Detection of SARS-CoV-2 RNA in Bivalve Mollusks by Droplet Digital RT-PCR (dd RT-PCR)
Abstract
:1. Introduction
2. Materials and Methods
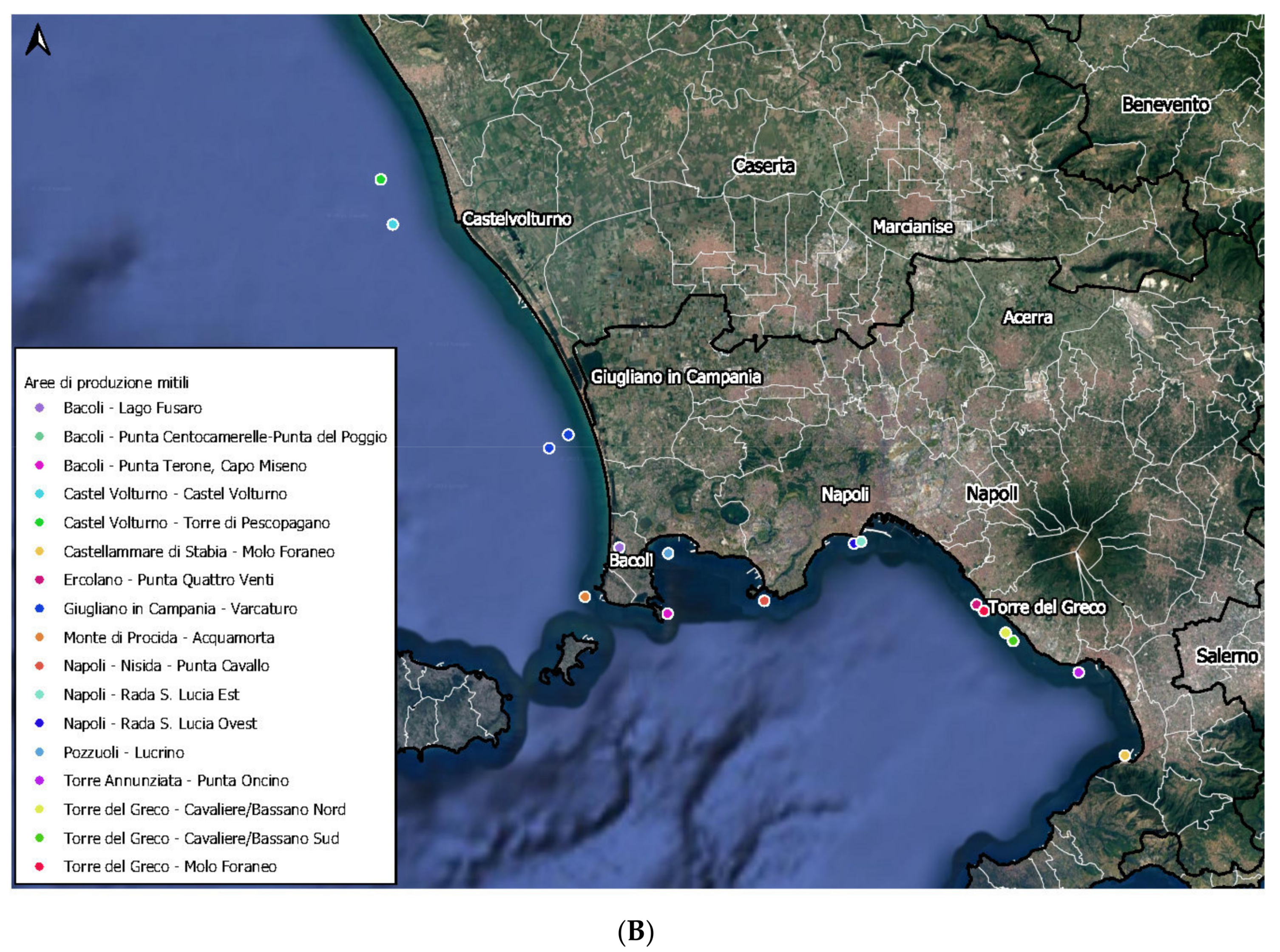
2.1. Sampling
2.2. Sample Preparation
2.3. Viral RNA Extraction
2.4. Droplet Digital RT-PCR (dd RT-PCR) for SARS-CoV-2 Detection
2.5. Real-Time RT-PCR Assay Using the Quanty COVID-19v2
2.6. Molecular Characterization of Positive Samples
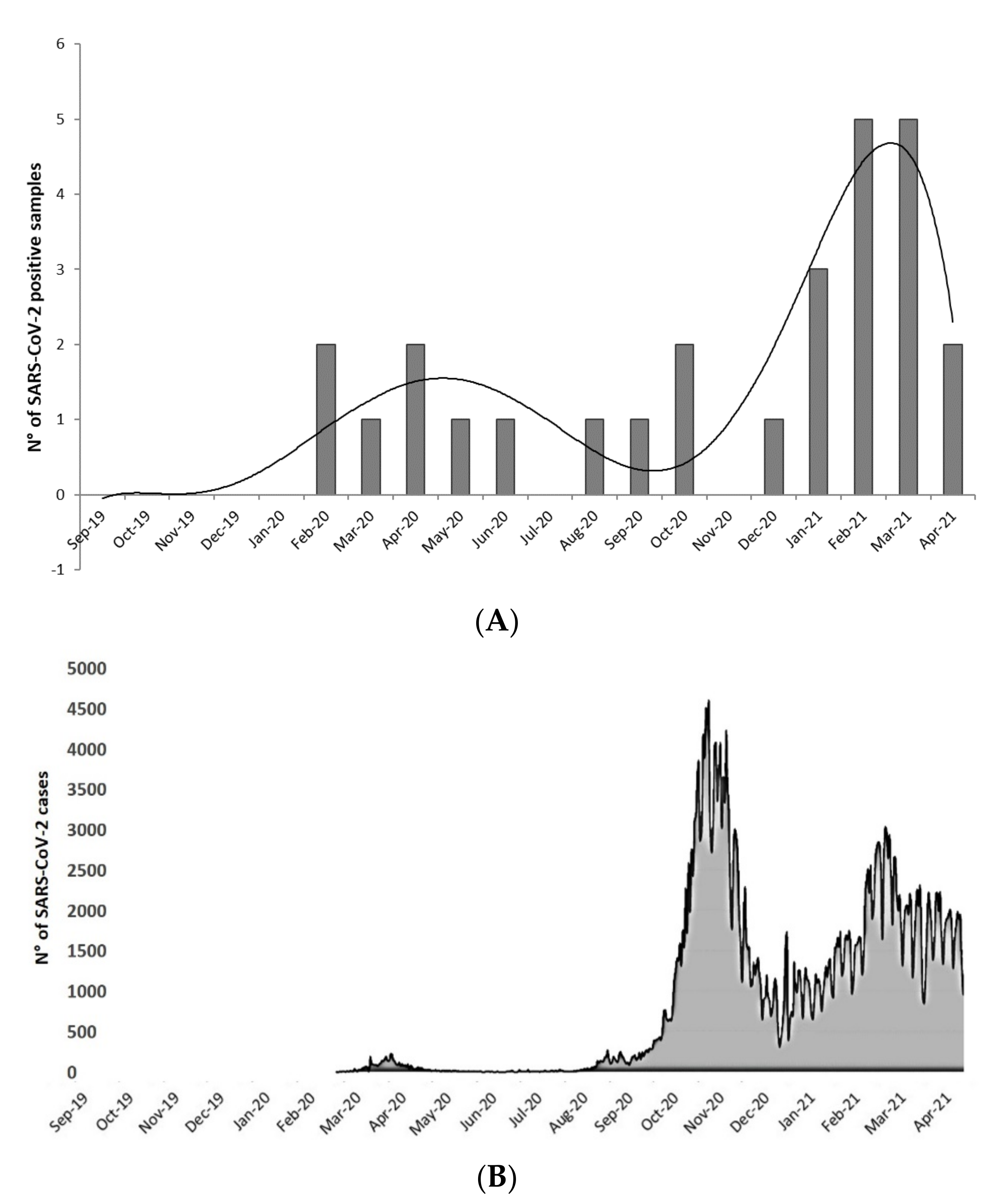
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chan, J.F.W.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.W.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Lo, I.L.; Lio, C.F.; Cheong, H.H.; Lei, C.I.; Cheong, T.H.; Zhong, X.; Tian, Y.; Sin, N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020, 16, 1698–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanno Ferraro, G.; Veneri, C.; Mancini, P.; Iaconelli, M.; Suffredini, E.; Bonadonna, L.; Lucentini, L.; Bowo-Ngandji, A.; Kengne-Nde, C.; Mbaga, D.S.; et al. A State-of-the-Art Scoping Review on SARS-CoV-2 in Sewage Focusing on the Potential of Wastewater Surveillance for the Monitoring of the COVID-19 Pandemic. Food Environ. Virol. 2021, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Amereh, F.; Negahban-Azar, M.; Isazadeh, S.; Dabiri, H.; Masihi, N.; Jahangiri-Rad, M.; Rafiee, M. Sewage Systems Surveillance for SARS-CoV-2: Identification of Knowledge Gaps, Emerging Threats, and Future Research Needs. Pathogens 2021, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Lundy, L.; Fatta-Kassinos, D.; Slobodnik, J.; Karaolia, P.; Cirka, L.; Kreuzinger, N.; Castiglioni, S.; Bijlsma, L.; Dulio, V.; Deviller, G.; et al. Making Waves: Collaboration in the time of SARS-CoV-2—Rapid development of an international co-operation and wastewater surveillance database to support public health decision-making. Water Res. 2021, 199, 117167. [Google Scholar] [CrossRef] [PubMed]
