Abstract
Staphylococcus aureus is a nosocomial bacterium causing different infectious diseases, ranging from skin and soft tissue infections to more serious and life-threatening infections such as septicaemia. S. aureus forms a complex structure of extracellular polymeric biofilm that provides a fully secured and functional environment for the formation of microcolonies, their sustenance and recolonization of sessile cells after its dispersal. Staphylococcus aureus biofilm protects the cells against hostile conditions, i.e., changes in temperature, limitations or deprivation of nutrients and dehydration, and, more importantly, protects the cells against antibacterial drugs. Drugs are increasingly becoming partially or fully inactive against S. aureus as they are either less penetrable or totally impenetrable due to the presence of biofilms surrounding the bacterial cells. Other factors, such as evasion of innate host immune system, genome plasticity and adaptability through gene evolution and exchange of genetic material, also contribute to the ineffectiveness of antibacterial drugs. This increasing tolerance to antibiotics has contributed to the emergence and rise of antimicrobial resistance (AMR), a serious problem that has resulted in increased morbidity and mortality of human and animal populations globally, in addition to causing huge financial losses to the global economy. The purpose of this review is to highlight different aspects of S. aureus biofilm formation and its overall architecture, individual biofilm constituents, clinical implications and role in pathogenesis and drug resistance. The review also discusses different techniques used in the qualitative and quantitative investigation of S. aureus biofilm and various strategies that can be employed to inhibit and eradicate S. aureus biofilm.
1. Introduction
Staphylococcus aureus (also denoted as Staph. aureus or S. aureus) is a Gram-positive pathogenic bacterium and is a major cause of different infectious illnesses in humans and animals [1,2]. These illnesses may range from simple skin and soft tissue infections to more serious and life threatening conditions such as blood infections (bacteraemia/septicaemia) [3]. Staphylococcus aureus secretes an extracellular polymeric substance (EPS), known as biofilm, that helps the microbe to resist and minimise the effect of antibacterial drugs [4].
Similar to any other bacterial biofilm, a Staphylococcus aureus biofilm also has two distinct components, i.e., water (about 97%) and the organic matter which includes EPS and microcolonies [5]. The EPS constitutes about 50 to 90% of the total organic matter of a biofilm and is a complex of different polymeric substances, such as extracellular DNA (eDNA), proteins and polysaccharides [6,7]. The remaining portion, 10–25%, consists of microcolonies [5].
In Staphylococcus aureus biofilm, the major component of EPS is the polysaccharide intercellular adhesin (PIA) [8]. The polysaccharide component of EPS has been given the name PIA due its function, i.e., intercellular adhesion of bacterial cells, and poly-β(1-6)-N-acetylglucosamine (PNAG), due to its chemical composition. PIA are cationic in nature and play a significant role in colonisation, biofilm formation and biofilm-related infections, immune evasion, resistance to antimicrobials and phagocytosis [9].
Staphylococcus aureus EPS also contains a range of proteins including accumulation associated proteins (Aap), surface binding protein A (Spa), fibrinogen binding protein (FnBP) A and B, extracellular matrix binding protein (Embp), amyloid fibres and S. aureus surface binding protein (SasG) [10,11]. Other S. aureus proteins that are found covalently attached to cell wall peptidoglycan (PG) by trans peptidases (sortases) are known as cell wall-anchored proteins (CWP) [12]. There are as many as 25 different CWPs, categorised as microbial surface component recognising adhesive matrix molecule (MSCRAMM), near iron transporter (NEAT), three-helical bundle and G5-E repeat proteins [12,13]. These S. aureus proteins perform different functions. For example, accumulation associated protein Aap interacts with PIA and plays a role in biofilm maturation [8]. SasG protein and surface binding protein A are responsible for surface attachment and causing infections [10]. CWA proteins facilitate adhesion to EPS, to host surface, and their interaction with CWA proteins on adjacent cells contributes to the accumulation of biofilm [13].
Similarly, amyloid fibres act as a scaffold that keeps S. aureus cells anchored to the biofilm matrix and thus maintain the stability of the biofilm [7,14]. Alongside PIA and EPS proteins, the third important component of S. aureus biofilm EPS is eDNA. eDNA has been reported to be involved in irreversible attachment, horizontal gene transfer, maintaining biofilm integrity, antimicrobial resistance and host immune system evasion [15].
The extra polymeric substance of a biofilm also contains charged (both positive and negative) groups and hydrophobic groups. The negatively charged groups found in EPS include carboxyl groups, phosphates, sulphates, glutamic acid and aspartic acid, while positively charged ones include amino sugars [16]. Despite of the presence of both positively and negatively charged species, the overall charge on the EPS surface is negative and thus can serve as a better target for positively charged moieties [17,18].
2. Biofilm Formation
The formation of biofilm proceeds through four different stages [19], which are
- Attachment of planktonic cells to the surface (either a biotic host or any abiotic surface);
- Colonisation and biofilm formation;
- Biofilm maturation;
- Biofilm dispersal.
Biofilm formation in S. aureus is initiated when free floating, planktonic cells attach to the available surface and start colonising [20]. S. aureus adherence to a surface is influenced by hydrophobic and hydrophilic interactions between the S. aureus cell surface and any biotic or abiotic surface [21,22,23]. It has been found that the S. aureus cell surface adheres to hydrophobic surfaces by the help of many weakly binding macromolecules, while its adherence to hydrophilic surfaces involves fewer but stronger binding macromolecules [23,24].
The formation of microcolonies is followed by the formation of an extracellular polymeric substance (EPS) that develops into a fully matured biofilm [19]. Once the biofilm is fully matured, the bacterial cells residing inside it release certain chemicals, i.e., D-amino acids and EPS-degrading enzymes such as alginate lyase, to break and disperse the biofilm [25]. These planktonic cells are ready to either recolonise the same site or attach to a different site and repeat the process to form a new biofilm [6]. Figure 1 depicts different stages involved in the formation of a bacterial biofilm.
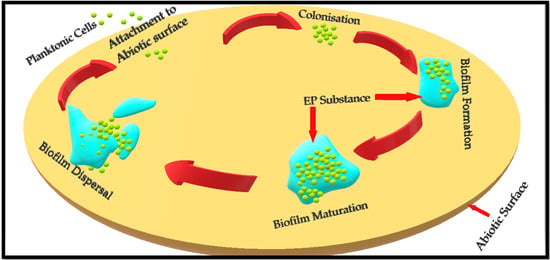
Figure 1.
Depiction of Staphylococcus aureus biofilm formation on an abiotic surface. Basic concept has been adopted from Idrees et al. (2020) and Paharik (2016) [7,26].
Staphylococcus aureus cells that are encased and protected by biofilms show different phenotypic characters compared to cells in their planktonic form. Biofilm-associated Staphylococcus aureus cells are more resistant to antibiotics and exhibit differences in cell size and growth, gene expression and protein production, compared to their free living counterparts [24].
Biofilm-associated S. aureus cells have been reported to have four different metabolic states, i.e., they can either be growing aerobically, can be fermentative, can be dormant, or can even be dead [27]. Besides the extracellular polymeric matrix that shelters the cells against antibiotics, the dormant and metabolically slow growing cells have also been reported to add to antimicrobial resistance [28]. Moormeier [29] reported that S. aureus cells encased in a biofilm grow at different rates, i.e., some cells grow at a faster rate as compared to other cells within the same biofilm. These cells are smaller in size and attain their normal size once released upon the dispersal of the biofilm.
Biofilm associated S. aureus cells exhibit altered gene expressions, i.e., up- and downregulation of genes has been witnessed in the cells residing inside a S. aureus biofilm. The differential gene expression accounts for the variation in cell sizes within a biofilm, their growth rates and protein production [6].
