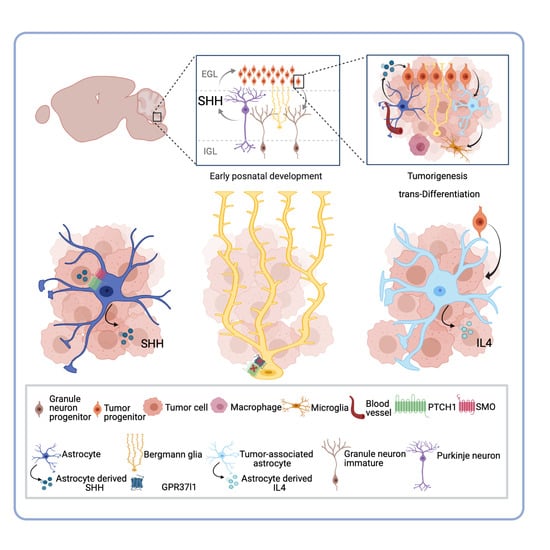
Decoding the Roles of Astrocytes and Hedgehog Signaling in Medulloblastoma
Abstract
:1. Molecular Signatures of Medulloblastoma Tumorigenesis
2. The Tumor Microenvironment and Its Roles in SHH Subgroup MB
3. Astrocytes and the Medulloblastoma Microenvironment: The New Player within the Complex Ecosystem
4. SHH-Activated Medulloblastoma: The Indispensable Role of Astrocyte-SHH Secretion in Tumor Progression
5. Therapeutic Approaches and the Intratumoral Heterogeneity of SHH Subgroup MB
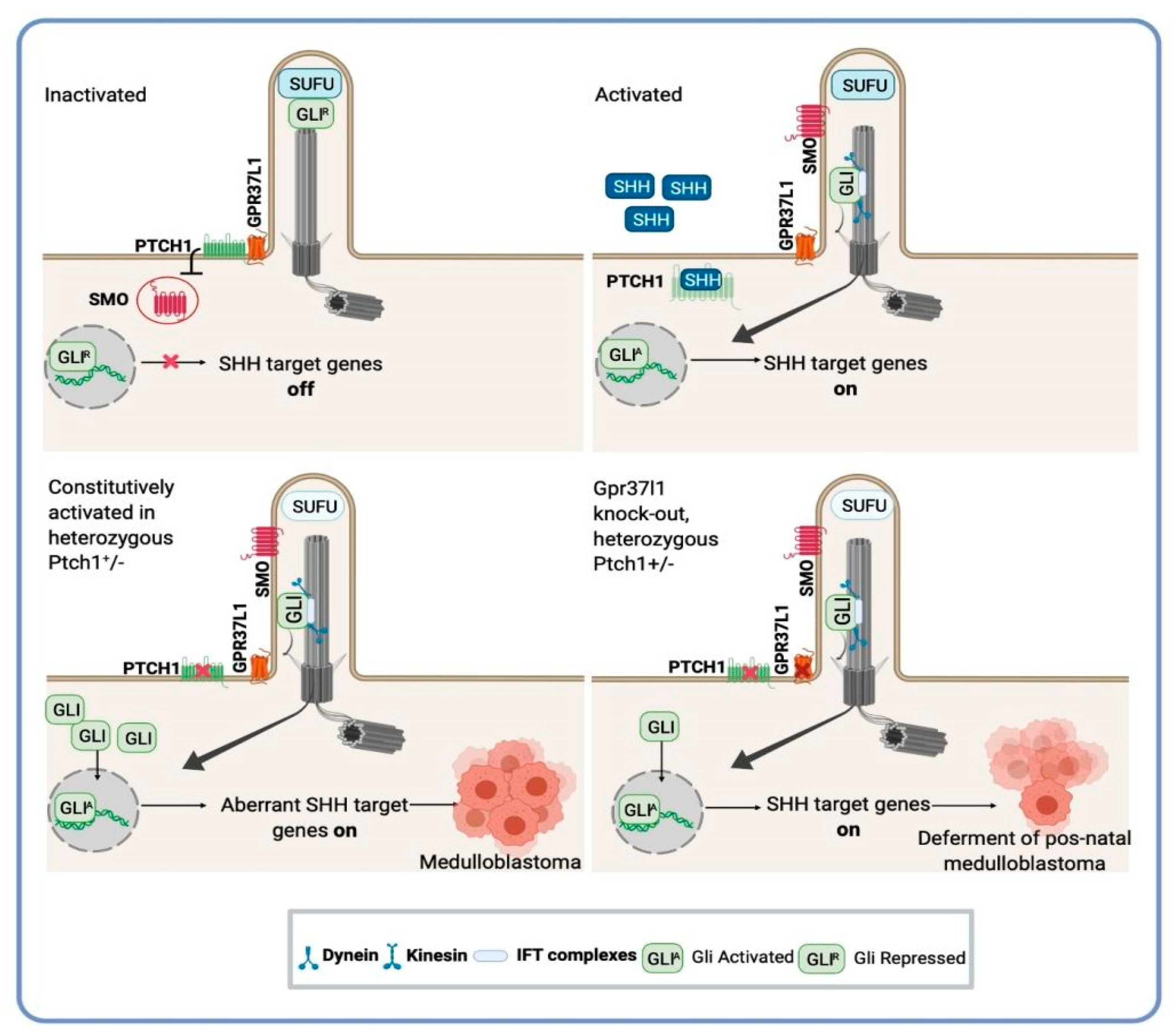
6. Novel Targets and Therapeutic Opportunities for Medulloblastoma: A Potential Application of Astrocytes-SHH Medulloblastoma Cross-Talk Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McNeil, D.E.; Coté, T.R.; Clegg, L.; Rorke, L.B. Incidence and trends in pediatric malignancies medulloblastoma/primitive neuroectodermal tumor: A SEER update. Med. Pediatr. Oncol. 2002, 39, 190–194. [Google Scholar] [CrossRef]
- Rutkowski, S.; Cohen, B.; Finlay, J.; Luksch, R.; Ridola, V.; Valteau-Couanet, D.; Hara, J.; Garrè, M.L.; Grill, J. Medulloblastoma in young children. Pediatr. Blood Cancer 2010, 54, 635–637. [Google Scholar] [CrossRef]
- Northcott, P.A.; Hielscher, T.; Dubuc, A.; Mack, S.C.; Shih, D.J.H.; Remke, M.; Al-Halabi, H.; Albrecht, S.; Jabado, N.; Eberhart, C.G.; et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011, 122, 231–240. [Google Scholar] [CrossRef]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.-J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2011, 123, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovestadt, V.; Ayrault, O.; Swartling, F.J.; Robinson, G.W.; Pfister, S.M.; Northcott, P.A. Medulloblastomics revisited: Biological and clinical insights from thousands of patients. Nat. Rev. Cancer 2019, 20, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Leal, L.F.; Evangelista, A.F.; De Paula, F.E.; Almeida, G.C.; Carloni, A.C.; Saggioro, F.; Stavale, J.N.; Malheiros, S.M.; Mançano, B.; De Oliveira, M.A.; et al. Reproducibility of the NanoString 22-gene molecular subgroup assay for improved prognostic prediction of medulloblastoma. Neuropathology 2018, 38, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Pomeroy, S.L.; Tamayo, P.; Gaasenbeek, M.; Sturla, L.M.; Angelo, M.; McLaughlin, M.E.; Kim, J.Y.H.; Goumnerova, L.C.; Black, P.M.; Lau, C.; et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002, 415, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.W.; Schlanstein, M.; Northcott, P.A.; Cho, Y.-J.; Koster, J.; Meeteren, A.S.-V.; Van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, F.M.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Liu, A.P.Y.; Northcott, P.A. Medulloblastoma genomics in the modern molecular era. Brain Pathol. 2019, 30, 679–690. [Google Scholar] [CrossRef]
- Garcia-Lopez, J.; Kumar, R.; Smith, K.S.; Northcott, P.A. Deconstructing Sonic Hedgehog Medulloblastoma: Molecular Subtypes, Drivers, and Beyond. Trends Genet. 2021, 37, 235–250. [Google Scholar] [CrossRef]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.; et al. Medulloblastoma. Nat. Rev. Dis. Prim. 2019, 5, 11. [Google Scholar] [CrossRef]
- Raffel, C.; Jenkins, R.B.; Frederick, L.; Hebrink, D.; Alderete, B.; Fults, D.W.; James, C.D. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997, 57, 842–845. [Google Scholar]
