An Example of Personalized Treatment in HR+ HER2+ Long Survivor Breast Cancer Patient (Case Report)
Abstract
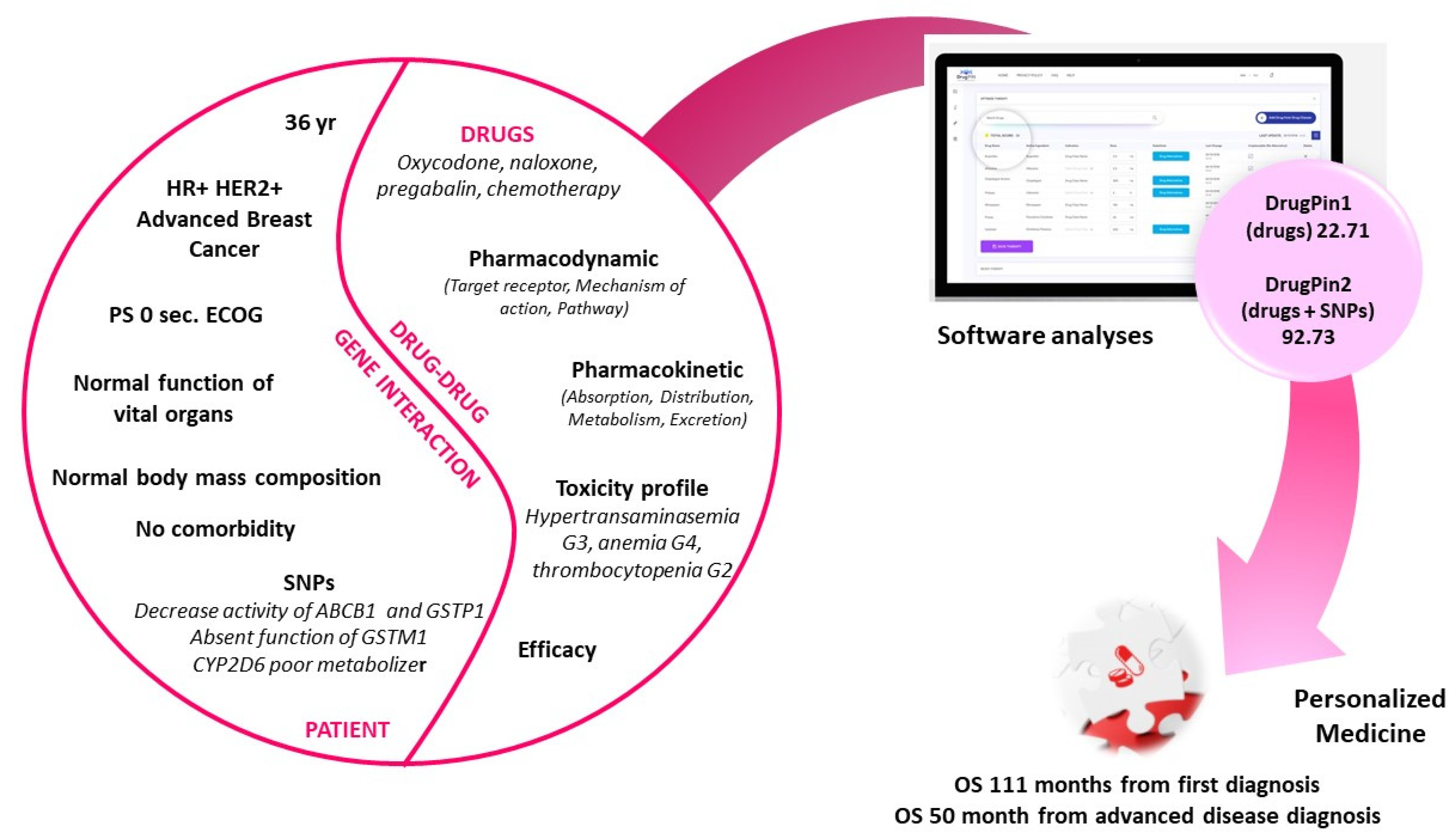
1. Introduction
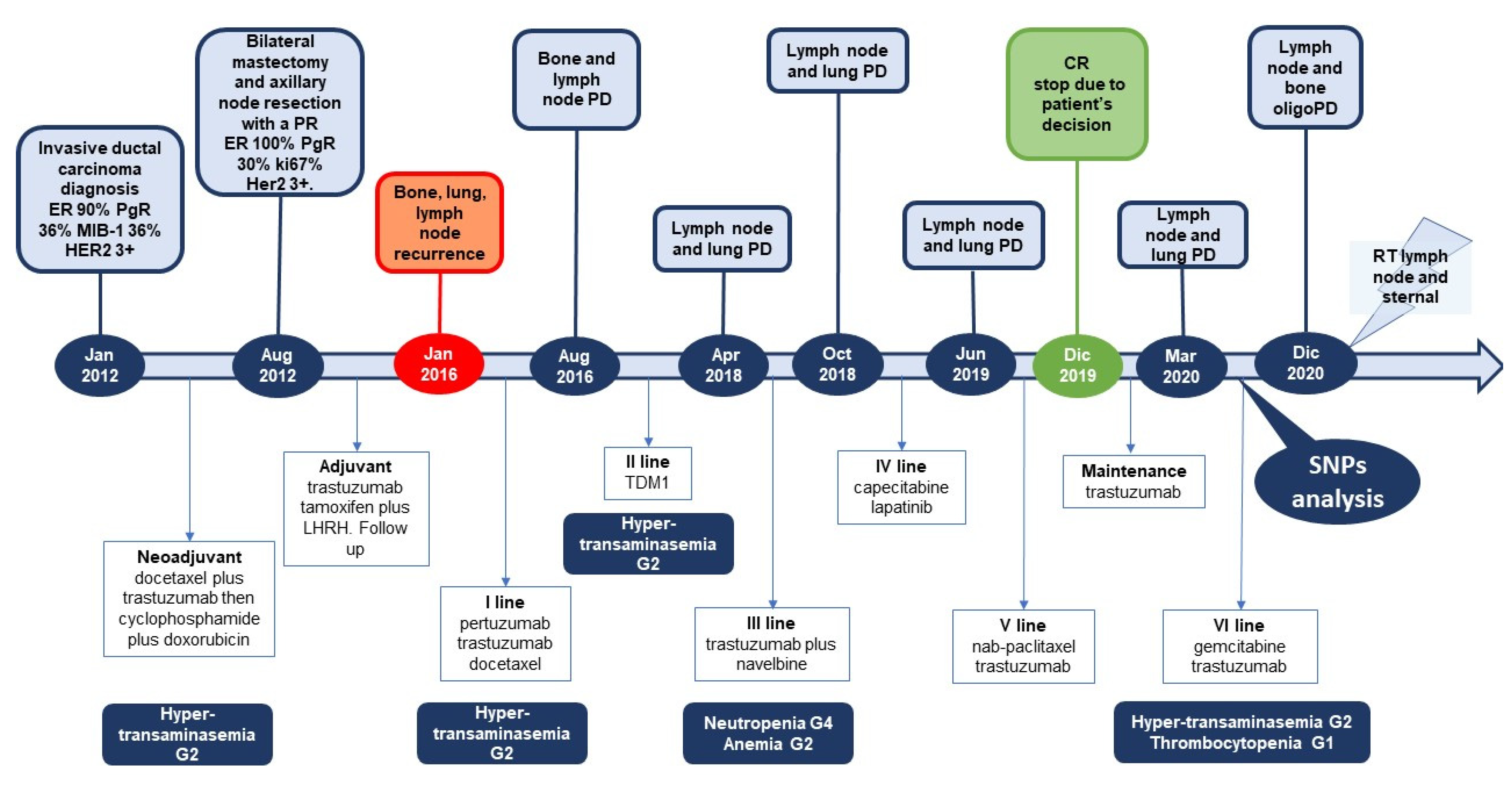
2. Case Presentation
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roberto, M.; Rossi, A.; Panebianco, M.; Pomes, L.; Arrivi, G.; Ierinò, D.; Simmaco, M.; Marchetti, P.; Mazzuca, F. Drug–Drug Interactions and Pharmacogenomic Evaluation in Colorectal Cancer Patients: The New Drug-PIN® System Comprehensive Approach. Pharmaceuticals 2021, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Tannenbaum, C.; Sheehan, N.L. Understanding and preventing drug–drug and drug–gene interactions. Expert Rev. Clin. Pharmacol. 2014, 7, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Danahey, K.; Knoebel, R.W.; Ratain, M.J.; Meltzer, D.O.; O’Donnell, P.H. Analysis of comprehensive pharmacogenomic profiling to impact in-hospital prescribing. Pharm. Genom. 2019, 29, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Verbeurgt, P.; Mamiya, T.; Oesterheld, J. How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics 2014, 15, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Evans, W.E.; McLeod, H.L. Pharmacogenomics—Drug Disposition, Drug Targets, and Side Effects. N. Engl. J. Med. 2003, 348, 538–549. [Google Scholar] [CrossRef]
- Erichsen, H.C.; Chanock, S.J. SNPs in cancer research and treatment. Br. J. Cancer 2004, 90, 747–751. [Google Scholar] [CrossRef]
- Palmirotta, R.; Carella, C.; Silvestris, E.; Cives, M.; Stucci, S.L.; Tucci, M.; Lovero, D.; Silvestris, F. SNPs in predicting clinical efficacy and toxicity of chemotherapy: Walking through the quicksand. Oncotarget 2018, 9, 25355–25382. [Google Scholar] [CrossRef]