3. Gene Expression during Staph. aureus Biofilm Formation and Dispersal
Like many other microbial functions, microbial biofilm formation is also encoded by certain biofilm-associated genes [30]. In S. aureus, biofilm formation is mainly encoded by 12 different genes, i.e., fibrinogen-binding proteins (fib) gene, fibronectin-binding proteins (fnbA and fnbB) genes, intercellular adhesion (icaA, B, C and D) genes, clumping factor (clfA and B), elastin binding protein (ebps), laminin binding protein (eno) and collagen binding protein (cna) gene [31]. Figure 2 lists different genes encoding the corresponding stages during S. aureus biofilm formation.
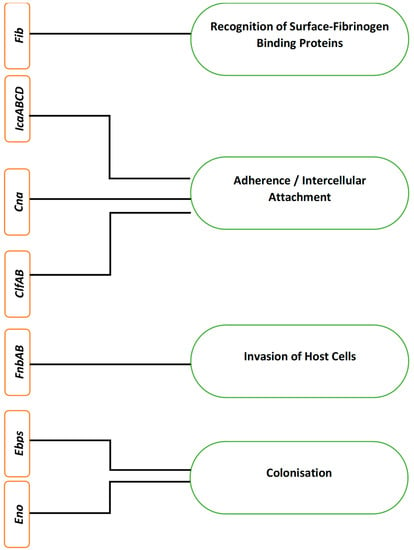
Figure 2.
Different genes and their corresponding encoding in Staphylococcus aureus biofilm formation.
The aforementioned genes encode different surface proteins to cause S. aureus adherence to the host, its penetration into the host and its colonisation, ultimately leading to biofilm formation and virulence. In S. aureus, the fib gene facilitates and encodes the recognition of surface-fibrinogen binding proteins, while the collagen binding proteins encoded by their corresponding cna genes promote adherence to the surface [26].
The intercellular adherence genes icaABCD encode the process of cell to cell adherence and initiate biofilm formation [32,33]. The co-expression of fnbAB genes also facilitates the formation of biofilm in S. aureus. It has also been found that although fnbA and fnbB genes in S. aureus are not involved in the process of adherence, they still contribute to the development of biofilm, i.e., fnAB act as invasins and facilitates S. aureus to penetrate into the host cells [34,35].
Clumping factor genes clfA and clfB encode cell wall-anchored proteins that attach to the surface fibrinogen of the host [36]. This attachment of clumping factors A and B encoded by clfAB genes facilitates S. aureus colonisation of the host, promotes biofilm formation and causes virulence by means of immune evasion through binding soluble fibrinogen [13,37]. Serine-aspartate repeat factors C and D (SdrCD) facilitate attachment to desquamated epithelial cells and nasal colonisation, while SdrE causes immune evasion by binding to complement factor H [13]. These proteins are encoded for their respective roles by the corresponding sdr genes [38]. The encoding of elastin-binding proteins and laminin-binding proteins by their respective genes, ebps and eno genes, facilitates colonisation of the host and biofilm formation [39].
Once the biofilm is fully matured, it disperses and releases the sessile cells which are ready to repopulate either the same site (primary site) or a new site (secondary). This dispersal of the Staphylococcus aureus biofilm is regulated by four different genes of the accessory gene regulatory (Agr) system [29]. The Agr genes encoding the dispersal of biofilm include AgrA, AgrB, AgrC and AgrD [40]. Agr-regulated dispersal of biofilm is brought about by the induction of different proteases and phenol-soluble modulins (PSMs) [28]. These proteases and PSMs disperse the biofilm by acting as surfactants [41,42].
3.1. Gene Expression and Quorum Sensing in Staphylococcus aureus
Cell to cell communication in Staphylococcus aureus is regulated by the accessory gene regulator (Agr), a system that prompts genetic adaptations for communication in bacterial cells [40]. Staphylococcus aureus has a distinctive Agr system that regulates the expression of different toxins and virulence factors and controls the bacterial-host interaction at the site of infection as well [43].
The Agr system consists of two pairs of genes, i.e., AgrA-AgrC and AgrB-AgrD, and activates in response to the secretion of an autoinducing peptide (AIP) [40,44]. Genes AgrB and AgrD regulate the expression and transportation of AIP. Once a sufficient amount of AIP is aggregated in the surrounding extracellular environment, the two-component system of AgrA and AgrD becomes activated, triggering the intracellular communication, quorum sensing [45,46].
3.2. Gene Expression and Staphylococcus aureus Pathogenesis
Pathogenesis in Staphylococcus aureus is a result of the expression of different virulence factors such as toxins, immunomodulators and exoenzymes [47]. The expression of these factors is regulated and encoded by particular genes [48]. Amongst the virulence factors in S. aureus, toxins are the most important factors that protect the bacterium by averting any possible elimination by its host’s defence system [49].
The expression of different toxins encoded by their corresponding genes causes different infectious morbidities. For instance, toxic shock syndrome toxin1 (TSST1), when encoded by tstH gene, causes toxic shock syndrome, while exfoliative toxin causes scalded skin syndrome when encoded by its corresponding eta and etb genes. Similarly, haemolysin and leukotoxin, regulated by hla and lukDE genes, are responsible for pore formation and membrane destruction in their hosts [49,50].
Exoenzymes such as lipases, proteases, nucleases, etc., encoded by genes associated with the Agr system, cause tissue destruction and metastatic infections [46,51]. The immunomodulators such as leucocidin, phenol-soluble modulins, etc., are responsible for skin invasive infections, abscesses and pneumonia when regulated by their corresponding genes, AgrA and AgrC [41].
Similarly, S. aureus pathogenesis involves evasion of the host’s innate immune system. S. aureus’s entry into subepidermal tissues or blood is encountered by the host immune system [52]. S. aureus counterattacks the host immune system and inactivates it by secreting different proteins, encoded by two immune evasion gene clusters, IEC1 and IEC2 [53].
Apart from the aforementioned virulence factors, there are other factors, such as attachment to the host and persistence factor (involved in biofilm formation), which, when regulated by their corresponding genes, cause different infectious diseases. Virulence factors involved in S. aureus attachment to the host surface, regulated by clfAB, fnbAB, cna and ica genes, and those involved in persistence, encoded by ica locus and hemB genes, are responsible for endocarditis, septic arthritis, cystic fibrosis and relapsing infections [47].
Various virulence factors, their corresponding regulatory genes and their clinical implications have been summarised in Table 1.

Table 1.
A list of different virulence factors, their corresponding regulatory genes and their resultant clinical implications. Data have been collected from Otto (2014), Gordon (2008) and Oogai (2011) [47,48,49].
4. Staphylococcus aureus Biofilm and Antimicrobial Resistance (AMR)
Antimicrobial resistance (AMR) is a serious threat to human and animal lives, as many of the traditional antimicrobial drugs are losing their efficacy (partial or full). According to World Health Organisation (WHO)’s report from 2019, AMR is causing 700,000 mortalities each year, and 230,000 people die as a direct consequence of resistance to antituberculosis drugs alone [54]. Apart from being a major cause of morbidity and mortality, both in humans and animals, AMR is also causing a huge financial loss to the global economy [55].
Similar to any other biofilm-forming bacteria, S. aureus also develops a biofilm to slow down or prevent the diffusion of antimicrobial drugs, hence hindering drugs’ access to the cells residing inside the biofilm [56]. The formation of biofilm is prompted by different factors such as nutrient deprivation, changes in temperature (too low or too high) and dehydration [57,58]. Since biofilm provides protection to the encased cells, S. aureus has therefore adopted it as one of its useful tools to resist antibiotics that are generally bactericidal to the cells in their free-floating planktonic form [59]. Bacterial cells that are covered in a biofilm have been reported to resist antibiotics 10–1000 times more compared to their corresponding planktonic forms [8].