- Taylor, M.D.; Liu, L.; Raffel, C.; Hui, C.C.; Mainprize, T.G.; Zhang, X.; Agatep, R.; Chiappa, S.; Gao, L.; Lowrance, A.; et al. Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 2002, 31, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Brugières, L.; Pierron, G.; Chompret, A.; Paillerets, B.B.-D.; Di Rocco, F.; Varlet, P.; Pierre-Kahn, A.; Caron, O.; Grill, J.; Delattre, O. Incomplete penetrance of the predisposition to medulloblastoma associated with germ-line SUFU mutations. J. Med. Genet. 2009, 47, 142–144. [Google Scholar] [CrossRef]
- Gibson, P.; Tong, Y.; Robinson, G.; Thompson, M.C.; Currle, D.S.; Eden, C.; Kranenburg, T.; Hogg, T.; Poppleton, H.; Martin, J.; et al. Subtypes of medulloblastoma have distinct developmental origins. Nat. Cell Biol. 2010, 468, 1095–1099. [Google Scholar] [CrossRef]
- Buczkowicz, P.; Ma, J.; Hawkins, C. GLI2 Is a Potential Therapeutic Target in Pediatric Medulloblastoma. J. Neuropathol. Exp. Neurol. 2011, 70, 430–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhukova, N.; Ramaswamy, V.; Remke, M.; Pfaff, E.; Shih, D.J.H.; Martin, D.C.; Castelo-Branco, P.; Baskin, B.; Ray, P.N.; Bouffet, E.; et al. Subgroup-Specific Prognostic Implications of TP53 Mutation in Medulloblastoma. J. Clin. Oncol. 2013, 31, 2927–2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramaswamy, V.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, S.C.; Doz, F.; Kool, M.; Dufour, C.; Vassal, G.; Milde, T.; et al. Risk stratification of childhood medulloblastoma in the molecular era: The current consensus. Acta Neuropathol. 2016, 131, 821–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, L.S.; Mançano, B.M.; de Paula, F.E.; dos Reis, M.B.; de Almeida, G.C.; Matsushita, M.; Junior, C.A.; Evangelista, A.F.; Saggioro, F.; Serafini, L.N.; et al. Expression of GNAS, TP53, and PTEN Improves the Patient Prognostication in Sonic Hedgehog (SHH) Medulloblastoma Subgroup. J. Mol. Diagn. 2020, 22, 957–966. [Google Scholar] [CrossRef]
- Viana-Pereira, M.; Almeida, G.C.; Stavale, J.N.; Malheiro, S.; Clara, C.; Lobo, P.; Pimentel, J.; Reis, R. Study of hTERT and Histone 3 Mutations in Medulloblastoma. Pathobiology 2016, 84, 108–113. [Google Scholar] [CrossRef]
- Remke, M.; Ramaswamy, V.; Peacock, J.; Shih, D.J.H.; Koelsche, C.; Northcott, P.A.; Hill, N.; Cavalli, F.M.G.; Kool, M.; Wang, X.; et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta Neuropathol. 2013, 126, 917–929. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, W.; Digon-Söntgerath, B.; Koch, A.; Waha, A.; Endl, E.; Dani, I.; Denkhaus, D.; Goodyer, C.G.; Sörensen, N.; Wiestler, O.D.; et al. Phosphatidylinositol 3′-Kinase/AKT Signaling Is Activated in Medulloblastoma Cell Proliferation and Is Associated with Reduced Expression ofPTEN. Clin. Cancer Res. 2006, 12, 3019–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodrich, L.V.; Milenković, L.; Higgins, K.M.; Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997, 277, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Stephen, D.; Joyner, A.; Curran, T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene 2005, 24, 4026–4036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellino, R.C.; Barwick, B.G.; Schniederjan, M.; Buss, M.C.; Becher, O.; Hambardzumyan, D.; Macdonald, T.J.; Brat, D.J.; Durden, D.L. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PLoS ONE 2010, 5, e10849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrd, T.; Grossman, R.G.; Ahmed, N. Medulloblastoma-Biology and microenvironment: A review. Pediatr. Hematol. Oncol. 2012, 29, 495–506. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yuelling, L.W.; Wang, Y.; Du, F.; Gordon, R.E.; O'Brien, J.A. Astrocytes promote medulloblastoma progression through hedgehog secretion. Cancer Res. 2017, 77, 6692–6703. [Google Scholar] [CrossRef] [Green Version]