- McClary-Gutierrez, J.S.; Mattioli, M.C.; Marcenac, P.; Silverman, A.I.; Boehm, A.B.; Bibby, K.; Balliet, M.; De Los Reyes, F.L.; Gerrity, D.; Griffith, J.F.; et al. SARS-CoV-2 Wastewater Surveillance for Public Health Action. Emerg. Infect. Dis. 2021, 27, E1–E9. [Google Scholar] [CrossRef]
- Johansson, M.A.; Vasconcelos, P.F.C.; Staples, J.E. The whole iceberg: Estimating the incidence of yellow fever virus infection from the number of severe cases. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 482–487. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.J.; Younger, A.D. Developing microbiological risk assessment for shellfish purification. Int. Biodeterior. Biodegrad. 2002, 50, 177–183. [Google Scholar] [CrossRef]
- McLeod, C.; Hay, B.; Grant, C.; Greening, G.; Day, D. Inactivation and elimination of human enteric viruses by Pacific oysters. J. Appl. Microbiol. 2009, 107, 1809–1818. [Google Scholar] [CrossRef]
- De Medici, D.; Ciccozzi, M.; Fiore, A.; Di Pasquale, S.; Parlato, A.; Ricci-Bitti, P.; Croci, L. Closed-circuit system for the depuration of mussels experimentally contaminated with hepatitis A virus. J. Food Prot. 2001, 64, 877–880. [Google Scholar] [CrossRef]
- Polo, D.; Lois, M.; Fernández-Núñez, M.T.; Romalde, J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021, 786, 147534. [Google Scholar] [CrossRef]
- Westhaus, S.; Weber, F.A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany—Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751, 141750. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Waldman, P.; Ferrier-Rembert, A.; Frenois-Veyrat, G.; Mouchel, J.M.; Boni, M.; Maday, Y.; Marechal, V.; Moulin, L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 2021, 198, 117183. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- Desdouits, M.; Piquet, J.C.; Wacrenier, C.; Le Mennec, C.; Parnaudeau, S.; Jousse, S.; Rocq, S.; Bigault, L.; Contrant, M.; Garry, P.; et al. Can shellfish be used to monitor SARS-CoV-2 in the coastal environment? Sci. Total Environ. 2021, 778. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Estes, M.K.; Jiang, X.; Metcalf, T.G. Concentration and detection of hepatitis A virus and rotavirus from shellfish by hybridization tests. Appl. Environ. Microbiol. 1991, 57, 2963–2968. [Google Scholar] [CrossRef] [Green Version]
- Kogawa, K.; Nakata, S.; Ukae, S.; Adachi, N.; Numata, K.; Matson, D.O.; Estes, M.K.; Chiba, S. Dot blot hybridization with a cDNA probe derived from the human calicivirus Sapporo 1982 strain. Arch. Virol. 1996, 141, 1949–1959. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Neill, F.H.; Woodley, C.M.; Manger, R.; Shay Fout, G.; Burkhardt, W.; Leja, L.; McGovern, E.R.; Guyader, F.L.E.; Metcalf, T.G.; et al. Collaborative evaluation of a method for the detection of Norwalk virus in shellfish tissues by PCR. Appl. Environ. Microbiol. 1996, 62, 254–258. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Wang, H.; Chen, Y.; He, J.; Chen, L.; Liu, Y.; Hu, X.; Li, A.; Liu, S.; Zhang, P.; et al. Clinical Data on Hospital Environmental Hygiene Monitoring and Medical Staff Protection during the Coronavirus Disease 2019 Outbreak. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.T.; Liu, Y.J.; Wang, J.; Xu, Z.G.; Yang, Y.; Shen, F.; Liu, X.H.; Zhou, X.; Liu, S.M. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin. Chem. 2015, 61, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costafreda, M.I.; Bosch, A.; Pintó, R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 2006, 72, 3846–3855. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, G.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021, 750, 141711. [Google Scholar] [CrossRef]