S. aureus is generally a commensal bacterium, colonising up to 30% of the human population (it colonises host skin, nostrils, armpits, and groin) globally, and many times, it causes little or no harm at all [60,61]. However, these niches of usual occurrence can sometimes become a primary cause of S. aureus infections [24]. Being a nosocomial pathogen, and due to its ability to form biofilms on both biotic and abiotic surfaces, Staphylococcus aureus infections are complicated and difficult to eradicate, particularly those caused by a methicillin-resistant strain (MRSA) [62,63]. Individuals with certain skin conditions, such as atopic dermatitis (AD), are highly susceptible to S. aureus colonisation and hence its biofilm formation [64,65]. Antibacterial drugs become less penetrable across the biofilm, allowing S. aureus cells to survive in the presence of drugs at reduced concentrations [56]. In addition to the major role of bacterial biofilms in drug resistance, other factors, such as evasion of innate host immune system, genome plasticity and adaptability through gene evolution and exchange of genetic material, also contribute to the development and rise of AMR [24,66].
Staphylococcus aureus biofilms on inanimate surfaces are equally problematic, particularly those found in health care facilities and food related industries. Medical devices need to be sterilised and free from all kinds of pathogens. S. aureus biofilm on medical devices (made of different materials such as steel, glass, etc.), if not sterilised, can be a major source of infections in hospitals, care homes and GP surgeries [67]. Sterilisation of such surfaces becomes difficult due to S. aureus’s resistance against surface cleaners and sanitisers [68].
Food industry-related biofilms may encase more than one type of microorganism, including S. aureus, and are therefore comparatively more resistant to sanitisers and surface cleaners than the biofilm that encases S. aureus cells alone [69]. This microbial resistance to surface cleaners and sanitisers results in food spoilage, contamination of freshly arrived food items and corrosion of storage surfaces, and more importantly, it increases the risk of transmission of infectious diseases [70].
Staphylococcus aureus biofilm-related infections cause serious life-threatening illnesses in human and animal populations and have incredibly challenging clinical implications that medical scientists have to deal with.
5. Clinical Implications of Staphylococcus aureus Biofilms
Staphylococcus aureus-related infections are problematic and are difficult to treat due to biofilm formation. The ineffectiveness of the host immune system and antibiotics against Staphylococcus aureus biofilm results in the development of chronic infectious diseases [8]. S. aureus can form biofilms on different surfaces and medical devices such as surgical instruments, implants, etc., in healthcare facilities. Such surface-related and medical device-associated S. aureus biofilms, if not properly sanitised and left untreated, can cause infections in patients [27]. Medical device-associated biofilms, those on implantable devices in particular, often result in the removal and replacement of these devices, thus posing surgical risks as well [8].
Staphylococcus aureus also develops biofilms inside the infected host tissues. Such infections are more problematic due to antimicrobial resistance, and their treatment often involves the removal of infected tissues [71]. This biofilm-related antimicrobial resistance is partly due to the presence of some dormant Staphylococcus aureus cells (also known as persister cells) encased by its biofilm. These cells maintain their dormancy during antimicrobial treatment and become active as soon as the treatment is withdrawn, thus causing a chronic recurrent infection [72].
Treatment of all such aforementioned S. aureus biofilm-related infections will require novel therapeutic strategies. The first and most important step in this regard is to study S. aureus biofilm and understand it thoroughly by employing suitable microbiological and physico-chemical techniques.
6. Techniques and Strategies Used in Studying Staphylococcus aureus Biofilm
Microbiologists adopt different approaches based on different techniques to study and evaluate Staphylococcus aureus biofilm. These techniques are either direct evaluation of biofilm, based on the measurement of thickness of a biofilm, or indirect techniques of measurement where a biofilm is quantified in terms of studying its different constituents or the activity of the bacterial cells within the biofilm [73]. These techniques are discussed in the following sections.
6.1. Direct Observation Techniques
Direct observation of the intricacy and changing aspects of biofilms can be studied by biofilm optical imaging technology, comprising light microscopy, scanning electron microscopy (SEM), transmission electron microscopy (TEM) and confocal laser scanning microscopy (CLSM). These methods are employed to examine the presence of biofilm and visualize its 3D structure [74].
6.1.1. Light Microscope and Transmission Electron Microscope
S. aureus adhered on cover slips, polystyrene Petri dishes and polymethacrylate films (acrylic sheets) is studied using a light microscope. Observation through a light microscope is incapable of creating a 3D structure but requires a planar and transparent surface for attachment of the microorganism. To improve the image clarity, fluorescent and epifluorescent dyes are used [75].
Cell components can be directly monitored with negative staining using TEM. Glycocalyx of Staphylococcal cells are stabilized using cationic reagents such as a combination of alcian blue, ruthenium red, lysine or paraformaldehyde. Following these steps, the sample is ready to be observed under TEM [76].
6.1.2. Profilometry and Scanning Electron Microscope
Sterile glass slides were incubated in medium seeded with the test culture. The slides were fixed using Bouin fixative and stained with Alcian blue. Under 100x oil immersion, pictures were taken by photomicroscope with red filter for blue photomicrographs. In order to study the effect of etchant on the biofilm formation and surface morphology of the substrate, this technique can be further modified [77].
The glass slides can be incised/etched chemically to change the surface roughness. The slides are treated with the required etchant at different time intervals followed by incubating the treated slides in overnight test culture. Optical microscope and profilometry were used to measure large fluctuations in thickness of bacterial biofilm and roughness of the substrate surface [78].
To examine the biofilm surface topography and morphology, the test culture was grown in biofilm-promoting agar plates. The mature colonies were removed and the footprints on the agar plates can be visualized under the profilometer [78].
Furthermore, the effect of increasing etching time on biofilm architecture, can be studied by scanning electron microscopy. Therefore, a distinctive correlation between the surface roughness and adherence of bacteria can be studied through this experiment [78].
As SEM is a high vacuum method, before imaging the samples, the biofilm plate was therefore dehydrated. The samples were mounted on SEM specimen stubs and splutter coated with heavy metals, and the electrons which were released from the metal coating were captured by SEM for imaging. The biofilm surface was navigated under the lens and scanned for the desired images [79].
This method is also efficient in studying bacterial adherence to submerged solid surfaces, which leads to biofilm formation and hence can be used as a simple model to explore S. aureus biofilm.
6.1.3. Scanning Transmission X-ray Microscopy (STXM)
STXM techniques help to explore the nature and extent of macromolecules and dense microbial communities in a biofilm. As a contrast mechanism, STXM uses near-edge X-ray adsorption spectroscopy (NEXAFS) as a new tool for hydrated biological samples [80]. This can be achieved due to the capacity of the X-ray to penetrate water, with decreased radiation damage as compared to electron beam approaches and the presence of appropriate analytic core edges in the soft X-ray sections [81,82]. Soft X-rays offer detection on a scale of 10 nm or less and greater penetration depth, which is appropriate for microorganisms and microbial biofilm imaging [76].
STXM can be coupled with transmission electron microscopy to examine biofilms for correlative mapping and better analysis of spatial distributions of biological macromolecules present in the biofilms [80]. These correlative approaches help in identifying the biochemical sources for the biofilm structure. The analytical ability of STXM answers questions concerning the distribution of carbohydrates, lipids and nucleic acids and the role of proteins in the extracellular matrix of the biofilm [76].
6.1.4. Fluorescent Tagging of Biofilm
Fluorescent tagging of biofilm includes the following:
- (a)
- Confocal laser scanning microscopy (CLSM)
Confocal microscopy has an advantage over SEM in that no dehydration of the sample is required, and the biofilm can be imaged in the hydrated state. CLSM is a great aid to study the physiology and 3D morphology of biofilm [79]. To observe the sample with a confocal microscope, the biofilm can be stained by fluorescence-labelled dyes or tagged with green fluorescent proteins. Biofilm-producing isolate can be engineered genetically by the tagging of microbial gene of interest by gene-cassette-encoding GFP as a reporter gene. These lectin stains are known to target the extracellular matrix, whereas some fluorophores go for eDNA (extracellular DNA), thus assisting us in visualizing the biofilm matrix content [83].