- Angelini, S.; Botticelli, A.; Onesti, C.E.; Giusti, R.; Sini, V.; Durante, V.; Strigari, L.; Gentile, G.; Cerbelli, B.; Pellegrini, P.; et al. Pharmacogenetic Approach to Toxicity in Breast Cancer Patients Treated with Taxanes. Anticancer Res. 2017, 37, 2633–2639. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Tulsyan, S.; Agarwal, G.; Lal, P.; Mittal, R.; Mittal, B. Influence of ABCB1 genetic variants in breast cancer treatment outcomes. Cancer Epidemiol. 2013, 37, 754–761. [Google Scholar] [CrossRef]
- Córdoba, E.E.; Abba, M.C.; Lacunza, E.; Fernandez, E.E.; Güerci, A.M. Polymorphic Variants in Oxidative Stress Genes and Acute Toxicity in Breast Cancer Patients Receiving Radiotherapy. Cancer Res. Treat. 2016, 48, 948–954. [Google Scholar] [CrossRef]
- Etienne-Grimaldi, M.-C.; Boyer, J.-C.; Beroud, C.; Mbatchi, L.; Van Kuilenburg, A.; Bobin-Dubigeon, C.; Thomas, F.; Chatelut, E.; Merlin, J.-L.; Pinguet, F.; et al. New advances in DPYD genotype and risk of severe toxicity under capecitabine. PLoS ONE 2017, 12, e0175998. [Google Scholar] [CrossRef] [PubMed]
- Pascual, T.; Apellániz-Ruiz, M.; Pernaut, C.; Cueto-Felgueroso, C.; Villalba, P.; Álvarez, C.; Manso, L.; Inglada-Pérez, L.; Robledo, M.; Rodriguez-Antona, C.; et al. Polymorphisms associated with everolimus pharmacokinetics, toxicity and survival in metastatic breast cancer. PLoS ONE 2017, 12, e0180192. [Google Scholar] [CrossRef] [PubMed]
- Boso, V.; Herrero, M.J.; Santaballa, A.; Palomar, L.; Megias, J.E.; De La Cueva, H.; Rojas, L.; Marqués, M.R.; Poveda, J.L.; Montalar, J.; et al. SNPs and taxane toxicity in breast cancer patients. Pharmacogenomics 2014, 15, 1845–1858. [Google Scholar] [CrossRef]
