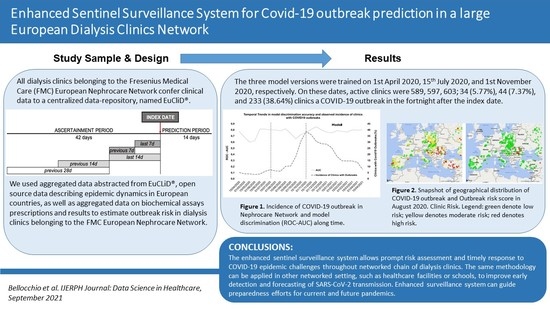
Enhanced Sentinel Surveillance System for COVID-19 Outbreak Prediction in a Large European Dialysis Clinics Network
Abstract
:1. Introduction
2. Materials and Methods
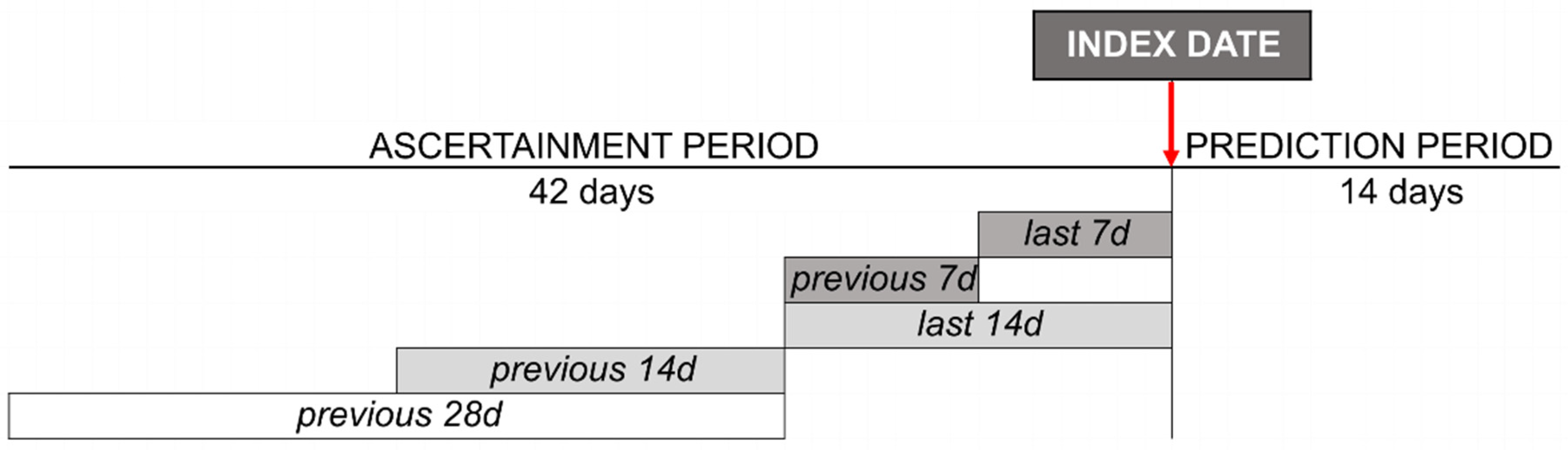
2.1. Design and Setting
2.2. Outcome Variable
2.3. Input Variables
- Open Source Data [24];
- Epidemic status in the clinical country/region (prefix: RG): 15 parameters;
- Aggregated Data abstracted from EuCLiD®:
- Epidemic status in the target clinic (prefix: CL): 5 variables;
- Distance-weighted information of the adjacent clinics (prefix: CLS); 5 variables. Adjacent clinics were defined as the 3 centers with shorter distance in terms of both latitude and longitude to the target clinic. Measures of the adjacent clinics, including cases and trends, were computed as the average value weighted for the inverse of the distance to the target clinic;
- Other parameters related to the target clinic (prefix: CL): 49 parameters.
2.4. Statistical Analysis
2.4.1. Model Derivation
2.4.2. Model Accuracy and Feature Importance
2.4.3. Descriptive Statistics
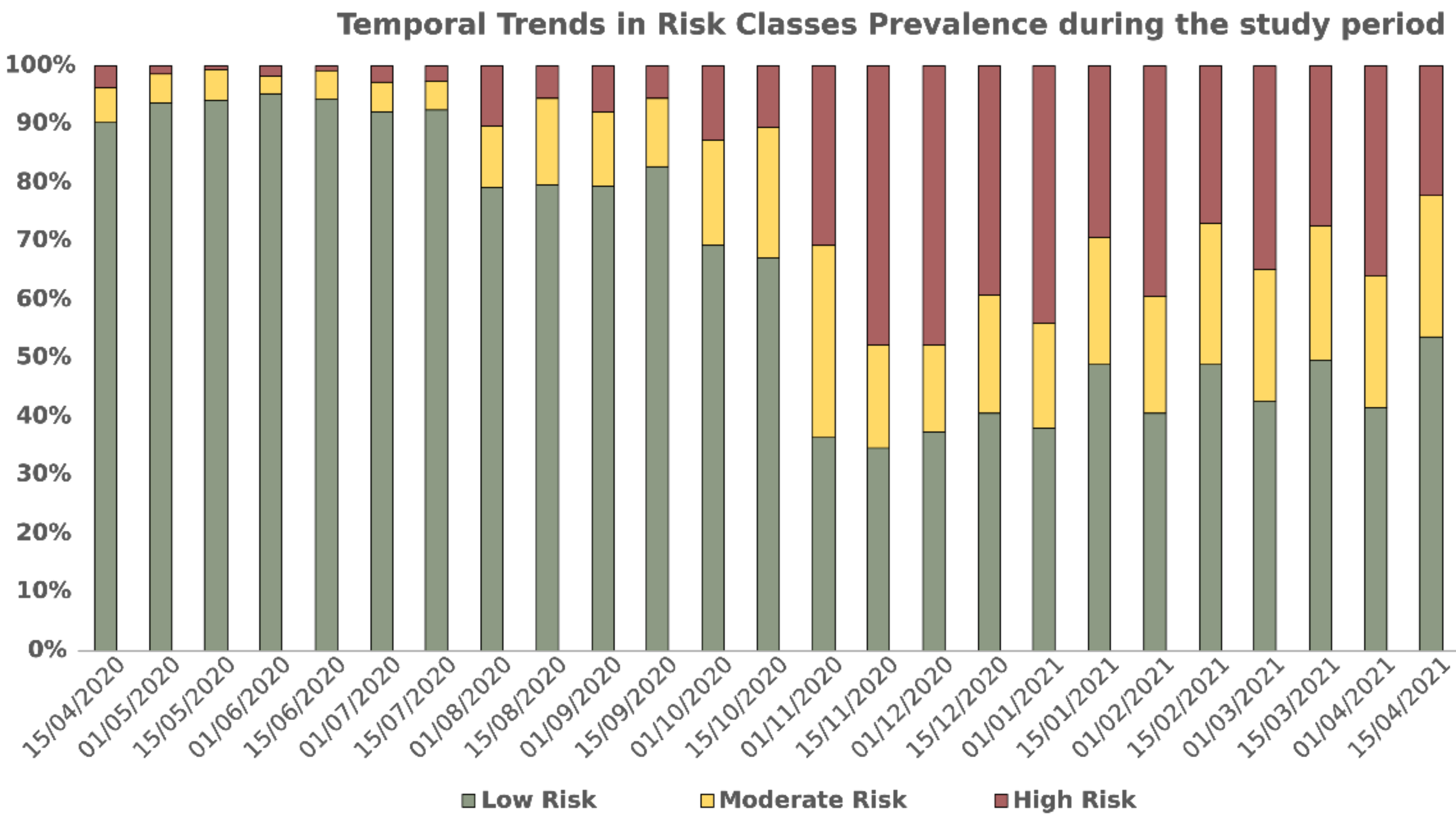
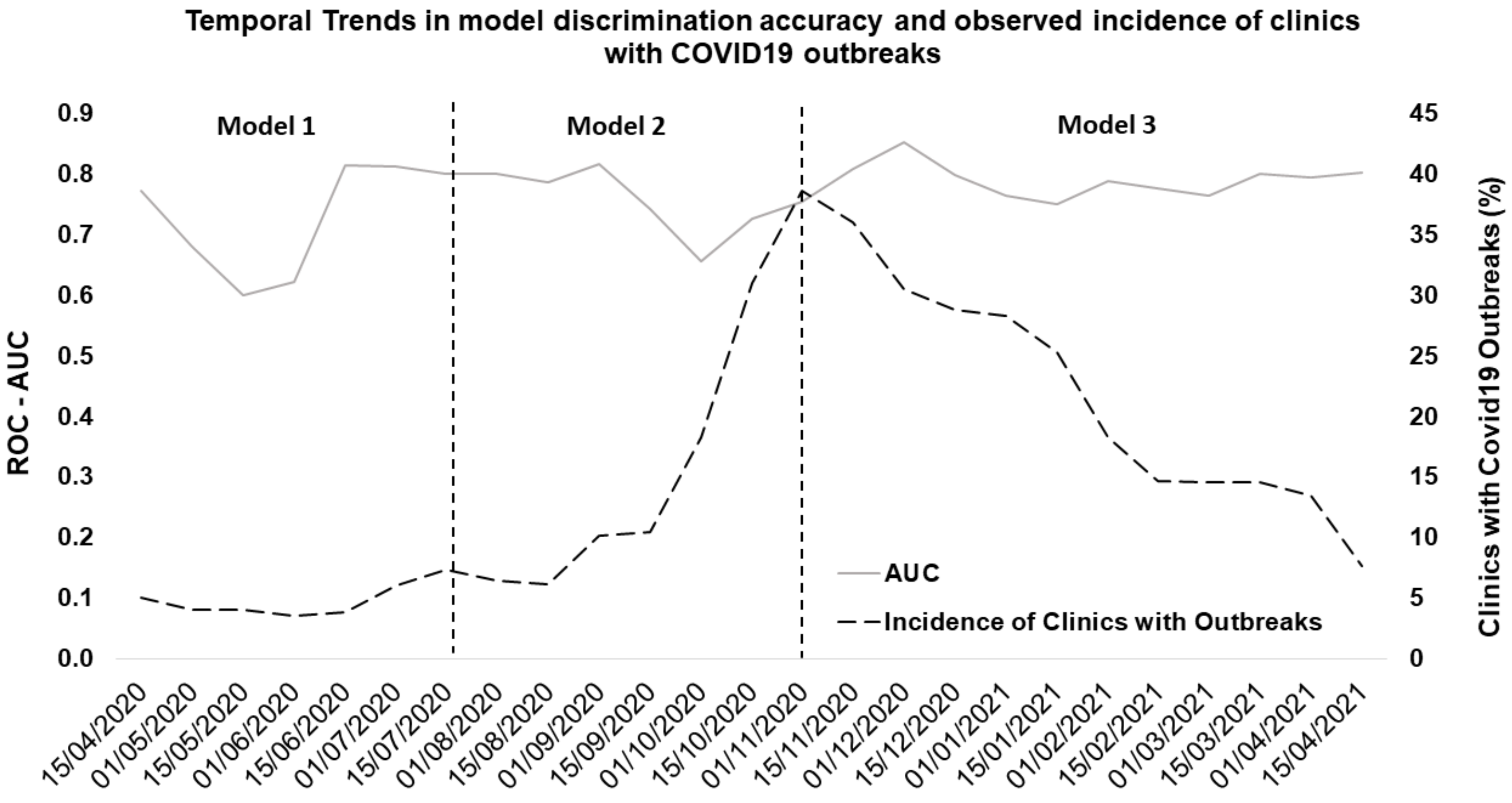
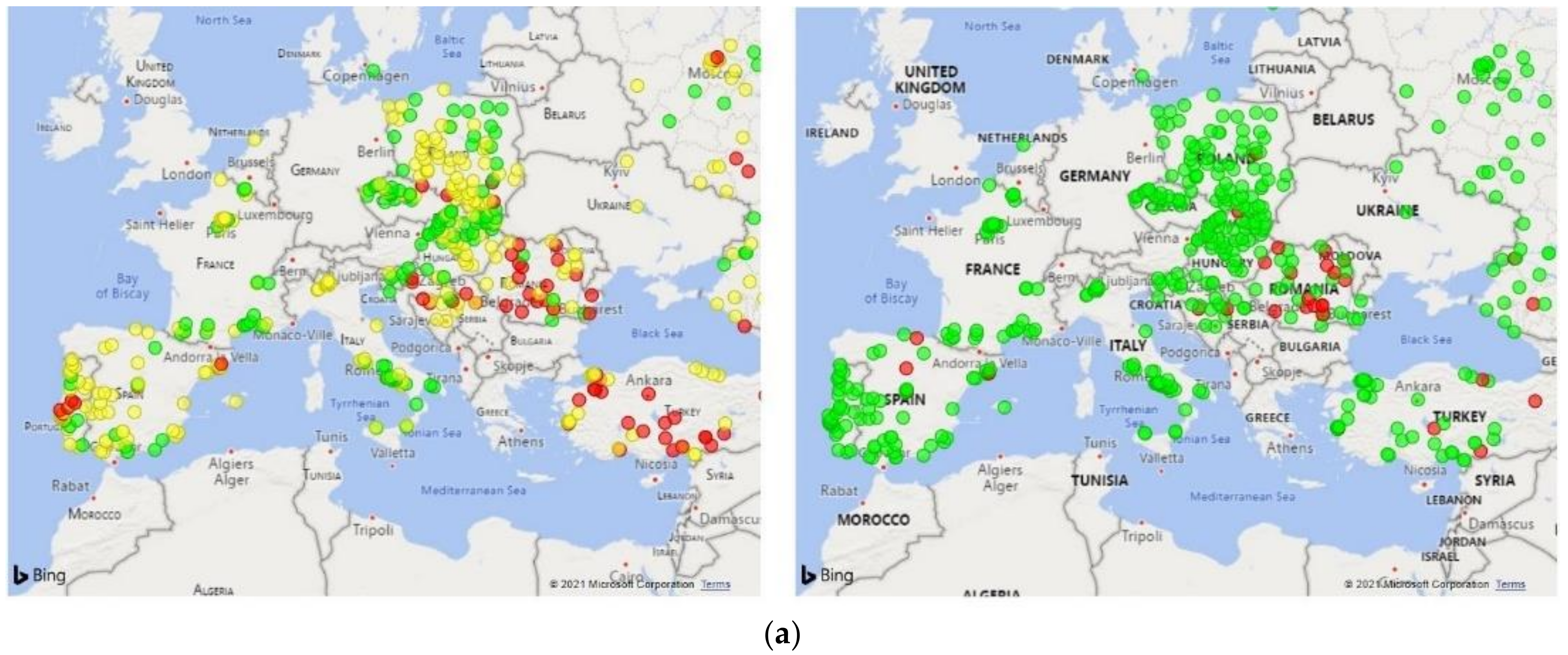
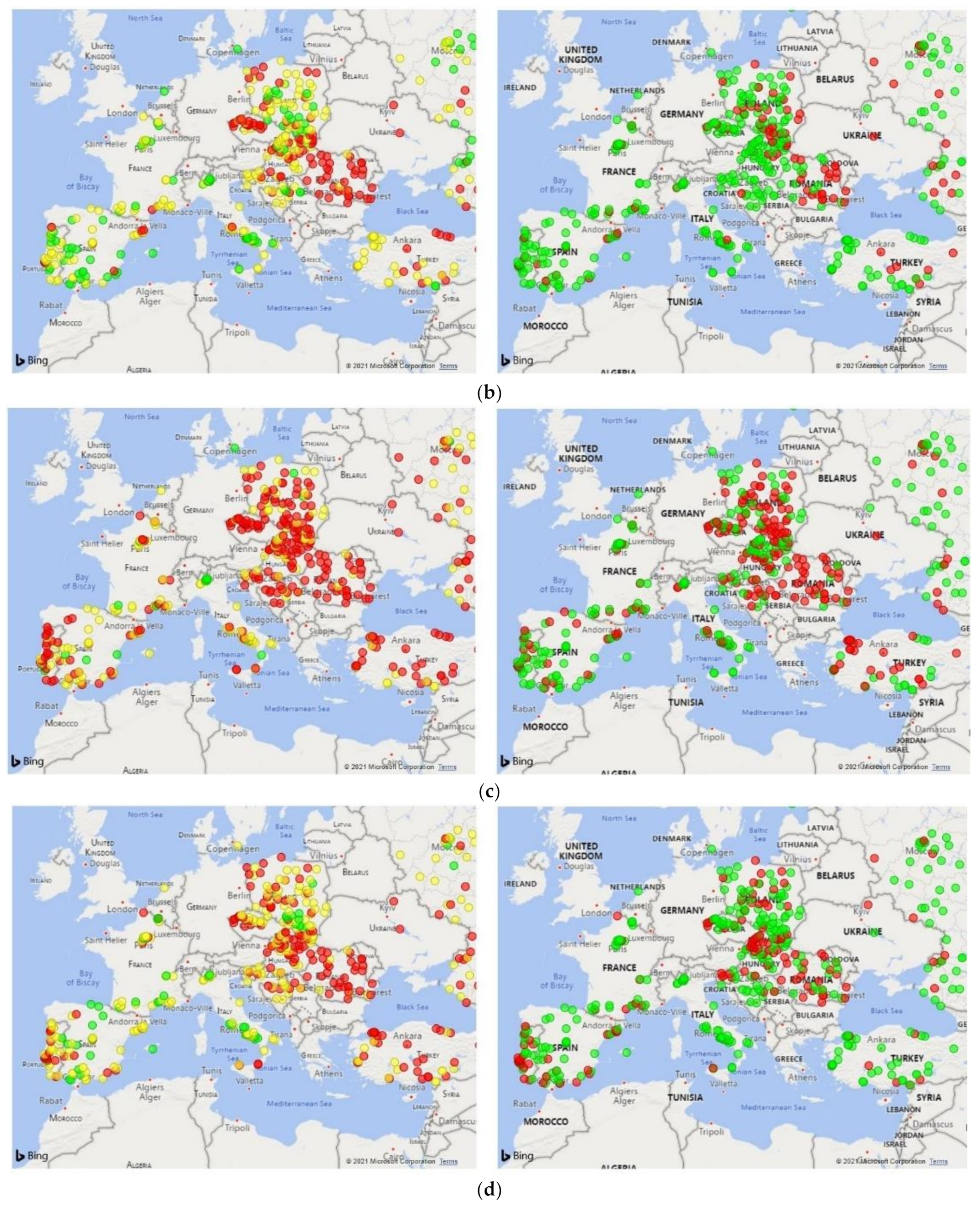
3. Results
3.1. Dialysis Clinic Characteristics
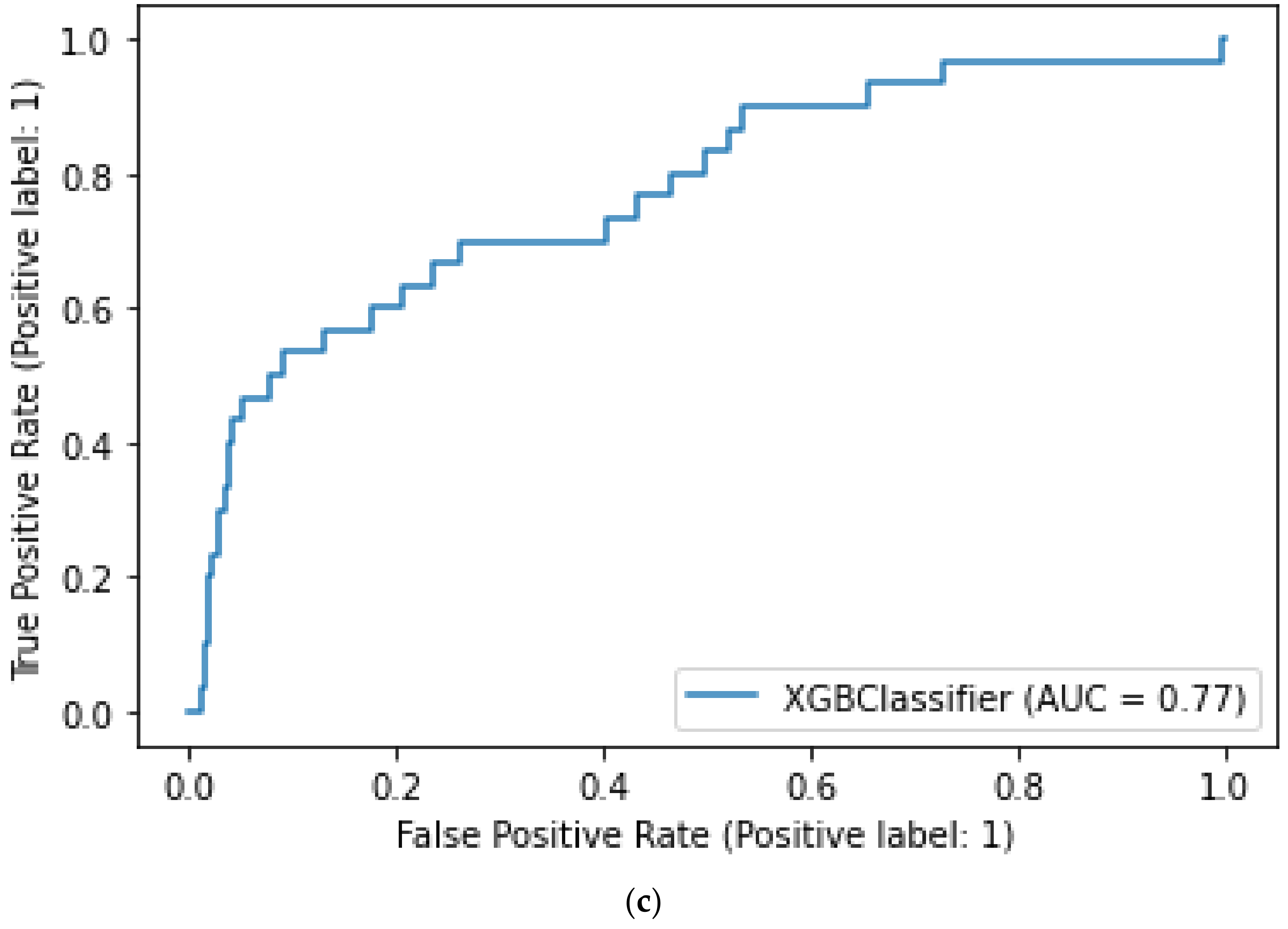
3.2. Model Performance
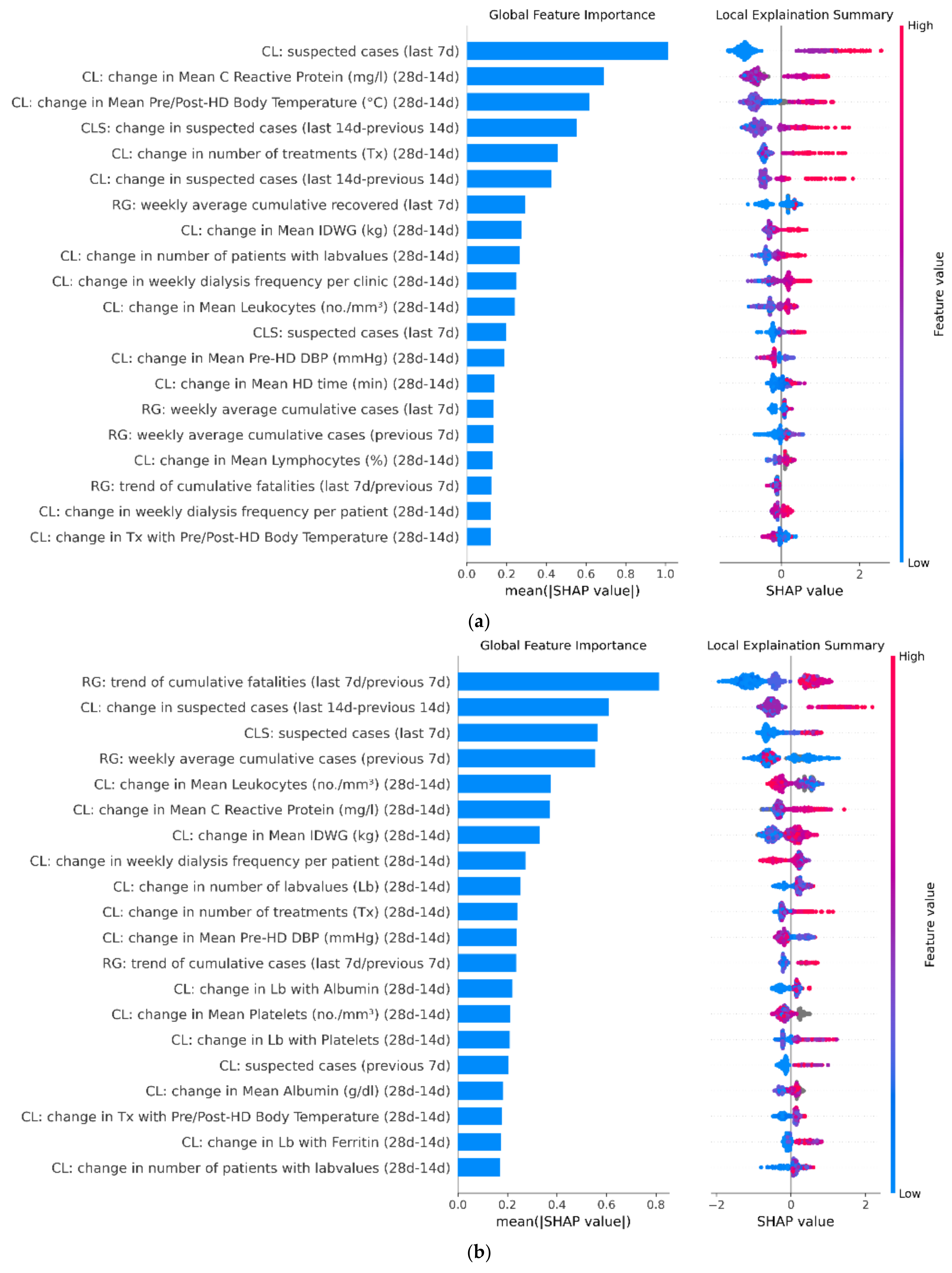
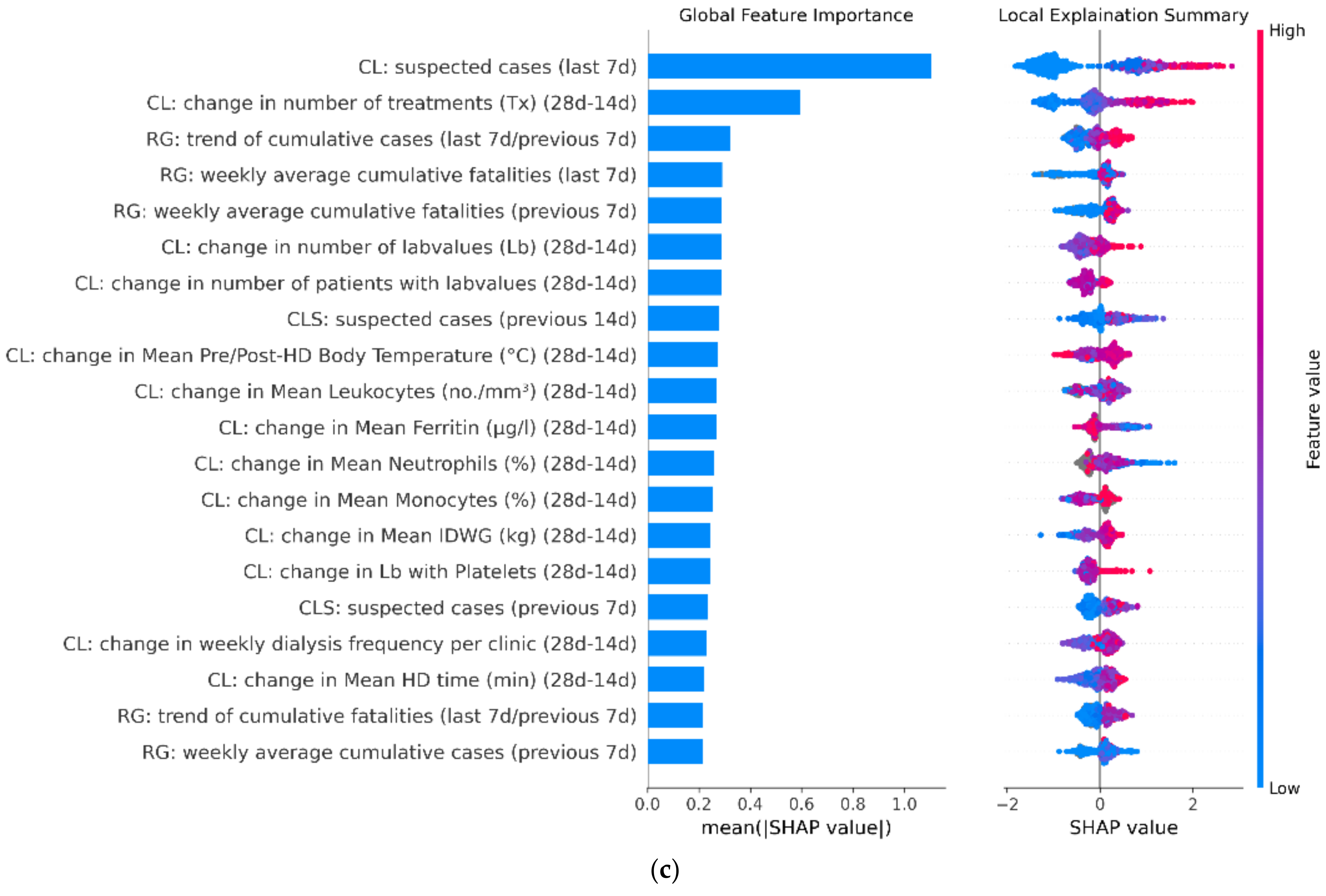
3.3. Model Feature Importance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Category | Variable | Reference Time |
|---|---|---|
| Epidemic Status in the Country/Region (prefix: RG) | ||
| cumulative cases | previous 7 days and last 7 days | |
| number of hospitalized | previous 7 days and last 7 days | |
| number of ICU patients | previous 7 days and last 7 days | |
| cumulative fatalities | previous 7 days and last 7 days | |
| cumulative recovered | previous 7 days and last 7 days | |
| trend of cumulative cases | last 7 days/previous 7 days | |
| trend of hospitalized patients | last 7 days/previous 7 days | |
| trend of ICU patients | last 7 days/previous 7 days | |
| trend of cumulative recovered in the last week | last 7 days/previous 7 days | |
| trend of cumulative fatalities | last 7 days/previous 7 days | |
