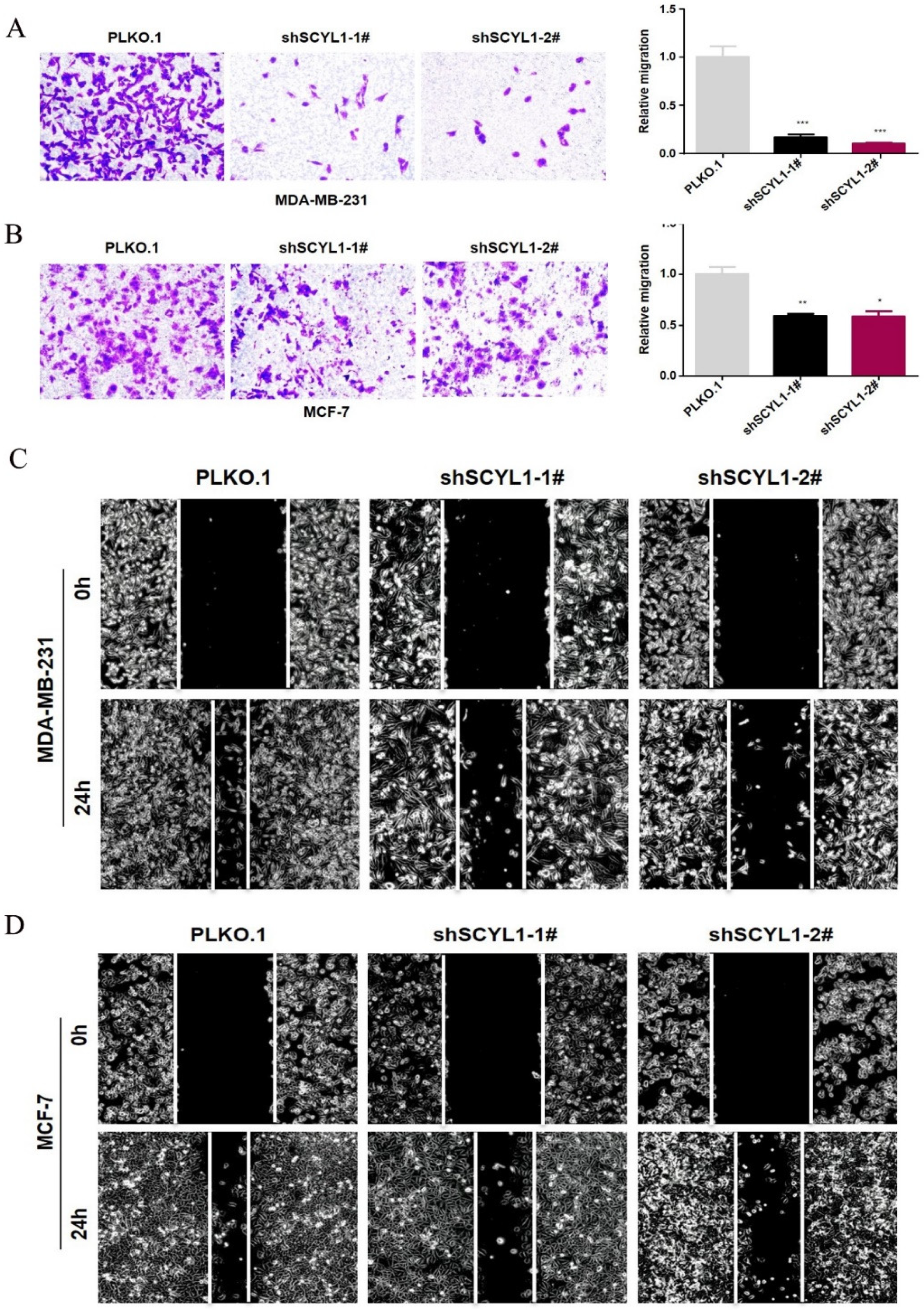
3.5. Downregulation of SCYL1 Inhibits Breast Cancer Cell Migration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA A Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Peart, O. Breast intervention and breast cancer treatment options. Radiol. Technol. 2015, 86, 535M–558M, quiz 559–562. [Google Scholar]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Guan, X. Cancer metastases: Challenges and opportunities. Acta Pharm. Sin. B 2015, 5, 402–418. [Google Scholar] [CrossRef]
- Chaudhary, L.N.; Wilkinson, K.H.; Kong, A. Triple-Negative Breast Cancer: Who Should Receive Neoadjuvant Chemotherapy? Surg. Oncol. Clin. N. Am. 2018, 27, 141–153. [Google Scholar] [CrossRef]
- Jones, S.E. Metastatic breast cancer: The treatment challenge. Clin. Breast Cancer 2008, 8, 224–233. [Google Scholar] [CrossRef]
- Li, G.; Hu, J.; Hu, G. Biomarker Studies in Early Detection and Prognosis of Breast Cancer. Adv. Exp. Med. Biol. 2017, 1026, 27–39. [Google Scholar] [CrossRef]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef]
- Liu, S.C.; Lane, W.S.; Lienhard, G.E. Cloning and preliminary characterization of a 105 kDa protein with an N-terminal kinase-like domain. Biochim. Biophys. Acta 2000, 1517, 148–152. [Google Scholar] [CrossRef]
- Kato, M.; Yano, K.; Morotomi-Yano, K.; Saito, H.; Miki, Y. Identification and characterization of the human protein kinase-like gene NTKL: Mitosis-specific centrosomal localization of an alternatively spliced isoform. Genomics 2002, 79, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006, 16, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, S. SCYL pseudokinases in neuronal function and survival. Neural Regen. Res. 2016, 11, 42–44. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.M.; Rutledge, S.L.; Schüle, R.; Mayerhofer, B.; Züchner, S.; Boltshauser, E.; Bittner, R.E. Disruptive SCYL1 Mutations Underlie a Syndrome Characterized by Recurrent Episodes of Liver Failure, Peripheral Neuropathy, Cerebellar Atrophy, and Ataxia. Am. J. Hum. Genet. 2015, 97, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Lenz, D.; McClean, P.; Kansu, A.; Bonnen, P.E.; Ranucci, G.; Thiel, C.; Straub, B.K.; Harting, I.; Alhaddad, B.; Dimitrov, B.; et al. SCYL1 variants cause a syndrome with low γ-glutamyl-transferase cholestasis, acute liver failure, and neurodegeneration (CALFAN). Genet. Med. 2018, 20, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Spagnoli, C.; Frattini, D.; Salerno, G.G.; Fusco, C. On CALFAN syndrome: Report of a patient with a novel variant in SCYL1 gene and recurrent respiratory failure. Genet. Med. 2019, 21, 1663–1664. [Google Scholar] [CrossRef]