- Pierri, B.; Mancusi, A.; Proroga, Y.T.R.; Capuano, F.; Cerino, P.; Girardi, S.; Vassallo, L.; Lo Conte, G.; Tafuro, M.; Cuomo, M.C.; et al. SARS-CoV-2 detection in nasopharyngeal swabs: Performance characteristics of a real-time RT-qPCR and a droplet digital RT-PCR assay based on the exonuclease region (ORF1b, nsp 14). J. Virol. Methods 2022, 300, 114420. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Iaconelli, M.; Lucentini, L.; Bonadonna, L.; Brusaferro, S.; Brandtner, D.; Fasanella, A.; et al. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021, 197, 117104. [Google Scholar] [CrossRef]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef] [PubMed]
- Beyer, J.; Green, N.W.; Brooks, S.; Allan, I.J.; Ruus, A.; Gomes, T.; Bråte, I.L.N.; Schøyen, M. Blue mussels (Mytilus edulis spp.) as sentinel organisms in coastal pollution monitoring: A review. Mar. Environ. Res. 2017, 130, 338–365. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, M.E. Can Environmental RNA Revolutionize Biodiversity Science? Trends Ecol. Evol. 2019, 34, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.A.; Biessy, L.; Latchford, J.L.; Zaiko, A.; von Ammon, U.; Audrezet, F.; Cristescu, M.E.; Pochon, X. Release and degradation of environmental DNA and RNA in a marine system. Sci. Total Environ. 2020, 704, 135314. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Sala-Comorera, L.; Reynolds, L.J.; Martin, N.A.; O’Sullivan, J.J.; Meijer, W.G.; Fletcher, N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021, 201, 117090. [Google Scholar] [CrossRef]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.N.; Sarmah, A.K.; Chao, H.P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Ge, Q.; Du, Y.; Li, G.; Li, W.; Zhang, T.; Tan, L.; Zhang, R.; Yuan, X.; et al. Detecting SARS-CoV-2 in the Breath of COVID-19 Patients. Front. Med. 2021, 8, 210. [Google Scholar] [CrossRef]