| epidemic status in the clinic (prefix: CL) | ||
| number of suspected COVID-19 cases | previous 14 days, previous 7 days, and last 7 days | |
| change in suspected cases | last 7 days–previous 7 days | |
| change in suspected cases | last 14 days–previous 14 days | |
| distance-weighted information of the adjacent clinics (prefix: CL) | ||
| number of COVID-19 suspected cases in the closest clinics | previous 14d, previous 7 days, and last 7 days | |
| change in COVID-19 suspected cases in the closest clinics | last 7 days–previous 7 days | |
| change in COVID-19 suspected cases in the closest clinics | last 14 days–previous 14 days | |
| other parameters related to the clinic (prefix: CL) | ||
| change in the number of treated patients | last 28 days–last 14 days | |
| change in the number of treatments | last 28 days–last 14 days | |
| change in the weekly dialysis frequency per clinic | last 28 days–last 14 days | |
| change in the weekly dialysis frequency per patient | last 28 days–last 14 days | |
| change in the number of treatments with pre/post-BT | last 28 days–last 14 days | |
| change in the number of treatments with pre/post-BT > 37 °C | last 28 days–last 14 days | |
| change in the percentage of treatments with pre/post-BT > 37 °C | last 28 days–last 14 days | |
| change in the mean value of pre/post-dialysis BT | last 28 days–last 14 days | |
| change in the number of treatments with pre-dyalisis diastolic BP | last 28 days–last 14 days | |
| change in the mean value of pre-dialysis diastolic BP | last 28 days–last 14 days | |
| change in the number of treatments with dialysis time | last 28 days–last 14 days | |
| change in the mean value of dialysis time | last 28 days–last 14 days | |
| change in the number of treatments with IDWG | last 28 days–last 14 days | |
| change in the mean value of IDWG | last 28 days–last 14 days | |
| change in the number of treatments with O2 sat | last 28 days–last 14 days | |
| change in the mean value of O2 sat | last 28 days–last 14 days | |
| change in the number of patients with lab tests | last 28 days–last 14 days | |
| change in the number of lab tests | last 28 days–last 14 days | |
| change in the number of lab tests with Albumin | last 28 days–last 14 days | |