- Hamlin, J.N.; Schroeder, L.K.; Fotouhi, M.; Dokainish, H.; Ioannou, M.S.; Girard, M.; Summerfeldt, N.; Melançon, P.; McPherson, P.S. Scyl1 scaffolds class II Arfs to specific subcomplexes of coatomer through the γ-COP appendage domain. J. Cell Sci. 2014, 127, 1454–1463. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, J.; Ling, Y.; Hou, L.; Zhang, B. Transcriptional upregulation of DNA polymerase beta by TEIF. Biochem. Biophys. Res. Commun. 2005, 333, 908–916. [Google Scholar] [CrossRef]
- Chafe, S.C.; Mangroo, D. Scyl1 facilitates nuclear tRNA export in mammalian cells by acting at the nuclear pore complex. Mol. Biol. Cell 2010, 21, 2483–2499. [Google Scholar] [CrossRef]
- Burman, J.L.; Hamlin, J.N.; McPherson, P.S. Scyl1 regulates Golgi morphology. PLoS ONE 2010, 5, e9537. [Google Scholar] [CrossRef] [PubMed]
- Witkos, T.M.; Chan, W.L.; Joensuu, M.; Rhiel, M.; Pallister, E.; Thomas-Oates, J.; Mould, A.P.; Mironov, A.A.; Biot, C.; Guerardel, Y.; et al. GORAB scaffolds COPI at the trans-Golgi for efficient enzyme recycling and correct protein glycosylation. Nat. Commun. 2019, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Sun, Y.; McNutt, M.A.; Sun, Q.; Hou, L.; Liu, H.; Shen, Q.; Ling, Y.; Chi, Y.; Zhang, B. Localization of TEIF in the centrosome and its functional association with centrosome amplification in DNA damage, telomere dysfunction and human cancers. Oncogene 2009, 28, 1549–1560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Liu, M.; Chen, L.; Chan, T.H.; Jiang, L.; Yuan, Y.F.; Guan, X.Y. Overexpression of N-terminal kinase like gene promotes tumorigenicity of hepatocellular carcinoma by regulating cell cycle progression and cell motility. Oncotarget 2015, 6, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Karlin, K.L.; Mondal, G.; Hartman, J.K.; Tyagi, S.; Kurley, S.J.; Bland, C.S.; Hsu, T.Y.; Renwick, A.; Fang, J.E.; Migliaccio, I.; et al. The oncogenic STP axis promotes triple-negative breast cancer via degradation of the REST tumor suppressor. Cell Rep. 2014, 9, 1318–1332. [Google Scholar] [CrossRef] [PubMed]
- Gingras, S.; Kuliyev, E.; Pelletier, S. SCYL1 does not regulate REST expression and turnover. PLoS ONE 2017, 12, e0178680. [Google Scholar] [CrossRef]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef]
- Sun, A.; Yu, G.; Dou, X.; Yan, X.; Yang, W.; Lin, Q. Nedd4-1 is an exceptional prognostic biomarker for gastric cardia adenocarcinoma and functionally associated with metastasis. Mol. Cancer 2014, 13, 248. [Google Scholar] [CrossRef]
- Shao, G.; Wang, R.; Sun, A.; Wei, J.; Peng, K.; Dai, Q.; Yang, W.; Lin, Q. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol. Cancer 2018, 17, 24. [Google Scholar] [CrossRef]
- Sun, A.; Wei, J.; Childress, C.; Shaw, J.H.t.; Peng, K.; Shao, G.; Yang, W.; Lin, Q. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy 2017, 13, 522–537. [Google Scholar] [CrossRef]