In order to study individual biofilm components, the stains used include SyproOrange for protein, Syto9 for nucleic acids, molecular probes of Nile red for lipid hydrophobic sites and specific lectins for sugar, glycoconjugate and carbohydrate [76,84]. Additional resolution is needed for multi-layered and 3D complex biofilm structures; imaging software is used to combine the layers of biofilms into a digital image.
- (b)
- Fluorescent in situ hybridization (FISH)
Probes of the fluorescent in situ hybridization (FISH) method can help in identifying the S. aureus biofilm present in a heterogeneous biofilm community. Fluorescent-labelled cells within biofilm can also be analysed by FISH. DNA probes aimed at hybridizing 16S rRNA of bacteria are incorporated with either an enzyme such as horseradish peroxidase or a fluorescent dye such as FITC or rhodamine [85].
An advantage of using probes conjugated with enzyme is that it does not destroy the microbe, and the growth activity within the biofilm can be determined with the number of ribosomes present in a microbe. Probes are intended to label the conserved region of only a specific species. FISH has been applied to explore the 3D distributions of different bacterial activities and examine the thickness of biofilms [86].
6.2. Indirect Observational Techniques
These include the following:
6.2.1. Tube Method
This is a qualitative technique described by Christensen et al. for the detection of biofilms. A solution of 1% crystal violet was used to stain the biofilm and observe the film around the wall and bottom of the test tube to confirm a biofilm producer. The extent of biofilm formed was recorded as 1: weak/none, 2: moderate and 3: high/strong. Ring formation at the liquid–air interface was not considered indicative of slime production [87].
This technique is frequently subjected to analytical restrictions and is incompetent for detecting bacterial adherence precisely. Therefore, to overcome these shortcomings, the following method can be adapted to modern approaches to obtain precise findings.
6.2.2. Congo Red Agar Method
The Congo red agar screening technique is used to detect whether the Staphylococcal isolates are biofilm producers. A positive result is shown by black colonies with a dry crystalline consistency. Weak slime producers are indicated as pink; however, darkening is seen at the centres of colonies occasionally. The blackening of the colonies devoid of dry crystalline morphology suggested moderate biofilm procedures [88].
6.2.3. Detection of Biofilm Production by Microtiter Plate Assay
The microtiter plate assay is a standard quantitative technique to detect biofilm production. The duration of incubation can be modified as needed, and the medium of choice for evaluating the biofilm production is TSB or MHB with 1% glucose. The technique was adapted for better quantification of the biofilms. Optical density (OD) was determined with a microplate reader at a wavelength of 570 nm (OD 570 nm) [89,90].
6.2.4. Biofilm-Associated Genes Detection by PCR
Biofilm-associated genes are amplified by PCR, such as multiplex, qualitative real-time PCR and conventional PCR, which can identify whether biofilm-associated genes are present in the sample. Instead of qualitative real-time PCR, multiplex and conventional PCR are applied for biofilm gene detection [91]. The isolated PCR product can be visualized on an agarose gel to confirm the amplified gene due to the presence of a DNA intercalating dye, ethidium bromide in the gel, whereas in qualitative real-time PCR, a pair of specific hybridization probes labelled with fluorescence dye is used for detection of the amplicon [92].
6.2.5. Mass Spectrometry
Since the extracellular polymeric substance consists of polysaccharides and proteins, such as extracellular enzymes, these proteins expressed within the EPS matrix are detected and categorized by mass spectrometry [93]. Chemicals and complex biologic molecules involved in the process of biofilm formation can be analysed in detail by MS. Matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) are the types of MS. By application of MALDI, the expression of proteins and enzymes in response to external factors and the bacterial growth can be monitored. Mass spectrometry is highly sensitivity and can be performed with a minimum amount of sample [75,94].
6.2.6. Atomic Force Microscopy (AFM)
Atomic force microscopy is a key technique for imaging samples at the subcellular level. AFM assists in investigating molecular interactions, detecting physiochemical properties such as surface hydrophobicity and quantifying macromolecules on the surface and within the cells [95,96]. The simple principle of AFM is to scan the surface of the sample with a sharp probe tip in a raster pattern; the interaction between the probe tip and the sample is measured on the flexible cantilever. The cantilever bends upon sensing an attractive force on the surface, and this force is measured by gauging the deflection of the cantilever using a photodiode and a laser beam [97,98].
AFM can quantify the adhesion force between the cells, surface and single molecules and is thus used to visualize biofilms. This tool has been applied in gaining an understanding of single and multispecies biofilm components and the core adhesion mechanisms. AFM images allow quantification of biofilm thickness, biomass roughness and EPS content [77]. A detailed insight into the viscoelasticity and adhesion of cells during biofilm formation can help with biofilm control strategies. The viscoelasticity of biofilm influences the penetration of antimicrobials and biofilm removal from the surface and thus plays an important role in protection against chemical and mechanical challenges. This approach can be used to combat antimicrobial resistance in biofilms [98,99]. The aforementioned techniques and their applications have been summarised and listed in Table 2.

Table 2.
Summary of techniques used for biofilm visualization and characterisation.
7. Strategies Used to Inhibit and Disrupt Staphylococcus aureus Biofilm
Biofilms are a major cause of antimicrobial resistance and complications in treating S. aureus-related infections [111]. It is, therefore, important to treat these biofilms to minimise and curtail the chances of antimicrobial resistance by adopting adequate antibiofilm strategies. Antibiofilm strategies are mainly of two types, i.e., the inhibition or prevention of new biofilm formation and the dispersal or eradication of existing biofilms [112].
Different antibiofilm agents including drugs, functional excipients and other antibiofilm molecules (both naturally occurring and synthetic) have been reported to serve the purpose of biofilm inhibition and its disruption in Staphylococcus aureus. These antibiofilm agents are discussed in the following sections.
7.1. Antibiofilm Drugs and Functional Excipients
Several publications have been reported to have provided information about antibacterial drugs with antibiofilm activities. Rifampin, for instance, an antibiotic used for the treatment of TB and other bacterial infections, has been reported to have inhibited and disrupted biofilm in S. aureus [113,114]. Rifampin has also been reported to have shown antibiofilm activity in combination with another antibiotic, ciprofloxacin, against S. aureus biofilm [115].
Similarly, vancomycin, an antibacterial drug used in the treatment of both methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible Staphylococcus aureus (MSSA), has been reported to effectively treat Staphylococcus aureus biofilms [112,116]. Vancomycin has been used in combination with other drugs to treat Staphylococcus aureus biofilm, e.g., a combination of vancomycin and rifampin as antibiofilm dual therapy in MRSA-related infections [117]. Hu et al. [118] reported the use of the antibiotics azithromycin and clindamycin for the inhibition and dispersal S. aureus biofilm. The antibiofilm mechanism of azithromycin is based on its ability to disrupt bacterial quorum sensing [119].
The mechanisms by which antibiotics inhibit and or disrupt Staphylococcus aureus biofilm are not fully known and have not been reported thus far. Therefore, the antibiofilm mechanisms of antibiotics still remain an area that needs to be explored in order to devise effective strategies for treating S. aureus biofilm-related infections.
Apart from antibiotics, other naturally existing compounds such as amino acids, antimicrobial proteins (AMP), polyether ionophores, essential oils and plant extracts have also been used to inhibit and eradicate Staphylococcus aureus biofilm.
Scientists have used amino acids as antibiofilm agents either (1) as individual amino acids, (2) as mixtures of different amino acids or (3) as combinations of amino acids and antibiotics. Sanchez et al. [120], for instance, used D-amino acids as individual amino acids, equimolar amounts of D-amino acids as a mixture and also a D-amino acids-antibiotic combination to inhibit and disperse Staphylococcus aureus biofilm. One similar study conducted by Warraich et al. [121] employed a combination of amino acids and the antibiotic ciprofloxacin, which not only successfully inhibited the formation of new biofilm but also disrupted the existing biofilm in Staphylococcus aureus.