- Coleman, K.K.; Tay, D.J.W.; Tan, K.S.; Ong, S.W.X.; Than, T.S.; Koh, M.H.; Chin, Y.Q.; Nasir, H.; Mak, T.M.; Chu, J.J.H.; et al. Viral Load of SARS-CoV-2 in Respiratory Aerosols Emitted by COVID-19 Patients while Breathing, Talking, and Singing. Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Edwards, D.A.; Ausiello, D.; Salzman, J.; Devlin, T.; Langer, R.; Beddingfield, B.J.; Fears, A.C.; Doyle-Meyers, L.A.; Redmann, R.K.; Killeen, S.Z.; et al. Exhaled aerosol increases with COVID-19 infection, age, and obesity. Proc. Natl. Acad. Sci. USA 2021, 118, e2021830118. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020, 743, 140444. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Zhu, S.; Shu, C.; Wang, D.; Song, J.; Song, Y.; Zhen, W.; Feng, Z.; Wu, G.; et al. Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19). China CDC Wkly. 2020, 2, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef]
- Dergham, J.; Delerce, J.; Bedotto, M.; La Scola, B.; Moal, V. Isolation of Viable SARS-CoV-2 Virus from Feces of an Immunocompromised Patient Suggesting a Possible Fecal Mode of Transmission. J. Clin. Med. 2021, 10, 2696. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, S.; Ruíz, A.; Pemán, J.; Salavert, M.; Domingo-Calap, P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Fumian, T.M.; Malta, F.C.; dos Santos, D.R.L.; Pauvolid-Corrêa, A.; Fialho, A.M.; Leite, J.P.G.; Miagostovich, M.P. SARS-CoV-2 RNA detection in stool samples from acute gastroenteritis cases, Brazil. J. Med. Virol. 2021, 93, 2543–2547. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
| Primer Name | Sequence | Concentrations | Reference |
|---|---|---|---|
| RdRp_SARSr-F | GTGARATGGTCATGTGTGGCGG | 600 nM | Corman et al., 2020 |
| RdRp_SARSr-R | CARATGTTAAASACACTATTAGCATA | 800 nM | |