- Hambardzumyan, D.; Becher, O.J.; Rosenblum, M.K.; Pandolfi, P.P.; Manova-Todorova, K.; Holland, E.C. PI3K pathway regulates survival of cancer stem cells residing in theperivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008, 22, 436–448. [Google Scholar] [CrossRef] [Green Version]
- Di Pietro, C.; La Sala, G.; Matteoni, R.; Marazziti, D.; Tocchini-Valentini, G.P. Genetic ablation of Gpr37l1 delays tumor occurrence in Ptch1 +/− mouse models of medulloblastoma. Exp. Neurol. 2019, 312, 33–42. [Google Scholar] [CrossRef]
- Chen, K.Y.; Chen, Y.J.; Cheng, C.J.; Jhan, K.Y.; Wang, L.C. Excretory/secretory products of Angiostrongyluscantonensis fifth-stage larvae induce endoplasmic reticulum stress via the Sonic hedgehog pathway in mouse astrocytes. Parasit. Vectors 2020, 13, 317. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Ventura, P.B.; Jiang, Y.; Rodriguez, F.J.; Wang, L.; Perry, J.S.A. Astrocytic trans-Differentiation Completes a Multicellular Paracrine Feedback Lop Required for Medulloblastoma Tumor Growth. Cell 2020, 180, 502–520.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Joshi, P.; Katsushima, K.; Liang, W.; Liu, W.; Goldenberg, N.A. The emerging field of noncoding RNAs and their importance in pediatric diseases. J. Pediatr. 2020, 221, S11–S19. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.S.; Ferrara, N. Targeting the tumor microenvironment with Src kinase inhibition. Clin. Cancer Res. 2010, 16, 775–777. [Google Scholar] [CrossRef] [Green Version]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amarante, M.K.; Vitiello, G.A.F.; Rosa, M.H.; Mancilla, I.A.; Watanabe, M.A.E. Potential use of CXCL12/CXCR4 and sonic hedgehog pathways as therapeutic targets in medulloblastoma. Acta Oncol. 2018, 57, 1134–1142. [Google Scholar] [CrossRef]
- Margol, A.S.; Robison, N.J.; Gnanachandran, J.; Hung, L.T.; Kennedy, R.J.; Vali, M.; Dhall, G.; Finlay, J.L.; Epstein, A.; Krieger, M.D.; et al. Tumor-Associated Macrophages in SHH Subgroup of Medulloblastomas. Clin. Cancer Res. 2015, 21, 1457–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, C.D.; Mitchell, D.A. Know your neighbors: Different tumor microenvironments have implications in immunotherapeutic targeting strategies across MB subgroups. Oncoimmunology 2016, 5, e1144002. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Franco-Barraza, J.; Wang, Y.; Zheng, C.; Zhang, L.; Qu, Y.; Long, Y.; Cukierman, E.; Yang, Z.-J. Sustained hedgehog signaling in medulloblastoma tumoroids is attributed to stromal astrocytes and astrocyte-derived extracellular matrix. Lab. Investig. 2020, 100, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Raviraj, R.; Nagaraja, S.S.; Selvakumar, I.; Mohan, S.; Nagarajan, D. The epigenetics of brain tumors and its modulation during radiation: A review. Life Sci. 2020, 256, 117974. [Google Scholar] [CrossRef]
- Hirata, E.; Sahai, E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Tamayo-Orrego, L.; Charron, F. Recent advances in SHH medulloblastoma progression: Tumor suppressor mechanisms and the tumor microenvironment. F1000Research 2019, 8, 1823. [Google Scholar] [CrossRef]
- Raza, M.; Prasad, P.; Gupta, P.; Kumar, N.; Sharma, T.; Rana, M.; Goldman, A.; Sehrawat, S. Perspectives on the role of brain cellular players in cancer-associated brain metastasis: Translational approach to understand molecular mechanism of tumor progression. Cancer Metastasis Rev. 2018, 37, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Ocasio, J.; Babcock, B.; Malawsky, D.; Weir, S.J.; Loo, L.; Simon, J.M.; Zylka, M.J.; Hwang, D.; Dismuke, T.; Sokolsky, M.; et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat. Commun. 2019, 10, 5829. [Google Scholar] [CrossRef] [Green Version]