- Sini, V.; Botticelli, A.; Lunardi, G.; Gori, S.; Marchetti, P. Pharmacogenetics and aromatase inhibitor induced side effects in breast cancer patients. Pharmacogenomics 2017, 18, 821–830. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, P.H.; Trubetskoy, V.; Nurhussein-Patterson, A.; Hall, J.P.; Nath, A.; Huo, D.; Fleming, G.F.; Ingle, J.N.; Abramson, V.G.; Morrow, P.K.; et al. Clinical evaluation of germline polymorphisms associated with capecitabine toxicity in breast cancer: TBCRC-015. Breast Cancer Res. Treat. 2020, 181, 623–633. [Google Scholar] [CrossRef]
- Jabir, R.S.; Naidu, R.; Annuar, M.A.B.A.; Ho, G.F.; Munisamy, M.; Stanslas, J. Pharmacogenetics of taxanes: Impact of gene polymorphisms of drug transporters on pharmacokinetics and toxicity. Pharmacogenomics 2012, 13, 1979–1988. [Google Scholar] [CrossRef]
- Priyadarshini, R.; Raj, G.M.; Kayal, S.; Ramesh, A.; Shewade, D.G. Influence of ABCB1 C3435T and C1236T gene polymorphisms on tumour response to docetaxel-based neo-adjuvant chemotherapy in locally advanced breast cancer patients of South India. J. Clin. Pharm. Ther. 2019, 44, 188–196. [Google Scholar] [CrossRef]
- Zhong, J.; Guo, Z.; Fan, L.; Zhao, X.; Zhao, B.; Cao, Z.; Cheng, L.; Shi, Y.; Li, X.; Zhang, Y.; et al. ABCB1 polymorphism predicts the toxicity and clinical outcome of lung cancer patients with taxane-based chemotherapy. Thorac. Cancer 2019, 10, 2088–2095. [Google Scholar] [CrossRef]
- Mir, O.; Alexandre, J.; Tran, A.; Durand, J.-P.; Pons, G.; Treluyer, J.-M.; Goldwasser, F. Relationship between GSTP1 Ile105Val polymorphism and docetaxel-induced peripheral neuropathy: Clinical evidence of a role of oxidative stress in taxane toxicity. Ann. Oncol. 2009, 20, 736–740. [Google Scholar] [CrossRef]
- Tran, A.; Jullien, V.; Alexandre, J.; Rabillon, F.; Girre, V.; Pons, G.; Goldwasser, F.; Rey, E.; Diéras, V.; Treluyer, J. Pharmacokinetics and toxicity of docetaxel: Role of CYP3A, MDR1, and GST polymorphisms. Clin. Pharmacol. Ther. 2006, 79, 570–580. [Google Scholar] [CrossRef]