- Zhu, X.; Niu, X.; Ge, C. Inhibition of LINC00994 represses malignant behaviors of pancreatic cancer cells: Interacting with miR-765-3p/RUNX2 axis. Cancer Biol. Ther. 2019, 20, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Iiizumi, M.; Liu, W.; Pai, S.K.; Furuta, E.; Watabe, K. Drug development against metastasis-related genes and their pathways: A rationale for cancer therapy. Biochim. Biophys. Acta 2008, 1786, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Burman, J.L.; Bourbonniere, L.; Philie, J.; Stroh, T.; Dejgaard, S.Y.; Presley, J.F.; McPherson, P.S. Scyl1, mutated in a recessive form of spinocerebellar neurodegeneration, regulates COPI-mediated retrograde traffic. J. Biol. Chem. 2008, 283, 22774–22786. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni-Gosavi, P.; Makhoul, C.; Gleeson, P.A. Form and function of the Golgi apparatus: Scaffolds, cytoskeleton and signalling. FEBS Lett. 2019, 593, 2289–2305. [Google Scholar] [CrossRef]
- Millarte, V.; Farhan, H. The Golgi in cell migration: Regulation by signal transduction and its implications for cancer cell metastasis. Sci. World J. 2012, 2012, 498278. [Google Scholar] [CrossRef]
- Liu, J.; Huang, Y.; Li, T.; Jiang, Z.; Zeng, L.; Hu, Z. The role of the Golgi apparatus in disease (Review). Int. J. Mol. Med. 2021, 47. [Google Scholar] [CrossRef]
- Halberg, N.; Sengelaub, C.A.; Navrazhina, K.; Molina, H.; Uryu, K.; Tavazoie, S.F. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell 2016, 29, 339–353. [Google Scholar] [CrossRef]
- Yu, R.Y.; Xing, L.; Cui, P.F.; Qiao, J.B.; He, Y.J.; Chang, X.; Zhou, T.J.; Jin, Q.R.; Jiang, H.L.; Xiao, Y. Regulating the Golgi apparatus by co-delivery of a COX-2 inhibitor and Brefeldin A for suppression of tumor metastasis. Biomater. Sci. 2018, 6, 2144–2155. [Google Scholar] [CrossRef]
- Li, H.; Zhang, P.; Luo, J.; Hu, D.; Huang, Y.; Zhang, Z.R.; Fu, Y.; Gong, T. Chondroitin Sulfate-Linked Prodrug Nanoparticles Target the Golgi Apparatus for Cancer Metastasis Treatment. ACS Nano 2019, 13, 9386–9396. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, A.; Tian, X.; Yang, W.; Lin, Q. Overexpression of SCYL1 Is Associated with Progression of Breast Cancer. Curr. Oncol. 2022, 29, 6922-6932. https://doi.org/10.3390/curroncol29100544
Sun A, Tian X, Yang W, Lin Q. Overexpression of SCYL1 Is Associated with Progression of Breast Cancer. Current Oncology. 2022; 29(10):6922-6932. https://doi.org/10.3390/curroncol29100544
Chicago/Turabian StyleSun, Aiqin, Xianyan Tian, Wannian Yang, and Qiong Lin. 2022. "Overexpression of SCYL1 Is Associated with Progression of Breast Cancer" Current Oncology 29, no. 10: 6922-6932. https://doi.org/10.3390/curroncol29100544
APA StyleSun, A., Tian, X., Yang, W., & Lin, Q. (2022). Overexpression of SCYL1 Is Associated with Progression of Breast Cancer. Current Oncology, 29(10), 6922-6932. https://doi.org/10.3390/curroncol29100544