- Sofroniew, M.V. Multiple roles for astrocytes as effectors of cytokines and inflammatory mediators. Neuroscientist 2014, 20, 160–172. [Google Scholar] [CrossRef]
- Khakh, B.S.; Sofroniew, M.V. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015, 18, 942–952. [Google Scholar] [CrossRef]
- Anderson, M.A.; Ao, Y.; Sofroniew, M.V. Heterogeneity of reactive astrocytes. Neurosci. Lett. 2014, 565, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Burda, J.E.; Bernstein, A.M.; Sofroniew, M.V. Astrocyte roles in traumatic brain injury. Exp. Neurol. 2016, 275, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Tong, X.; Ao, Y.; Faas, G.C.; Nwaobi, S.E.; Xu, J.; Haustein, M.D.; Anderson, M.A.; Mody, I.; Olsen, M.; Sofroniew, M.V.; et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci. 2014, 17, 694–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robel, S.; Berninger, B.; Götz, M. The stem cell potential of glia: Lessons from reactive gliosis. Nat. Rev. Neurosci. 2011, 12, 88–104. [Google Scholar] [CrossRef]
- Silver, J.; Miller, J.H. Regeneration beyond the glial scar. Nat. Rev. Neurosci. 2004, 5, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pekny, M.; Pekna, M. Reactive gliosis in the pathogenesis of CNS diseases. Biochim. Biophys Acta. 2016, 1862, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, D.; Priego, N.; Fustero-Torre, C.; Valiente, M. Reactive astrocytes in brain metastasis. Front. Oncol. 2017, 7, 298. [Google Scholar] [CrossRef] [Green Version]
- Placone, A.L.; Quiñones-Hinojosa, A.; Searson, P.C. The role of astrocytes in the progression of brain cancer: Complicating the picture of the tumor microenvironment. Tumor Biol. 2016, 37, 61–69. [Google Scholar] [CrossRef]
- Brandao, M.; Simon, T.; Critchley, G.; Giamas, G. Astrocytes, the rising stars of the glioblastoma microenvironment. Glia 2019, 67, 779–790. [Google Scholar] [CrossRef]
- Gronseth, E.; Gupta, A.; Koceja, C.; Kumar, S.; Kutty, R.G.; Rarick, K.; Wang, L.; Ramchandran, R. Astrocytes influence medulloblastoma phenotypes and CD133 surface expression. PLoS ONE 2020, 15, e0235852. [Google Scholar] [CrossRef]
- Nedergaard, M.; Ransom, B.; Goldman, S.A. New roles for astrocytes: Redefining the functional architecture of the brain. Trends Neurosci. 2003, 26, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Barres, B.A. The Mystery and Magic of Glia: A perspective on their roles in health and disease. Neuron 2008, 60, 430–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sofroniew, M.V. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009, 32, 638–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sofroniew, M.V. Astrocyte reactivity: Subtypes, states, and functions in cns innate immunity. Trends Immunol. 2020, 41, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Traiffort, E.; Charytoniuk, D.; Watroba, L.; Faure, H.; Sales, N.; Ruat, M. Discrete localizations of hedgehog signalling components in the developing and adult rat nervous system. Eur. J. Neurosci. 1999, 11, 3199–3214. [Google Scholar] [CrossRef]
- Garcia, A.D.; Petrova, R.; Eng, L.; Joyner, A.L. Sonic Hedgehog regulates discrete populations of astrocytes in the adult muse forebrain. J. Neurosci. 2010, 30, 13597–13608. [Google Scholar] [CrossRef]