The antibiofilm activity of D-amino acids involves their ability to inhibit the formation of biofilm-related proteins and disruption of eDNA. Hochbaum et al. [122] experimentally assessed the role of D-amino acids in inhibiting the formation of biofilm-related proteins in S. aureus. Similarly, Warraich et al. [121] used confocal microscopy and showed that the antibiofilm mechanism of D-amino acids involved their ability to disrupt eDNA in the EPS matrix of S. aureus biofilm.
Amino acids, when linked together, form protein polymers. Different proteins have been reported with antimicrobial properties. These antimicrobial proteins (AMP) have also been the target of scientific exploration for their antibiofilm activity, either alone or in combination with other antibiotics. For example, Nair et al. [123] have used a bactericidal protein P128 in synergy with different antibiotics, such as vancomycin, gentamicin, linezolid and daptomycin, to inhibit the formation of S. aureus biofilm.
The antibiofilm mechanism of AMPs involves (i) the prevention of gene expression encoding the formation of biofilm-related proteins and (ii) AMPs’ attachment to eDNA [17].
Scientists have also been exploring the antibiofilm activity of naturally occurring polyether ionophores. One such study carried out by Hickey et al. [124] used four different polyether ionophores, lasalocid, monensin, narasin and salinomycin, and successfully eradicated Staphylococcus aureus biofilm. Although, the antibiofilm activity of polyether ionophores has been reported, the exact mechanism of action needs to be investigated.
Alongside the aforementioned antibiofilm approaches, recent years have seen scientific work focussing on the use of essential oils and plant extracts amongst the future’s effective antibiofilm therapies. Bazargani and Rohloff [125] have reported the use of different essential oils and plant extracts to inhibit and disperse Staphylococcus aureus biofilm.
Essential oils and plant extracts have also been used in combination with antibiotics against S. aureus biofilm, e.g., Monteiro-Neto et al. [126] studied Cuminaldehyde, a natural compound found in essential oils, in combination with ciprofloxacin and reported its synergetic antibiofilm activity against S. aureus biofilm. The antibiofilm mechanism through which plant extracts and essential oils target S. aureus biofilm involves the disruption of bacterial quorum sensing [119,127].
7.2. Other Antibiofilm Molecules
Antibiofilm molecules are small molecules (other than the drugs and functional excipients discussed in Section 6.1) exhibiting antibiofilm activity. Unlike the functional excipients (most of which are antibacterial as well), these molecules are not antibacterial but are antibiofilm only [128]. These molecules include imidazoles, indoles, pyrazoles, carbazoles, pyrroles, oxazolidinones and furanones, derivatives of phenazine, cinnamide and quinoline, etc. [129]. The molecular mechanism of antibiofilm activity of these small molecules is not yet fully known.
Apart from these small molecules, enzymes, such as DNA and exopolysaccharide degrading enzymes, and nanoparticles of heavy metals, such as silver and zinc, have also been used to prevent Staphylococcus aureus biofilm formation. The enzymes DNase I and Dispersin B have been reported to have degraded eDNA, while the enzyme a-amylase has been reported for its degradation of exopolysaccharides in S. aureus biofilm [119].
Nanoparticles have also been used either in isolation or in combination with other antibacterial drugs for the inhibition and dispersal of S. aureus biofilms. For instance, Fontecha-Umaña et al. [130] used silver and zinc oxide nanoparticles on their own, while Hamida et al. [131] reported a combination of silver nanoparticles and antibiotic ampicillin as an antibiofilm strategy for Staphylococcus aureus biofilm. The antibiofilm activity of nanoparticles has been reported in numerous studies; however, their mechanism of action has yet to be explored.
The antibiofilm agents mentioned in Section 6.1 and Section 6.2 have different mechanisms of actions for the inhibition and/or dispersal of Staphylococcus aureus biofilm. These mechanisms include the obstruction of bacterial adhesion and quorum sensing; disruption of extracellular DNA (eDNA), proteins, lipopolysaccharides, exopolysaccharides; and inhibition of signalling molecules, i.e., N-acyl homo-serine lactones [74,129]. Table 3 summarises different kinds of antibiofilm agents and their mechanisms of action.

Table 3.
Different categories of antibiofilm agents with examples and reported mechanism of action against S. aureus biofilm.
8. Conclusions and Future Perspectives
Microorganisms including S. aureus have the remarkable ability to enclose themselves in a protective environment, i.e., biofilms, to survive and cause recalcitrant infections. The primary focus of this review was to discuss the different genetic and morphological factors that influence biofilm formation in S. aureus specifically and their clinical implications. As highlighted in this review, techniques such as microtiter plate assay coupled with crystal violet staining, CLSM and mass spectroscopy provide high throughput detailed information on biofilm morphology in the normal hydrated state and aid in the characterisation of individual EPS components, respectively.
Despite the advances made in studying the composition, architecture and dynamics of biofilm formation in S. aureus, little is known about the mechanism of action of antibiofilm agents used to inhibit or disrupt these biofilms. There is also a dearth of information on the effect of antibiofilm agents and their mode of action on the physiology of cells residing in different layers of the biofilm. Further research is needed to fill in these knowledge gaps. The most interesting question to answer would be whether these antibiofilm agents have specific targets (EPS proteins/polysaccharides, eDNA, persister cells, etc.) or whether they have multiple mechanisms for biofilm inhibition and dispersal. Deciphering the precise mechanism will predict if nosocomial pathogens, such as S. aureus, will develop resistance to these novel agents or not.
Novel antibiofilm and antimicrobial combinations have the potential to hinder the development of resistance in microorganisms if they have multiple modes of action. Therefore, elucidating the multimodal effects of functional excipients, such as amino acids and antibiofilm agents, will inform the design and development of these drug combinations. This will lead to the targeted use of these agents to treat a wide range of biofilm-related infections caused by this notorious pathogen and eventually strengthen the fight against AMR.
Author Contributions
Conceptualization, M.I. and A.R.; methodology, M.I., S.S. and A.R.; investigation, M.I. and S.S.; writing—original draft preparation, M.I. and S.S.; writing—review and editing, M.I., N.K. and A.R.; visualization, A.R. and N.K.; supervision, A.R. and N.K.; project administration, A.R. and N.K. All authors have read and agreed to the published version of the manuscript.
Funding
M.I. was funded by faculty of science and engineering, university of wolverhampton.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis or interpretation of the data; in the writing of the manuscript or in the decision to publish the results.