| change in the mean value of Albumin | last 28 days–last 14 days | |
| change in the number of lab tests with lymphocytes | last 28 days–last 14 days | |
| change in the mean value of lymphocytes | last 28 days–last 14 days | |
| change in the number of lab tests with monocytes | last 28 days–last 14 days | |
| change in the mean value of monocytes | last 28 days–last 14 days | |
| change in the number of lab tests with neutrophils | last 28 days–last 14 days | |
| change in the mean value of neutrophils | last 28 days–last 14 days | |
| change in the number of lab tests with platelets | last 28 days–last 14 days | |
| change in the mean value of platelets | last 28 days–last 14 days | |
| change in the number of lab tests with PDW | last 28 days–last 14 days | |
| change in the mean value of PDW | last 28 days–last 14 days | |
| change in the number of lab tests with leukocytes | last 28 days–last 14 days | |
| change in the mean value of leukocytes | last 28 days–last 14 days | |
| change in the number of lab tests with D-dimer | last 28 days–last 14 days | |
| change in the mean value of D-dimer | last 28 days–last 14 days | |
| change in the number of lab tests with CRP | last 28 days–last 14 days | |
| change in the mean value of CRP | last 28 days–last 14 days | |
| change in the number of lab tests with IL-6 | last 28 days–last 14 days | |
| change in the mean value of IL-6 | last 28 days–last 14 days | |
| change in the number of lab tests with ANP | last 28 days–last 14 days | |
| change in the mean value of ANP | last 28 days–last 14 days | |
| change in the number of lab tests with BNP | last 28 days–last 14 days | |
| change in the mean value of BNP | last 28 days–last 14 days | |
| change in the number of lab tests with Ferritin | last 28 days–last 14 days | |
| change in the mean value of Ferritin | last 28 days–last 14 days | |
| Number of patients with at least one hospitalization | last 14 days | |
| Number of hospitalizations | last 14 days | |
References
- Li, J.; Huang, D.Q.; Zou, B.; Yang, H.; Hui, W.Z.; Rui, F.; Yee, N.T.S.; Liu, C.; Nerurkar, S.N.; Kai, J.C.Y.; et al. Epidemiology of COVID-19: A systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J. Med. Virol. 2021, 93, 1449–1458. [Google Scholar] [CrossRef]