- Jiao, J.; Chen, D.F. Induction of neurogenesis in nonconventional neurogenic regions of the adult central nervous systeby niche astrocyte-produced signals. Stem Cells 2008, 26, 1221–1230. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Reyes, L.E.; Verbitsky, M.; Blesa, J.; Jackson-Lewis, V.; Paredes, D.; Tillack, K.; Phani, S.; Kramer, E.; Przedborski, S.; Kottmann, A. Sonic Hedgehog Maintains Cellular and Neurochemical Homeostasis in the Adult Nigrostriatal Circuit. Neuron 2012, 75, 306–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Reyes, L.E.; Chiang, C.-C.; Zhang, M.; Johnson, J.; Arrillaga-Tamez, M.; Couturier, N.H.; Reddy, N.; Starikov, L.; Capadona, J.R.; Kottmann, A.H.; et al. Sonic Hedgehog is expressed by hilar mossy cells and regulates cellular survival and neurogenesis in the adult hippocampus. Sci. Rep. 2019, 9, 17402–17420. [Google Scholar] [CrossRef]
- Amankulor, N.M.; Hambardzumyan, D.; Pyonteck, S.M.; Becher, O.J.; Joyce, J.A.; Holland, E.C. Sonic hedgehog pathway activation is induced by acute brain injury and regulated by injury-related inflammation. J. Neurosci. 2009, 29, 10299–10308. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, J.I.; Dodelet-Devillers, A.; Kebir, H.; Ifergan, I.; Fabre, P.J.; Terouz, S.; Sabbagh, M.; Wosik, K.; Bourbonnière, L.; Bernard, M.; et al. The Hedgehog Pathway Promotes Blood-Brain Barrier Integrity and CNS Immune Quiescence. Science 2011, 334, 1727–1731. [Google Scholar] [CrossRef] [Green Version]
- Sirko, S.; Behrendt, G.; Johansson, P.; Tripathi, P.; Costa, M.; Bek, S.; Heinrich, C.; Tiedt, S.; Colak, D.; Dichgans, M.; et al. Reactive Glia in the Injured Brain Acquire Stem Cell Properties in Response to Sonic Hedgehog. Cell Stem Cell 2013, 12, 426–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitter, K.; Tamagno, I.; Feng, X.; Ghosal, K.; Amankulor, N.; Holland, E.C.; Hambardzumyan, D. The SHH/Gli pathway is reactivated in reactive glia and drives proliferation in response to neurodegeneration-induced lesions. Glia 2014, 62, 1595–1607. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.Y.; Fleming, J.T.; Chiang, C. Bergmann glial Sonic hedgehog signaling activity is required for proper cerebellar cortical expansion and architecture. Dev. Biol. 2018, 440, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Priego, N.; Valiente, M. The potential of astrocytes as immune modulators in brain tumors. Front. Immunol. 2019, 10, 1314. [Google Scholar] [CrossRef]
- Zhang, G.; Rich, J.N. Reprogramming the microenvironment: Tricks of tumor-derived astrocytes. Cell Res. 2020, 30, 633–634. [Google Scholar] [CrossRef]
- Gronseth, E.; Wang, L.; Harder, D.R.; Ramchandran, R. The Role of Astrocytes in Tumor Growth and Progression. In Astrocyte-Physiology and Pathology; IntechOpen: London, United Kingdom, 2018. [Google Scholar]
- Liu, H.; Sun, Y.; O’Brien, J.; Franco-Barraza, J.; Qi, X.; Yuan, H.; Jin, W.; Zhang, J.; Gu, C.; Zhao, Z.; et al. Necroptotic astrocytes contribute to maintaining stemness of disseminated medulloblastoma through CCL2 secretion. Neuro. Oncol. 2020, 22, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Shiga, T.; Ichikawa, M.; Hirata, Y. Spatial and temporal pattern of postnatal proliferation of Bergmann glial cells in rat cerebellum: An autoradiographic study. Anat. Embryol. 1983, 167, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Buffo, A.; Rossi, F. Origin, lineage and function of cerebellar glia. Prog. Neurobiol. 2013, 109, 42–63. [Google Scholar] [CrossRef]
- Terry, T.T.; Cheng, T.; Mahjoub, M.; Zong, H. Mosaic Analysis with Double Markers reveals IGF1R function in granule cell progenitors during cerebellar development. Dev. Biol. 2020, 465, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.P.B.; Carpi-Santos, R.; Gomes, F.C.A. The role of astrocytes in the development of the cerebellum. Cerebellum 2019, 18, 1017–1035. [Google Scholar] [CrossRef] [PubMed]