- Sugishita, M.; Imai, T.; Kikumori, T.; Mitsuma, A.; Shimokata, T.; Shibata, T.; Morita, S.; Inada-Inoue, M.; Sawaki, M.; Hasegawa, Y.; et al. Pharmacogenetic association between GSTP1 genetic polymorphism and febrile neutropenia in Japanese patients with early breast cancer. Breast Cancer 2014, 23, 195–201. [Google Scholar] [CrossRef]
- Zhang, B.L.; Tong, S.U.; Zhang, B.N.; Zheng, S.; Ning, L.Ü.; Xu, B.H.; Xiang, W.A.; Chen, G.J.; Yu, D.K.; Lin, D.X. Polymorphisms of GSTP1 is associated with differences of chemotherapy response and toxicity in breast cancer. Chin. Med. J. 2011, 124, 199–204. [Google Scholar] [PubMed]
- Yoshihama, T.; Fukunaga, K.; Hirasawa, A.; Nomura, H.; Akahane, T.; Kataoka, F.; Yamagami, W.; Aoki, D.; Mushiroda, T. GSTP1 rs1695 is associated with both hematological toxicity and prognosis of ovarian cancer treated with paclitaxel plus carboplatin combination chemotherapy: A comprehensive analysis using targeted resequencing of 100 pharmacogenes. Oncotarget 2018, 9, 29789–29800. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kinnunen, M.; Piirainen, P.; Kokki, H.; Lammi, P.; Kokki, M. Updated Clinical Pharmacokinetics and Pharmacodynamics of Oxycodone. Clin. Pharmacokinet. 2019, 58, 705–725. [Google Scholar] [CrossRef] [PubMed]
- Heiskanen, T.; Olkkola, K.T.; Kalso, E. Effects of blocking CYP2D6 on the pharmacokinetics and pharmacodynamics of oxycodone*. Clin. Pharmacol. Ther. 1998, 64, 603–611. [Google Scholar] [CrossRef]
- Susce, M.T.; Murray-Carmichael, E.; De Leon, J. Response to hydrocodone, codeine and oxycodone in a CYP2D6 poor metabolizer. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2006, 30, 1356–1358. [Google Scholar] [CrossRef]
- Foster, A.; Mobley, E.; Wang, Z. Complicated Pain Management in a CYP450 2D6 Poor Metabolizer. Pain Pract. 2007, 7, 352–356. [Google Scholar] [CrossRef]