| RdRP_SARSr-P1 | FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ | 100 nM | |
| RdRP_SARSr-P2 | FAM-CAGGTGGAACCTCATCAGGAGATGC-BBQ | 100 nM | |
| N_Sarbeco_F | CACATTGGCACCCGCAATC | 600 nM | Corman et al., 2020 |
| N_Sarbeco_R | GAGGAACGAGAAGAGGCTTG | 800 nM | |
| N_Sarbeco_P | FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ | 200 nM | |
| 2297-CoV-2-F | ACATGGCTTTGAGTTGACATCT | 500 nM | La Rosa et al., 2021 |
| 2298-CoV-2-R | AGCAGTGGAAAAGCATGTGG | 900 nM | |
| 2299-CoV-2-P | FAM-CATAGACAACAGGTGCGCTC-MGBEQ | 250 nM | |
| E_Sarbeco_F | ACAGGTACGTTAATAGTTAATAGCGT | 400 nM | Corman et al., 2020 |
| E_Sarbeco_R | ATATTGCAGCAGTACGCACACA | 400 nM | |
| E_Sarbeco_P1 | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ | 200 nM |
| (A) | |||||
| dd RT-PCR Gene Target | N° of Sequenced Samples | ||||
| N° of Samples | Orf1b nsp 14 Region | RdRp Gene | E Gene | N Gene | |
| 1 | + | + | + | - | 1 |
| 1 | + | + | - | - | 0 |
| 3 | - | + | + | - | 0 |
| 9 | + | - | - | - | 1 |
| 5 | - | + | - | - | 2 |
| 160 | - | - | - | - | 1 |
| 179 | 11 | 10 | 4 | 0 | 5 |
| (B) | |||||
| Real-Time RT-PCR Gene Target | N° of Sequenced Samples | ||||
| N° of Samples | N1 | N2 | |||
| 1 | + | + | 0 | ||
| 5 | + | - | 3 | ||
| 5 | - | + | 0 | ||
| 168 | - | - | 2 | ||
| 179 | 6 | 6 | 5 | ||
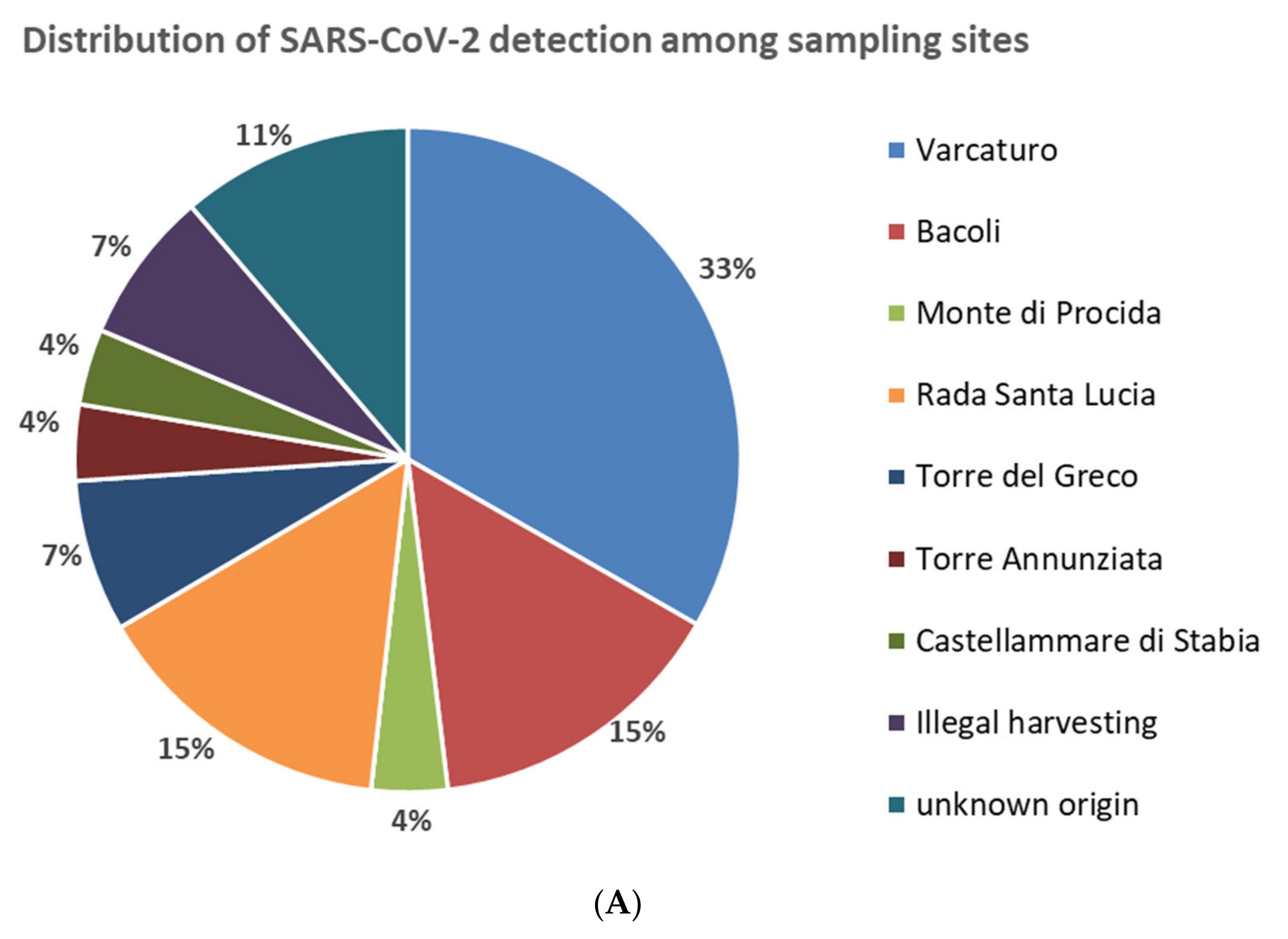
| Sampling Site | Position in the Coastline | SARS-CoV-2 Detection | ||
|---|---|---|---|---|
| N° of Positive Samples | % of Positive in the Site | % of Positive in the Total | ||
| Varcaturo | North | 9/22 | 41 | 5.0 |
| Bacoli | North | 4/27 | 15 | 2.2 |
| Monte di Procida | North | 1/14 | 7 | 0.6 |
| Pozzuoli | North | 0/8 | 0 | 0 |
| Nisida | North | 0/10 | 0 | 0 |
| Subtotal North | 14/81 | 17 | 7.8 | |
| Rada Santa Lucia | Center | 4/16 | 25 | 2.2 |
| Subtotal Center | 4/16 | 25 | 2.2 | |
| Torre del Greco | South | 2/32 | 6 | 1.1 |