- Wallace, V.A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999, 9, 445–448. [Google Scholar] [CrossRef] [Green Version]
- Wechsler-Reya, R.J.; Scott, M.P. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [Green Version]
- McMahon, A.P.; Ingham, P.W.; Tabin, C.J. Developmental roles and clinical significance of Hedgehog signaling. Curr. Top. Dev. Biol. 2003, 53, 1–114. [Google Scholar] [CrossRef]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of smoothened. Nature 2002, 418, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef] [Green Version]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef] [Green Version]
- Oliver, T.; Read, T.-A.; Kessler, J.D.; Mehmeti, A.; Wells, J.F.; Huynh, T.T.T.; Lin, S.M.; Wechsler-Reya, R.J. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 2005, 132, 2425–2439. [Google Scholar] [CrossRef] [Green Version]
- Romer, J.; Curran, T. Targeting medulloblastoma: Small-molecule inhibitors of the Sonic Hedgehog pathway as potential cancer therapeutics. Cancer Res. 2005, 65, 4975–4978. [Google Scholar] [CrossRef] [Green Version]
- Merk, D.J.; Segal, R.A. Sonic hedgehog signaling is blue: Insights from the patched mutant mice. Trends Neurosci. 2018, 41, 870–872. [Google Scholar] [CrossRef] [PubMed]
- Schüller, U.; Heine, V.; Mao, J.; Kho, A.T.; Dillon, A.K.; Han, Y.-G.; Huillard, E.; Sun, T.; Ligon, A.H.; Qian, Y.; et al. Acquisition of Granule Neuron Precursor Identity Is a Critical Determinant of Progenitor Cell Competence to Form Shh-Induced Medulloblastoma. Cancer Cell 2008, 14, 123–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurawel, R.H.; Allen, C.; Wechsler-Reya, R.; Scott, M.P.; Raffel, C. Evidence that haploinsufficiency of Ptch leads to medulloblastoma in mice. Genes Chromosomes Cancer 2000, 28, 77–81. [Google Scholar] [CrossRef]
- Mao, J.; Ligon, K.L.; Rakhlin, E.Y.; Thayer, S.P.; Bronson, R.T.; Rowitch, D.; McMahon, A.P. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006, 66, 10171–10178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- YYang, Z.-J.; Ellis, T.; Markant, S.L.; Read, T.-A.; Kessler, J.D.; Bourboulas, M.; Schüller, U.; Machold, R.; Fishell, G.; Rowitch, D.; et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008, 14, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuler, D. A kemoterápiaszerepe a gyermekkori medulloblastoma kezelésében. [The role of chemotherapy in pediatric medulloblastoma]. Magy. Onkol. 2010, 54, 145–152. [Google Scholar] [CrossRef]
- Ayrault, O.; Zhao, H.; Zindy, F.; Qu, C.; Sherr, C.J.; Roussel, M.F. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010, 70, 5618–5627. [Google Scholar] [CrossRef] [Green Version]
- Pazzaglia, S.; Tanori, M.; Mancuso, M.; Gessi, M.; Pasquali, E.; Leonardi, S.; Oliva, M.A.; Rebessi, S.; Di Majo, V.; Covelli, V.; et al. Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene 2006, 25, 5575–5580. [Google Scholar] [CrossRef] [Green Version]