- Smith, H.S. Opioid Metabolism. Mayo Clin. Proc. 2009, 84, 613–624. [Google Scholar] [CrossRef]
- Wong, M.; Balleine, R.L.; Blair, E.Y.; McLachlan, A.J.; Ackland, S.P.; Garg, M.B.; Evans, S.; Farlow, D.; Collins, M.; Rivory, L.P.; et al. Predictors of Vinorelbine Pharmacokinetics and Pharmacodynamics in Patients With Cancer. J. Clin. Oncol. 2006, 24, 2448–2455. [Google Scholar] [CrossRef]
- Gusella, M.; Pasini, F.; Caruso, D.; Barile, C.; Modena, Y.; Fraccon, A.P.; Bertolaso, L.; Menon, D.; Crepaldi, G.; Bononi, A.; et al. Clinical outcomes of oral metronomic vinorelbine in advanced non-small cell lung cancer: Correlations with pharmacokinetics and MDR1 polymorphisms. Cancer Chemother. Pharmacol. 2019, 83, 493–500. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panebianco, M.; Taurelli Salimbeni, B.; Roberto, M.; Marchetti, P. An Example of Personalized Treatment in HR+ HER2+ Long Survivor Breast Cancer Patient (Case Report). Curr. Oncol. 2021, 28, 1980-1987. https://doi.org/10.3390/curroncol28030184
Panebianco M, Taurelli Salimbeni B, Roberto M, Marchetti P. An Example of Personalized Treatment in HR+ HER2+ Long Survivor Breast Cancer Patient (Case Report). Current Oncology. 2021; 28(3):1980-1987. https://doi.org/10.3390/curroncol28030184
Chicago/Turabian StylePanebianco, Martina, Beatrice Taurelli Salimbeni, Michela Roberto, and Paolo Marchetti. 2021. "An Example of Personalized Treatment in HR+ HER2+ Long Survivor Breast Cancer Patient (Case Report)" Current Oncology 28, no. 3: 1980-1987. https://doi.org/10.3390/curroncol28030184
APA StylePanebianco, M., Taurelli Salimbeni, B., Roberto, M., & Marchetti, P. (2021). An Example of Personalized Treatment in HR+ HER2+ Long Survivor Breast Cancer Patient (Case Report). Current Oncology, 28(3), 1980-1987. https://doi.org/10.3390/curroncol28030184