References
- Adams, M. Staphylococcus aureus and other pathogenic Gram-positive cocci. In Foodborne Pathogens; de Blackburn, C.W., McClure, P.J., Eds.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Elsevier: Cambridge, UK, 2009; pp. 802–819. ISBN 9781845693626. [Google Scholar]
- Schaumburg, F.; Pauly, M.; Anoh, E.; Mossoun, A.; Wiersma, L.; Schubert, G.; Flammen, A.; Alabi, A.S.; Muyembe-Tamfum, J.-J.; Grobusch, M.P.; et al. Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 2015, 21, 345.e1–345.e8. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.D.; Malachowa, N.; Deleo, F.R. Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol. 2015, 185, 1518–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, J.B.; Mlynek, K.D.; Hettiarachchi, H.; Alamneh, Y.A.; Biggemann, L.; Zurawski, D.V.; Black, C.C.; Bane, C.E.; Kim, R.K.; Granick, M.S. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS ONE 2018, 13, e0205526. [Google Scholar] [CrossRef] [PubMed]
- Nazir, R.; Zaffar, M.R.; Amin, I. Bacterial biofilms: The remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes. In Freshwater Microbiology: Perspectives of Bacterial Dynamics in Lake Ecosystems; Elsevier: Amsterdam, The Netherlands, 2019; pp. 307–340. ISBN 9780128174951. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Idrees, M.; Mohammad, A.R.; Karodia, N.; Rahman, A. Multimodal Role of Amino Acids in Microbial Control and Drug Development. Antibiotics 2020, 9, 330. [Google Scholar] [CrossRef]
- Reffuveille, F.; Josse, J.; Vallé, Q.; Gangloff, C.M.; Gangloff, S.C. Staphylococcus aureus Biofilms and their Impact on the Medical Field. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; InTech: Rijeka, Croatia, 2017; Chapter 11; p. 187. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.T.; Nguyen, T.H.; Otto, M. The staphylococcal exopolysaccharide PIA–Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 2020, 18, 3324–3334. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, A.; Bhattacharyya, S.; Kundu, A.; Dutta, D.; Das, A.K. Macroscopic amyloid fiber formation by staphylococcal biofilm associated SuhB protein. Biophys. Chem. 2016, 217, 32–41. [Google Scholar] [CrossRef]
- Lacey, K.A.; Leech, J.M.; Lalor, S.J.; McCormack, N.; Geoghegan, J.A.; McLoughlin, R.M. The Staphylococcus aureus cell wall-anchored protein clumping factor A is an important T cell antigen. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [Green Version]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Lin, S.; Soteyome, T.; Peters, B.M.; Li, Y.; Chen, H.; Su, J.; Li, L.; Li, B.; Xu, Z.; et al. Biofilm Formation of Staphylococcus aureus under Food Heat Processing Conditions: First Report on CML Production within Biofilm. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Extracellular Polymeric Substances in Microbial Biofilms. In Microbial Glycobiology; Elsevier Inc.: Amsterdam, The Netherlands, 2010; pp. 733–758. ISBN 9780123745460. [Google Scholar]
- Algburi, A.; Comito, N.; Kashtanov, D.; Dicks, L.M.T.; Chikindas, M.L. Control of biofilm formation: Antibiotics and beyond. Appl. Environ. Microbiol. 2017, 83, e02508-16. [Google Scholar] [CrossRef] [Green Version]
- Guangyin, Z.; Youcai, Z. Harvest of Bioenergy From Sewage Sludge by Anaerobic Digestion. In Pollution Control and Resource Recovery; Elsevier: Amsterdam, The Netherlands, 2017; pp. 181–273. [Google Scholar]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef] [Green Version]
- Hamadi, F.; Latrache, H.; Mabrrouki, M.; Elghmari, A.; Outzourhit, A.; Ellouali, M.; Chtaini, A. Effect of pH on distribution and adhesion of Staphylococcus aureus to glass. J. Adhes. Sci. Technol. 2005, 19, 73–85. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maikranz, E.; Spengler, C.; Thewes, N.; Thewes, A.; Nolle, F.; Jung, P.; Bischoff, M.; Santen, L.; Jacobs, K. Different binding mechanisms of: Staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale 2020, 12, 19267–19275. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [Green Version]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Archer, N.K.; Mazaitis, M.J.; William Costerton, J.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [Green Version]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, K. The genomics and proteomics of biofilm formation. Genome Biol. 2003, 4, 219. [Google Scholar] [CrossRef] [Green Version]
- Nourbakhsh, F.; Namvar, A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg. Infect. Control 2016, 11, Doc07. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. Microbiologyopen 2020, 9, e00946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmi, K.; Qrei, W.; Abdeen, Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genom. 2019, 20, 571–578. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A Novel Staphylococcus aureus Biofilm Phenotype Mediated by the Fibronectin-Binding Proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835. [Google Scholar] [CrossRef] [Green Version]
- Sinha, B.; Francois, P.; Que, Y.-A.; Hussain, M.; Heilmann, C.; Moreillon, P.; Lew, D.; Krause, K.-H.; Peters, G.; Herrmann, M. Heterologously Expressed Staphylococcus aureusFibronectin-Binding Proteins Are Sufficient for Invasion of Host Cells. Infect. Immun. 2000, 68, 6871. [Google Scholar] [CrossRef] [Green Version]
- Wolz, C.; Goerke, C.; Landmann, R.; Zimmerli, W.; Fluckiger, U. Transcription of Clumping Factor A in Attached and Unattached Staphylococcus aureus In Vitro and during Device-Related Infection. Infect. Immun. 2002, 70, 2758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman-Bausier, P.; Labate, C.; Towell, A.M.; Derclaye, S.; Geoghegan, J.A.; Dufrêne, Y.F. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc. Natl. Acad. Sci. USA 2018, 115, 5564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabat, A.; Melles, D.C.; Martirosian, G.; Grundmann, H.; Van Belkum, A.; Hryniewicz, W. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J. Clin. Microbiol. 2006, 44, 1135–1138. [Google Scholar] [CrossRef] [Green Version]
- Kot, B.; Sytykiewicz, H.; Sprawka, I. Expression of the Biofilm-Associated Genes in Methicillin-Resistant Staphylococcus aureus in Biofilm and Planktonic Conditions. Int. J. Mol. Sci. 2018, 19, 3487. [Google Scholar] [CrossRef] [Green Version]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef] [Green Version]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Saggu, S.K.; Jha, G.; Mishra, P.C. Enzymatic Degradation of Biofilm by Metalloprotease From Microbacterium sp. SKS10. Front. Bioeng. Biotechnol. 2019, 7, 192. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Zhou, R.; Wang, H.; Zeng, Z. Accessory gene regulator in Staphylococcus biofilm formation and infection. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2008, 33, 1066–1070. [Google Scholar]
- Grundstad, M.L.; Parlet, C.P.; Kwiecinski, J.M.; Kavanaugh, J.S.; Crosby, H.A.; Cho, Y.-S.; Heilmann, K.; Diekema, D.J.; Horswill, A.R. Quorum Sensing, Virulence, and Antibiotic Resistance of USA100 Methicillin-Resistant Staphylococcus aureus Isolates. mSphere 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Lepp, D.; Gohari, I.M.; Wu, T.; Zhou, H.; Yin, X.; Yu, H.; Prescott, J.F.; Nie, S.P.; Xie, M.Y.; et al. The Agr-like quorum sensing system is required for pathogenesis of necrotic enteritis caused by Clostridium perfringens in poultry. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [Green Version]
- Rajasree, K.; Fasim, A.; Gopal, B. Conformational features of the Staphylococcus aureus AgrA-promoter interactions rationalize quorum-sensing triggered gene expression. Biochem. Biophys. Rep. 2016, 6, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oogai, Y.; Matsuo, M.; Hashimoto, M.; Kato, F.; Sugai, M.; Komatsuzawa, H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 2011, 77, 8097–8105. [Google Scholar] [CrossRef] [Green Version]
- Gordon, R.J.; Lowy, F.D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008, 46, S350–S359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Kolar, S.L.; Antonio Ibarra, J.; Rivera, F.E.; Mootz, J.M.; Davenport, J.E.; Stevens, S.M.; Horswill, A.R.; Shaw, L.N. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2013, 2, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Thammavongsa, V.; Schneewind, O.; Missiakas, D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr. Opin. Microbiol. 2012, 15, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; Van Kessel, K.P.M.; Van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [Green Version]
- WHO. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis; WHO: Geneva, Switzerland, 2019; Volume 29, pp. 2019–2021. [Google Scholar]
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Thuy, N.D.T.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control 2018, 7, 98. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M. Role of biofilms in antimicrobial resistance. ASAIO J. 2000, 46, S47–S52. [Google Scholar] [CrossRef]
- Rizzato, C.; Torres, J.; Kasamatsu, E.; Camorlinga-Ponce, M.; Bravo, M.M.; Canzian, F.; Kato, I. Potential role of biofilm formation in the development of digestive tract cancer with special reference to helicobacter pylori infection. Front. Microbiol. 2019, 10, 846. [Google Scholar] [CrossRef] [PubMed]
- Fadeeva, E.; Schlie-Wolter, S.; Chichkov, B.N.; Paasche, G.; Lenarz, T. Structuring of biomaterial surfaces with ultrashort pulsed laser radiation. In Laser Surface Modification of Biomaterials: Techniques and Applications; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 145–172. ISBN 9780081009420. [Google Scholar]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of multidrug-resistant Staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in Shanghai. Food Control 2020, 112, 107106. [Google Scholar] [CrossRef]
- Staph Can Lurk Deep within Nose, Study Finds|News Center|Stanford Medicine. Available online: https://med.stanford.edu/news/all-news/2013/12/staph-can-lurk-deep-within-nose-study-finds.html (accessed on 8 July 2021).