- Sasai, K.; Romer, J.T.; Lee, Y.; Finkelstein, D.; Fuller, C.; McKinnon, P.J.; Curran, T. Shh pathway activity is down-regulated in cultured medulloblastoma cells: Implications for preclinical studies. Cancer Res. 2006, 66, 4215–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimkus, T.K.; Carpenter, R.L.; Qasem, S.; Chan, M.; Lo, H.W. Targeting the sonic hedgehog signaling pathway: Review of smoothened and GLI inhibitors. Cancers 2016, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhang, Y.; Sun, B.; McMahon, A.P.; Wang, Y. Hedgehog Signaling: From basic biology to cancer therapy. Cell Chem. Biol. 2017, 24, 252–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severini, L.L.; Quaglio, D.; Basili, I.; Ghirga, F.; Bufalieri, F.; Caimano, M.; Balducci, S.; Moretti, M.; Romeo, I.; Loricchio, E.; et al. A Smo/Gli Multitarget Hedgehog Pathway Inhibitor Impairs Tumor Growth. Cancers 2019, 11, 1518. [Google Scholar] [CrossRef] [Green Version]
- Packer, R.J.; Gajjar, A.; Vezina, G.; Rorke-Adams, L.; Burger, P.C.; Robertson, P.L.; Bayer, L.; LaFond, D.; Donahue, B.R.; Marymont, M.H.; et al. Phase III Study of Craniospinal Radiation Therapy Followed by Adjuvant Chemotherapy for Newly Diagnosed Average-Risk Medulloblastoma. J. Clin. Oncol. 2006, 24, 4202–4208. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.K.; Porter, J.A.; Young, K.E.; Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 1998, 280, 1603–1607. [Google Scholar] [CrossRef]
- Incardona, J.P.; Gaffield, W.; Kapur, R.P.; Roelink, H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development 1998, 125, 3553–3562. [Google Scholar] [CrossRef]
- Lou, E.; Nelson, A.C.; Kool, M. Differential response of SHH-expressing adult medulloblastomas to the sonic hedgehog inhibitor vismodegib: Whole-genome analysis. Cancer Biol. Ther. 2019, 20, 1398–1402. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Qu, P.-R.; Zhang, S.; Li, S.-W.; Zhang, J.; Wang, B.; Liu, P.; Li, C.-D.; Zhao, F. The clinical treatment and outcome of cerebellopontine angle medulloblastoma: A retrospective study of 15 cases. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kieran, M.W.; Chisholm, J.; Casanova, M.; Brandes, A.A.; Aerts, I.; Bouffet, E.; Bailey, S.; Leary, S.; Macdonald, T.J.; Mechinaud, F.; et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro-Oncology 2017, 19, 1542–1552. [Google Scholar] [CrossRef]
- Lorusso, P.M.; Rudin, C.; Reddy, J.C.; Tibes, R.; Weiss, G.J.; Borad, M.J.; Hann, C.L.; Brahmer, J.R.; Chang, I.; Darbonne, W.C.; et al. Phase I Trial of Hedgehog Pathway Inhibitor Vismodegib (GDC-0449) in Patients with Refractory, Locally Advanced or Metastatic Solid Tumors. Clin. Cancer Res. 2011, 17, 2502–2511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Song, Q.; Day, B.W. Phase I and phase II sonidegib and vismodegib clinical trials for the treatment of paediatric and adult MB patients: A systemic review and meta-analysis. Acta Neuropathol. Commun. 2019, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Atwood, S.; Sarin, K.; Whitson, R.J.; Li, J.R.; Kim, G.; Rezaee, M.; Ally, M.S.; Kim, J.; Yao, C.; Chang, A.L.S.; et al. Smoothened Variants Explain the Majority of Drug Resistance in Basal Cell Carcinoma. Cancer Cell 2015, 27, 342–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danial, C.; Sarin, K.Y.; Oro, A.E.; Chang, A.L. An investigator-initiated open-label trial of sonidegib in advanced basal cell carcinoma patients resistant to vismodegib. Clin. Cancer Res. 2016, 22, 1325–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharpe, H.J.; Wang, W.; Hannoush, R.N.; de Sauvage, F.J. Regulation of the oncoprotein Smoothened by small molecules. Nat. Chem. Biol. 2015, 11, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Lau, K.M.; Ng, H.K. Signaling pathway and molecular subgroups of medulloblastoma. Int. J. Clin. Exp. Pathol. 2013, 6, 1211–1222, Erratum in: Int. J. Clin. Exp. Pathol. 2015, 8, 11945. [Google Scholar]
- Xin, M.; Ji, X.; De La Cruz, L.K.; Thareja, S.; Wang, B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med. Res. Rev. 2018, 38, 870–913. [Google Scholar] [CrossRef]