| Torre Annunziata | South | 1/7 | 14 | 0.6 |
| Castellammare di Stabia | South | 1/14 | 7 | 0.6 |
| Subtotal South | 4/53 | 8 | 2.2 | |
| Other origins | 0/2 | 0 | 0 | |
| Illegal harvesting | - | 2/3 | 67 | 1.1 |
| Unknown origin | 3/24 | 13 | 1.7 | |
| Total | 27/179 | - | 15.1% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancusi, A.; Capuano, F.; Girardi, S.; Di Maro, O.; Suffredini, E.; Di Concilio, D.; Vassallo, L.; Cuomo, M.C.; Tafuro, M.; Signorelli, D.; et al. Detection of SARS-CoV-2 RNA in Bivalve Mollusks by Droplet Digital RT-PCR (dd RT-PCR). Int. J. Environ. Res. Public Health 2022, 19, 943. https://doi.org/10.3390/ijerph19020943
Mancusi A, Capuano F, Girardi S, Di Maro O, Suffredini E, Di Concilio D, Vassallo L, Cuomo MC, Tafuro M, Signorelli D, et al. Detection of SARS-CoV-2 RNA in Bivalve Mollusks by Droplet Digital RT-PCR (dd RT-PCR). International Journal of Environmental Research and Public Health. 2022; 19(2):943. https://doi.org/10.3390/ijerph19020943
Chicago/Turabian StyleMancusi, Andrea, Federico Capuano, Santa Girardi, Orlandina Di Maro, Elisabetta Suffredini, Denise Di Concilio, Lucia Vassallo, Maria Concetta Cuomo, Maria Tafuro, Daniel Signorelli, and et al. 2022. "Detection of SARS-CoV-2 RNA in Bivalve Mollusks by Droplet Digital RT-PCR (dd RT-PCR)" International Journal of Environmental Research and Public Health 19, no. 2: 943. https://doi.org/10.3390/ijerph19020943
APA StyleMancusi, A., Capuano, F., Girardi, S., Di Maro, O., Suffredini, E., Di Concilio, D., Vassallo, L., Cuomo, M. C., Tafuro, M., Signorelli, D., Pierri, A., Pizzolante, A., Cerino, P., La Rosa, G., Proroga, Y. T. R., & Pierri, B. (2022). Detection of SARS-CoV-2 RNA in Bivalve Mollusks by Droplet Digital RT-PCR (dd RT-PCR). International Journal of Environmental Research and Public Health, 19(2), 943. https://doi.org/10.3390/ijerph19020943