- Muller, C.P. Do asymptomatic carriers of SARS-COV-2 transmit the virus? Lancet Reg. Health 2021, 4, 100082. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byambasuren, O.; Cardona, M.; Bell, K.; Clark, J.; McLaws, M.-L.; Glasziou, P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: Systematic review and meta-analysis. JAMMI Off. J. Assoc. Med. Microbiol. Infect. Dis. Can. 2020, 5, 223–234. [Google Scholar] [CrossRef]
- Galvani, A.P.; May, R.M. Epidemiology: Dimensions of superspreading. Nature 2005, 438, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, J.; Siddle, K.J.; Shaw, B.M.; Loreth, C.; Schaffner, S.; Gladden-Young, A.; Adams, G.; Fink, T.; Tomkins-Tinch, C.H.; Krasilnikova, L.A.; et al. Phylogenetic analysis of SARS-CoV-2 in the Boston area highlights the role of recurrent importation and superspreading events. medRxiv 2020, PMC7457619. [Google Scholar] [CrossRef]
- Lewis, D. Superspreading drives the COVID pandemic-and could help to tame it. Nature 2021, 590, 544–546. [Google Scholar] [CrossRef]
- Chu, D.K.; Akl, E.A.; Duda, S.; Solo, K.; Yaacoub, S.; Schünemann, H.J.; El-harakeh, A.; Bognanni, A.; Lotfi, T.; Loeb, M.; et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: A systematic review and meta-analysis. Lancet 2020, 395, 1973–1987. [Google Scholar] [CrossRef]
- Regmi, K.; Lwin, C.M. Impact of non-pharmaceutical interventions for reducing transmission of COVID-19: A systematic review and meta-analysis protocol. BMJ Open 2020, 10, e041383. [Google Scholar] [CrossRef]
- Apata, I.W.; Cobb, J.; Navarrete, J.; Burkart, J.; Plantinga, L.; Lea, J.P. COVID-19 infection control measures and outcomes in urban dialysis centers in predominantly African American communities. BMC Nephrol. 2021, 22, 81. [Google Scholar] [CrossRef]
- Weiner, D.E.; Watnick, S.G. Hemodialysis and COVID-19: An Achilles’ Heel in the Pandemic Health Care Response in the United States. Kidney Med. 2020, 2, 227–230. [Google Scholar] [CrossRef]
- Meijers, B.; Messa, P.; Ronco, C. Safeguarding the Maintenance Hemodialysis Patient Population during the Coronavirus Disease 19 Pandemic. Blood Purif. 2020, 49, 259–264. [Google Scholar] [CrossRef]
- Li, S.Y.; Tang, Y.S.; Chan, Y.J.; Tarng, D.C. Impact of the COVID-19 pandemic on the management of patients with end-stage renal disease. J. Chin. Med. Assoc. 2020, 83, 628–633. [Google Scholar] [CrossRef]