- Sakr, A.; Brégeon, F.; Mège, J.L.; Rolain, J.M.; Blin, O. Staphylococcus aureus nasal colonization: An update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 2018, 9, 2419. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.V.; Fowler, V.G., Jr.; Skov, R.; Bruun, N.E. Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA. Future Microbiol. 2011, 6, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Amyes, S.G. Treatment of staphylococcal infection. BMJ 2005, 330, 976–977. [Google Scholar] [CrossRef]
- Min, S.H.; Kang, M.H.; Sur, J.H.; Park, H.M. Staphylococcus pseudintermedius infection associated with nodular skin lesions and systemic inflammatory response syndrome in a dog. Can. Vet. J. 2014, 55, 480–483. [Google Scholar] [PubMed]
- Sonesson, A.; Przybyszewska, K.; Eriksson, S.; Mörgelin, M.; Kjellström, S.; Davies, J.; Potempa, J.; Schmidtchen, A. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci. Rep. 2017, 7, 8682–8689. [Google Scholar] [CrossRef]
- Verma, J.; Bag, S.; Saha, B.; Kumar, P.; Ghosh, T.S.; Dayal, M.; Senapati, T.; Mehra, S.; Dey, P.; Desigamani, A.; et al. Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2019, 116, 6226–6231. [Google Scholar] [CrossRef] [Green Version]
- Manzur, A.; Gavalda, L.; de Gopegui, E.R.; Mariscal, D.; Dominguez, M.A.; Perez, J.L.; Segura, F.; Pujol, M. Spanish Network for Research in, I.D. Prevalence of methicillin-resistant Staphylococcus aureus and factors associated with colonization among residents in community long-term-care facilities in Spain. Clin. Microbiol. Infect. 2008, 14, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Marques, S.C.; Rezende, J.D.G.O.S.; Alves, L.A.D.F.; Silva, B.C.; Alves, E.; de Abreu, L.R.; Piccoli, R.H. Formation of biofilms by Staphylococcus aureus on stainless steel and glass surfaces and its resistance to some selected chemical sanitizers. Braz. J. Microbiol. 2007, 38, 538–543. [Google Scholar] [CrossRef] [Green Version]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Chmielewski, R.A.N.; Frank, J.F. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, 1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Characklis, W.G.; Trulear, M.G.; Bryers, J.D.; Zelver, N. Dynamics of biofilm processes: Methods. Water Res. 1982, 16, 1207–1216. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Kırmusaoğlu, S. The Methods for Detection of Biofilm and Screening Antibiofilm Activity of Agents. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: London, UK, 2019. [Google Scholar]
- Lawrence, J.R.; Swerhone, G.D.W.; Leppard, G.G.; Araki, T.; Zhang, X.; West, M.M.; Hitchcock, A.P. Scanning transmission X-ray, laser scanning, and transmission electron microscopy mapping of the exopolymeric matrix of microbial biofilms. Appl. Environ. Microbiol. 2003, 69, 5543–5554. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Biswas, N.; Datta, A.; Dey, R.; Maiti, P. Atomic force microscopy in biofilm study. Microscopy 2014, 63, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Biswas, N.; Datta, A.; Maiti, P.K. Periodicities in the roughness and biofilm growth on glass substrate with etching time: Hydrofluoric acid etchant. PLoS ONE 2019, 14, e0214192. [Google Scholar] [CrossRef]
- Jhass, A.K.; Johnston, D.A.; Gulati, A.; Anand, R.; Stoodley, P.; Sharma, S. A scanning electron microscope characterisation of biofilm on failed craniofacial osteosynthesis miniplates. J. Cranio-Maxillofac. Surg. 2014, 42, e372–e378. [Google Scholar] [CrossRef] [PubMed]
- Warwick, T.; Franck, K.; Kortright, J.B.; Meigs, G.; Moronne, M.; Myneni, S.; Rotenberg, E.; Seal, S.; Steele, W.F.; Ade, H.; et al. A scanning transmission x-ray microscope for materials science spectromicroscopy at the advanced light source. Rev. Sci. Instrum. 1998. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Balhorn, R.; Mazrimas, J.; Kirz, J. Mapping and measuring DNA to protein ratios in mammalian sperm head by XANES imaging. J. Struct. Biol. 1996, 116, 335–344. [Google Scholar] [CrossRef]
- Loo, B.W.; Sauerwald, I.M.; Hitchcock, A.P.; Rothman, S.S. A new sample preparation method for biological soft X-ray microscopy: Nitrogen-based contrast and radiation tolerance properties of glycol methacrylate-embedded and sectioned tissue. J. Microsc. 2001, 204, 69–86. [Google Scholar] [CrossRef] [Green Version]
- Neu, T.R.; Swerhone, G.D.W.; Lawrence, J.R. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neu, T.R.; Lawrence, J.R. In situ characterization of extracellular polymeric substances (EPS) in biofilm systems. In Microbial Extracellular Polymeric Substances; Springer: Heidelberg/Berlin, Germany, 1999; pp. 21–47. [Google Scholar]
- Van Gijlswijk, R.P.M.; Zijlmans, H.J.M.A.A.; Wiegant, J.; Bobrow, M.N.; Erickson, T.J.; Adler, K.E.; Tanke, H.J.; Raap, A.K. Fluorochrome-labeled tyramides: Use in immunocytochemistry and fluorescence in situ hybridization. J. Histochem. Cytochem. 1997, 45, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, A.; Bishop, P.L.; Okabe, S.; Lee, S.G.; Kim, I.S. Effect of dissolved oxygen concentration on the biofilm and in situ analysis by fluorescence in situ hybridization (FISH) and microelectrodes. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2003, 47, 49–57. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rattan, A. Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods. Indian J. Med. Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar] [CrossRef]
- Sanchez, C.J.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoramrooz, S.S.; Mansouri, F.; Marashifard, M.; Malek Hosseini, S.A.A.; Akbarian Chenarestane-Olia, F.; Ganavehei, B.; Gharibpour, F.; Shahbazi, A.; Mirzaii, M.; Darban-Sarokhalil, D. Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 2016. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.; Martinko, J.; Bender, K.; Buckley, D.; Stahl, D. Brock Biology of Microorganisms; Pearson Education Ltd.: Edinburgh Gate, UK, 2015. [Google Scholar]
- Park, A.J.; Murphy, K.; Krieger, J.R.; Brewer, D.; Taylor, P.; Habash, M.; Khursigara, C.M. A temporal examination of the planktonic and biofilm proteome of whole cell pseudomonas aeruginosa pao1 using quantitative mass spectrometry. Mol. Cell. Proteom. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shunmugaperumal, T. Biofilm Eradication and Prevention: A Pharmaceutical Approach to Medical Device Infections; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; ISBN 9780470479964. [Google Scholar]
- Dufrêne, Y.F. Atomic force microscopy in microbiology: New structural and functional insights into the microbial cell surface. MBio 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Dufrêne, Y.F. Refining our perception of bacterial surfaces with the atomic force microscope. J. Bacteriol. 2004, 186, 3283–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, G.; Müller, T.; Meyer, R.L. Single-cell force spectroscopy of bacteria enabled by naturally derived proteins. Langmuir 2014, 30, 4019–4025. [Google Scholar] [CrossRef] [PubMed]
- Relucenti, M.; Familiari, G.; Donfrancesco, O.; Taurino, M.; Li, X.; Chen, R.; Artini, M.; Papa, R.; Selan, L. Microscopy methods for biofilm imaging: Focus on sem and VP-SEM pros and cons. Biology 2021, 10, 51. [Google Scholar] [CrossRef]