- Mille, F.; Tamayo-Orrego, L.; Lévesque, M.; Remke, M.; Korshunov, A.; Cardin, J.; Bouchard, N.; Izzi, L.; Kool, M.; Northcott, P.A.; et al. The Shh Receptor Boc Promotes Progression of Early Medulloblastoma to Advanced Tumors. Dev. Cell 2014, 31, 34–47. [Google Scholar] [CrossRef] [Green Version]
- Thompson, E.M.; Ashley, D.; Landi, D. Current medulloblastoma subgroup specific clinical trials. Transl. Pediatr. 2020, 9, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Maximov, V.; Chen, Z.; Wei, Y.; Robinson, M.H.; Herting, C.J.; Shanmugam, N.; Rudneva, V.A.; Goldsmith, K.C.; Macdonald, T.J.; Northcott, P.A.; et al. Tumour-associated macrophages exhibit anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, V.; Yeerna, H.; Nohata, N.; Chiou, J.; Harismendy, O.; Raimondi, F.; Inoue, A.; Russell, R.B.; Tamayo, P.; Gutkind, J.S. Illuminating the Onco-GPCRome: Novel G protein–coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019, 294, 11062–11086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DI Pietro, C.; Marazziti, D.; Lasala, G.; Abbaszadeh, Z.; Golini, E.; Matteoni, R.; Tocchini-Valentini, G.P. Primary Cilia in the Murine Cerebellum and in Mutant Models of Medulloblastoma. Cell. Mol. Neurobiol. 2017, 37, 145–154. [Google Scholar] [CrossRef]
- Marazziti, D.; Di Pietro, C.; Golini, E.; Mandillo, S.; Lasala, G.; Matteoni, R.; Tocchini-Valentini, G.P. Precocious cerebellum development and improved motor functions in mice lacking the astrocyte cilium-, patched 1-associated Gpr37l1 receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 16486–16491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.G.; Kim, H.J.; Dlugosz, A.A.; Ellison, D.W.; Gilbertson, R.J.; Alvarez-Buylla, A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009, 15, 1062–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.G.; Alvarez-Buylla, A. Role of primary cilia in brain development and cancer. Curr. Opin. Neurobiol. 2010, 20, 58–67. [Google Scholar] [CrossRef] [Green Version]
- La Sala, G.; Di Pietro, C.; Matteoni, R.; Bolasco, G.; Marazziti, D.; Tocchini-Valentini, G.P. Gpr37l1/prosaposin receptor regulates Ptch1 trafficking, Shh production, and cell proliferation in cerebellar primary astrocytes. J. Neurosci. Res. 2021, 99, 1064–1083, Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Snuderl, M.; Batista, A.; Kirkpatrick, N.D.; de Almodovar, C.R.; Riedemann, L.; Walsh, E.C.; Anolik, R.; Huang, Y.; Martin, J.; Kamoun, W.; et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell 2013, 152, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, T.T.; Teixeira, S.A.; Gonzalez-Reyes, L.; Reis, R.M. Decoding the Roles of Astrocytes and Hedgehog Signaling in Medulloblastoma. Curr. Oncol. 2021, 28, 3058-3070. https://doi.org/10.3390/curroncol28040267
Duarte TT, Teixeira SA, Gonzalez-Reyes L, Reis RM. Decoding the Roles of Astrocytes and Hedgehog Signaling in Medulloblastoma. Current Oncology. 2021; 28(4):3058-3070. https://doi.org/10.3390/curroncol28040267
Chicago/Turabian StyleDuarte, Terence Teixeira, Silvia Aparecida Teixeira, Luis Gonzalez-Reyes, and Rui Manuel Reis. 2021. "Decoding the Roles of Astrocytes and Hedgehog Signaling in Medulloblastoma" Current Oncology 28, no. 4: 3058-3070. https://doi.org/10.3390/curroncol28040267
APA StyleDuarte, T. T., Teixeira, S. A., Gonzalez-Reyes, L., & Reis, R. M. (2021). Decoding the Roles of Astrocytes and Hedgehog Signaling in Medulloblastoma. Current Oncology, 28(4), 3058-3070. https://doi.org/10.3390/curroncol28040267