- Lamarche, C.; Iliuta, I.A.; Kitzler, T. Infectious Disease Risk in Dialysis Patients: A Transdisciplinary Approach. Can. J. Kidney Health Dis. 2019, 6, 2054358119839080. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.L.; Prendecki, M.; Dhutia, A.; Gan, J.; Edwards, C.; Prout, V.; Lightstone, L.; Parker, E.; Marchesin, F.; Griffith, M.; et al. Longevity of SARS-CoV-2 immune responses in hemodialysis patients and protection against reinfection. Kidney Int. 2021, 99, 1470–1477. [Google Scholar] [CrossRef]
- Lisowska, K.A.; Pindel, M.; Pietruczuk, K.; Kuźmiuk-Glembin, I.; Storoniak, H.; Dębska-Ślizień, A.; Witkowski, J.M. The influence of a single hemodialysis procedure on human T lymphocytes. Sci. Rep. 2019, 9, 5041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betjes, M.G.H. Immune cell dysfunction and inflammation in end-stage renal disease. Nat. Rev. Nephrol. 2013, 9, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.; Dubey, P.; Benitez, J.; Torres, J.P.; Reddy, S.; Shokar, N.; Aung, K.; Mukherjee, D.; Dwivedi, A.K. A systematic review and meta-analysis of geographic differences in comorbidities and associated severity and mortality among individuals with COVID-19. Sci. Rep. 2021, 11, 8562. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Jager, K.J.; Kramer, A.; Chesnaye, N.C.; Couchoud, C.; Sánchez-Álvarez, J.E.; Garneata, L.; Collart, F.; Hemmelder, M.H.; Ambühl, P.; Kerschbaum, J.; et al. Results from the ERA-EDTA Registry indicate a high mortality due to COVID-19 in dialysis patients and kidney transplant recipients across Europe. Kidney Int. 2020, 98, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Steil, H.; Amato, C.; Carioni, C.; Kirchgessner, J.; Marcelli, D.; Mitteregger, A.; Moscardo, V.; Orlandini, G.; Gatti, E. EuCliD®—A Medical Registry. Methods Inf. Med. 2004, 43, 83–88. [Google Scholar] [PubMed]
- Merello Godino, J.I.; Rentero, R.; Orlandini, G.; Marcelli, D.; Ronco, C. Results from EuCliD® (European Clinical Dialysis Database): Impact of shifting treatment modality. Int. J. Artif. Organs 2002, 25, 1049–1060. [Google Scholar] [CrossRef]
- European Commission Joint Research Centre—ISPRA-Space, Security and Migration Directorate (JRC). Available online: https://covid-statistics.jrc.ec.europa.eu/ (accessed on 12 July 2020).