- Rutering, J.; Ilmer, M.; Recio, A.; Coleman, M.; Vykoukal, J.; Alt, E.; Orleans, N. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-review Christina. Nat. Rev Drug Discov. 2016, 5, 1–8. [Google Scholar]
- Wille, G.; Hellal, J.; Ollivier, P.; Richard, A.; Burel, A.; Jolly, L.; Crampon, M.; Michel, C. Cryo-Scanning Electron Microscopy (SEM) and Scanning Transmission Electron Microscopy (STEM)-in-SEM for Bio- and Organo-Mineral Interface Characterization in the Environment. Microsc. Microanal. 2017, 23, 1159–1172. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, O.P.; ka, P. Scanning Electron Microscope: Advantages and Disadvantages in Imaging Components. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1877–1882. [Google Scholar] [CrossRef]
- Magana, M.; Sereti, C.; Ioannidis, A.; Mitchell, C.A.; Ball, A.R.; Magiorkinis, E.; Chatzipanagiotou, S.; Hamblin, M.R.; Hadjifrangiskou, M.; Tegos, G.P. Options and limitations in clinical investigation of bacterial biofilms. Clin. Microbiol. Rev. 2018, 31, e00084-16. [Google Scholar] [CrossRef] [Green Version]
- Gomes, L.C.; Moreira, J.M.R.; Simões, M.; Melo, L.F.; Mergulhão, F.J. Biofilm Localization in the Vertical Wall of Shaking 96-Well Plates. Scientifica 2014, 2014, 1–6. [Google Scholar] [CrossRef]
- Hassan, A.; Usman, J.; Kaleem, F.; Omair, M.; Khalid, A.; Iqbal, M. 05/03/2011 Evaluation of Different Detection Methods of Biofilm Formation in the Clinical Isolates; Elsevier BV: Amsterdam, The Netherlands, 2011; Volume 15, pp. 305–311. [Google Scholar]
- Kaiser, T.D.L.; Pereira, E.M.; dos Santos, K.R.N.; Maciel, E.L.N.; Schuenck, R.P.; Nunes, A.P.F. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagn. Microbiol. Infect. Dis. 2013, 75, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latka, A.; Drulis-Kawa, Z. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zegaer, B.H.; Ioannidis, A.; Babis, G.C.; Ioannidou, V.; Kossyvakis, A.; Bersimis, S.; Papaparaskevas, J.; Petinaki, E.; Pliatsika, P.; Chatzipanagiotou, S. Detection of Bacteria Bearing Resistant Biofilm Forms, by Using the Universal and Specific PCR is Still Unhelpful in the Diagnosis of Periprosthetic Joint Infections. Front. Med. 2014, 1, 30. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Chen, Y.-P.P.; Qiu, J.-H.H.; Dai, Y.-Z.Z.; Feng, B. Imaging the Microprocesses in Biofilm Matrices; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 37, pp. 214–226. [Google Scholar]
- Agostinho, A.; James, G.; Wazni, O.; Citron, M.; Wilkoff, B.D. Inhibition of Staphylococcus aureus Biofilms by a Novel Antibacterial Envelope for Use with Implantable Cardiac Devices. Clin. Transl. Sci. 2009, 2, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev. Anti. Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef] [Green Version]
- Holstege, C.P. Rifampin. In Encyclopedia of Toxicology: Third Edition; Elsevier: Amsterdam, The Netherlands, 2014; pp. 134–136. ISBN 9780123864543. [Google Scholar]
- Rees, C.E.D.; Green, L.H.; Goldman, E.; Loessner, M.J. Practical Handbook of Microbiology; Goldman, E., Green, L.H., Eds.; CRC Press: Boca Raton, FL, USA, 2015; ISBN 9780429168932. [Google Scholar]
- Wells, C.M.; Beenken, K.E.; Smeltzer, M.S.; Courtney, H.S.; Jennings, J.A.; Haggard, W.O. Ciprofloxacin and Rifampin Dual Antibiotic-Loaded Biopolymer Chitosan Sponge for Bacterial Inhibition. Mil. Med. 2018, 183, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vancomycin—StatPearls—NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK459263/ (accessed on 23 May 2021).
- Salem, A.H.; Elkhatib, W.F.; Noreddin, A.M. Pharmacodynamic assessment of vancomycin-rifampicin combination against methicillin resistant Staphylococcus aureus biofilm: A parametric response surface analysis. J. Pharm. Pharmacol. 2011, 63, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ramezanpour, M.; Hayes, A.J.; Liu, S.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Sub-inhibitory clindamycin and azithromycin reduce S. aureus exoprotein induced toxicity, inflammation, barrier disruption and invasion. J. Clin. Med. 2019, 8, 1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.J.; Akers, K.S.; Romano, D.R.; Woodbury, R.L.; Hardy, S.K.; Murray, C.K.; Wenke, J.C. D-amino acids enhance the activity of antimicrobials against biofilms of clinical wound isolates of Staphylococcus aureus and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 4353–4361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warraich, A.A.; Mohammed, A.R.; Perrie, Y.; Hussain, M.; Gibson, H.; Rahman, A. Evaluation of anti-biofilm activity of acidic amino acids and synergy with ciprofloxacin on Staphylococcus aureus biofilms. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Hochbaum, A.I.; Kolodkin-Gal, I.; Foulston, L.; Kolter, R.; Aizenberg, J.; Losick, R. Inhibitory Effects of d-Amino Acids on Staphylococcus aureus Biofilm Development. J. Bacteriol. 2011, 193, 5616. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.; Desai, S.; Poonacha, N.; Vipra, A.; Sharma, U. Antibiofilm activity and synergistic inhibition of Staphylococcus aureus biofilms by bactericidal protein P128 in combination with antibiotics. Antimicrob. Agents Chemother. 2016, 60, 7280–7289. [Google Scholar] [CrossRef] [Green Version]
- Hickey, E.E.; Wong, H.S.; Khazandi, M.; Ogunniyi, A.D.; Petrovski, K.R.; Garg, S.; Page, S.W.; O’Handley, R.; Trott, D.J. Repurposing Ionophores as novel antimicrobial agents for the treatment of bovine mastitis caused by Gram-positive pathogens. J. Vet. Pharmacol. Ther. 2018, 41, 746–754. [Google Scholar] [CrossRef]
- Bazargani, M.M.; Rohloff, J. Antibiofilm activity of essential oils and plant extracts against Staphylococcus aureus and Escherichia coli biofilms. Food Control 2016, 61, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Monteiro-Neto, V.; de Souza, C.D.; Gonzaga, L.F.; da Silveira, B.C.; Sousa, N.C.F.; Pontes, J.P.; Santos, D.M.; Martins, W.C.; Pessoa, J.F.V.; Carvalho Júnior, A.R.; et al. Cuminaldehyde potentiates the antimicrobial actions of ciprofloxacin against Staphylococcus aureus and Escherichia coli. PLoS ONE 2020, 15. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; d’Acierno, A.; Coppola, R.; Jesus Ayala-Zavala, F.; Gomez da Cruz, A.; De Feo, V. Essential Oils and Microbial Communication. In Essential Oils–Oils of Nature; IntechOpen: Lobdon, UK, 2020. [Google Scholar]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Agents that inhibit bacterial biofilm formation. Future Med. Chem. 2015, 7, 647–671. [Google Scholar] [CrossRef]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef] [PubMed]
- Fontecha-Umaña, F.; Ríos-Castillo, A.G.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Antimicrobial activity and prevention of bacterial biofilm formation of silver and zinc oxide nanoparticle-containing polyester surfaces at various concentrations for use. Foods 2020, 9, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Al-Zaban, M.I. Novel Biogenic Silver Nanoparticle-Induced Reactive Oxygen Species Inhibit the Biofilm Formation and Virulence Activities of Methicillin-Resistant Staphylococcus aureus (MRSA) Strain. Front. Bioeng. Biotechnol. 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).