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Available online: https://github.com/dmlc/xgboost (accessed on 21 December 2019).
- Van Rossum, G. Python Tutorial; CreateSpace Independent Publishing Platform; (online publisher); 1995. [Google Scholar]
- Ling, C.X.; Huang, J.; Zhang, H. AUC: A better measure than accuracy in comparing learning algorithms. In Advances in Artificial Intelligence. Canadian AI 2003; Lecture Notes in Computer Science (Lecture Notes in Artificial Intelligence); Springer: Berlin/Heidelberg, Germany, 2003; Volume 2671, pp. 329–341. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; 2017. [Google Scholar]
- Abbas, M.; Robalo Nunes, T.; Martischang, R.; Zingg, W.; Iten, A.; Pittet, D.; Harbarth, S. Nosocomial transmission and outbreaks of coronavirus disease 2019: The need to protect both patients and healthcare workers. Antimicrob. Resist. Infect. Control 2021, 10, 7. [Google Scholar] [CrossRef]
- Leeds, C. COVID 19: Health care workers, risks, protection and transmission. Lancet Reg. Health 2021, 1, 100022. [Google Scholar] [CrossRef]
- La Milia, V.; Bacchini, G.; Bigi, M.C.; Casartelli, D.; Cavalli, A.; Corti, M.; Crepaldi, M.; Limardo, M.; Longhi, S.; Manzoni, C.; et al. COVID-19 Outbreak in a Large Hemodialysis Center in Lombardy, Italy. Kidney Int. Rep. 2020, 5, 1095–1099. [Google Scholar] [CrossRef]
- Kliger, A.S.; Silberzweig, J. Mitigating risk of COVID-19 in dialysis facilities. Clin. J. Am. Soc. Nephrol. 2020, 15, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Basile, C.; Combe, C.; Pizzarelli, F.; Covic, A.; Davenport, A.; Kanbay, M.; Kirmizis, D.; Schneditz, D.; Van Der Sande, F.; Mitra, S. Recommendations for the prevention, mitigation and containment of the emerging SARS-CoV-2 (COVID-19) pandemic in haemodialysis centres. Nephrol. Dial. Transplant. 2020, 35, 737–741. [Google Scholar] [CrossRef]
- Kliger, A.S.; Cozzolino, M.; Jha, V.; Harbert, G.; Ikizler, T.A. Managing the COVID-19 pandemic: International comparisons in dialysis patients. Kidney Int. 2020, 98, 12–16. [Google Scholar] [CrossRef]
- Marcelli, D. EuCliD (European Clinical Database): A database comparing different realities. J. Nephrol. 2001, 14, S94–S100. [Google Scholar]
- Richards, N.; Ayala, J.A.; Cesare, S.; Chazot, C.; Di Benedetto, A.; Gassia, J.P.; Merello, J.I.; Rentero, R.; Scatizzi, L.; Marcelli, D. Assessment of quality guidelines implementation using a continuous quality improvement programme. Blood Purif. 2007, 25, 221–228. [Google Scholar] [CrossRef]
- Marcelli, D.; Moscardó, V.; Steil, H.; Day, M.; Kirchgessner, J.; Mitteregger, A.; Orlandini, G.; Gatti, E. Data management and quality assurance for dialysis network. Contrib. Nephrol. 2002, 137, 293–299. [Google Scholar] [CrossRef]
- Delamater, P.L.; Street, E.J.; Leslie, T.F.; Yang, Y.T.; Jacobsen, K.H. Complexity of the basic reproduction number (R0). Emerg. Infect. Dis. 2019, 25, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Corbett, R.W.; Blakey, S.; Nitsch, D.; Loucaidou, M.; McLean, A.; Duncan, N.; Ashby, D.R.; Appelbe, M.; Brown, E.; Cairns, T.; et al. Epidemiology of COVID-19 in an urban dialysis center. J. Am. Soc. Nephrol. 2020, 31, 1815–1823. [Google Scholar] [CrossRef]
- Meyers, L.A.; Pourbohloul, B.; Newman, M.E.J.; Skowronski, D.M.; Brunham, R.C. Network theory and SARS: Predicting outbreak diversity. J. Theor. Biol. 2005, 232, 71–81. [Google Scholar] [CrossRef]
- Frieden, T.R.; Lee, C.T. Identifying and interrupting superspreading events-implications for control of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.L.; Srinivasan, R.; Brownstein, J.S.; Galvani, A.P.; Meyers, L.A. Disease Surveillance on Complex Social Networks. PLoS Comput. Biol. 2016, 12, e1004928. [Google Scholar] [CrossRef] [PubMed]
- Zwald, M.L.; Lin, W.; Sondermeyer Cooksey, G.L.; Weiss, C.; Suarez, A.; Fischer, M.; Bonin, B.J.; Jain, S.; Langley, G.E.; Park, B.J.; et al. Rapid Sentinel Surveillance for COVID-19—Santa Clara County, California, March 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 419–421. [Google Scholar] [CrossRef]
- Colman, E.; Holme, P.; Sayama, H.; Gershenson, C. Efficient sentinel surveillance strategies for preventing epidemics on networks. PLoS Comput. Biol. 2019, 15, e1007517. [Google Scholar] [CrossRef]
- Monaghan, C.; Larkin, J.; Chaudhuri, S.; Han, H.; Jiao, Y.; Bermudez, K.; Weinhandl, E.; Dahne-Steuber, I.; Belmonte, K.; Neri, L.; et al. Artificial Intelligence for COVID-19 Risk Classification in Kidney Disease: Can Technology Unmask an Unseen Disease? medRxiv 2020. [Google Scholar] [CrossRef]

| Low Risk Group. P(Class = L) = 0.648 | |||
|---|---|---|---|
| P(Class = L|Outbreak = Yes) | P(Class ≠ L|Outbreak = Yes) | P(Class = L|Outbreak = No) | P(Class ≠ L|Outbreak = No) |
| 0.23 | 0.77 | 0.73 | 0.27 |
| P(Outbreak = Yes|Class = L) | P(Outbreak = No|Class = L) | P(Outbreak = Yes|Class ≠ L) | P(Outbreak = No|Class ≠ L) |
| 0.06 | 0.94 | 0.37 | 0.63 |
| High Risk Group P(Class = H) = 0.197 | |||
|---|---|---|---|
| P(Class = H|Outbreak = Yes) | P(Class ≠ H|Outbreak = Yes) | P(Class = H|Outbreak = No) | P(Class ≠ H|Outbreak = No) |
| 0.51 | 0.49 | 0.14 | 0.86 |
| P(Outbreak = Yes|Class = H) | P(Outbreak = No|Class = H) | P(Outbreak = Yes|Class ≠ H) | P(Outbreak = No|Class ≠ H) |
| 0.40 | 0.60 | 0.09 | 0.91 |
| Risk Class | RR |
|---|---|
| L | −ref |
| M | 3.45 |
| H | 5.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellocchio, F.; Carioni, P.; Lonati, C.; Garbelli, M.; Martínez-Martínez, F.; Stuard, S.; Neri, L. Enhanced Sentinel Surveillance System for COVID-19 Outbreak Prediction in a Large European Dialysis Clinics Network. Int. J. Environ. Res. Public Health 2021, 18, 9739. https://doi.org/10.3390/ijerph18189739
Bellocchio F, Carioni P, Lonati C, Garbelli M, Martínez-Martínez F, Stuard S, Neri L. Enhanced Sentinel Surveillance System for COVID-19 Outbreak Prediction in a Large European Dialysis Clinics Network. International Journal of Environmental Research and Public Health. 2021; 18(18):9739. https://doi.org/10.3390/ijerph18189739
Chicago/Turabian StyleBellocchio, Francesco, Paola Carioni, Caterina Lonati, Mario Garbelli, Francisco Martínez-Martínez, Stefano Stuard, and Luca Neri. 2021. "Enhanced Sentinel Surveillance System for COVID-19 Outbreak Prediction in a Large European Dialysis Clinics Network" International Journal of Environmental Research and Public Health 18, no. 18: 9739. https://doi.org/10.3390/ijerph18189739
APA StyleBellocchio, F., Carioni, P., Lonati, C., Garbelli, M., Martínez-Martínez, F., Stuard, S., & Neri, L. (2021). Enhanced Sentinel Surveillance System for COVID-19 Outbreak Prediction in a Large European Dialysis Clinics Network. International Journal of Environmental Research and Public Health, 18(18), 9739. https://doi.org/10.3390/ijerph18189739