Spatiotemporal Patterns of Cholera Hospitalization in Vellore, India
Abstract
1. Introduction
2. Materials and Methods
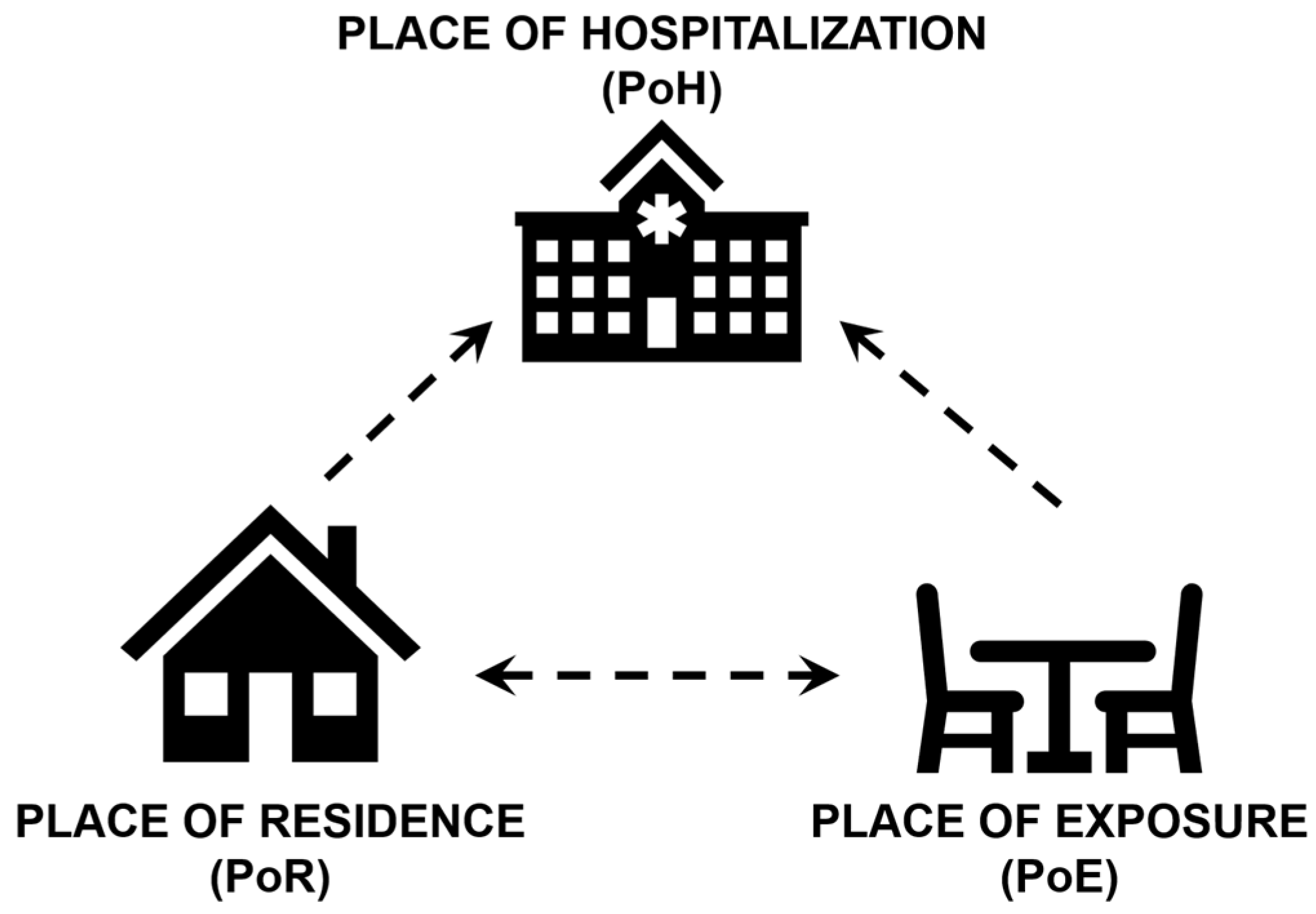
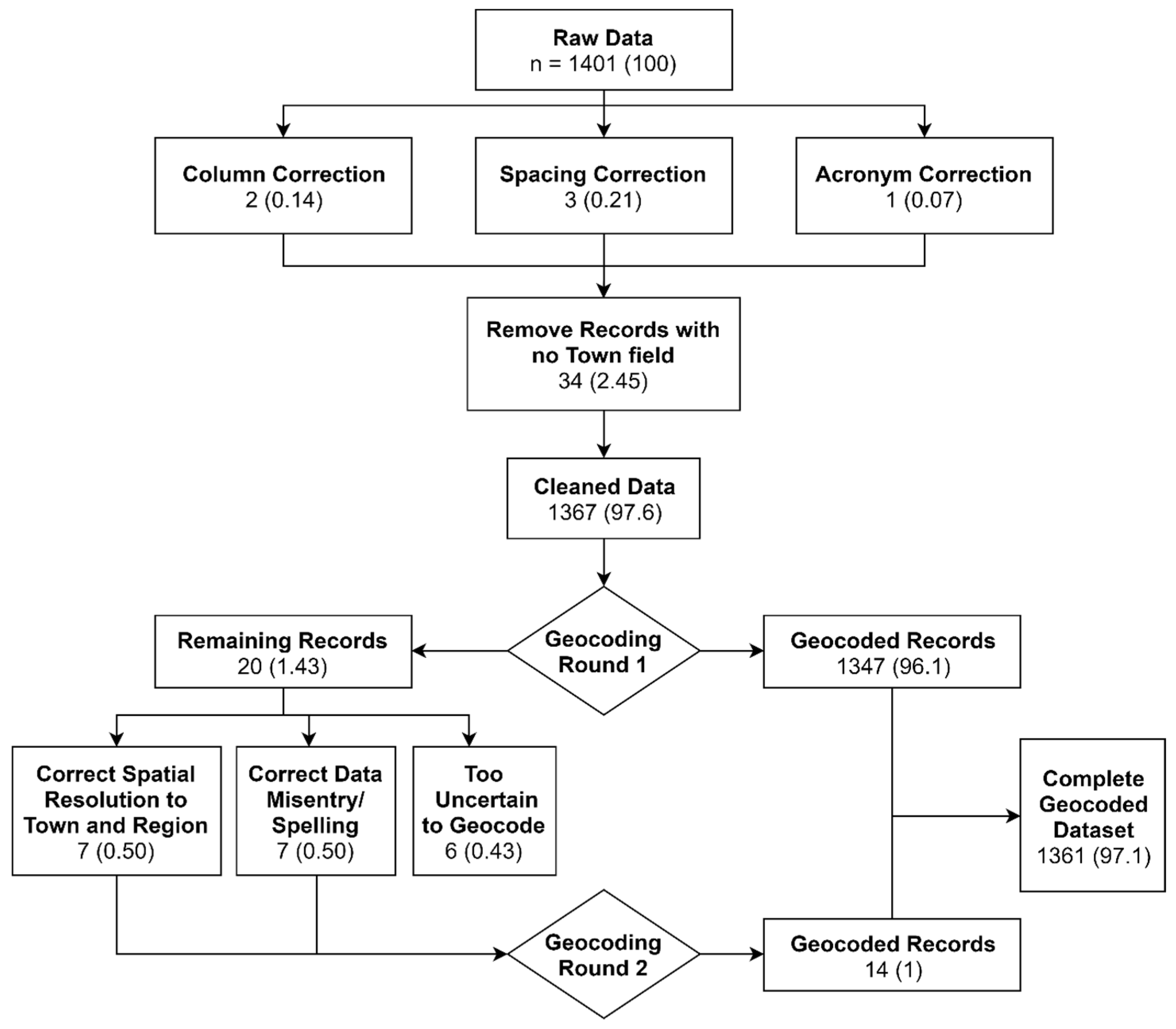
2.1. Data Abstraction and Geocoding
2.2. Examination of Point Patterns
2.3. Point Process Modeling
2.4. Temporal Modeling
3. Results
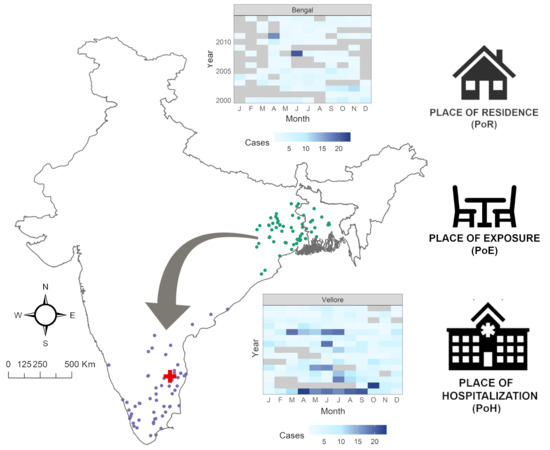
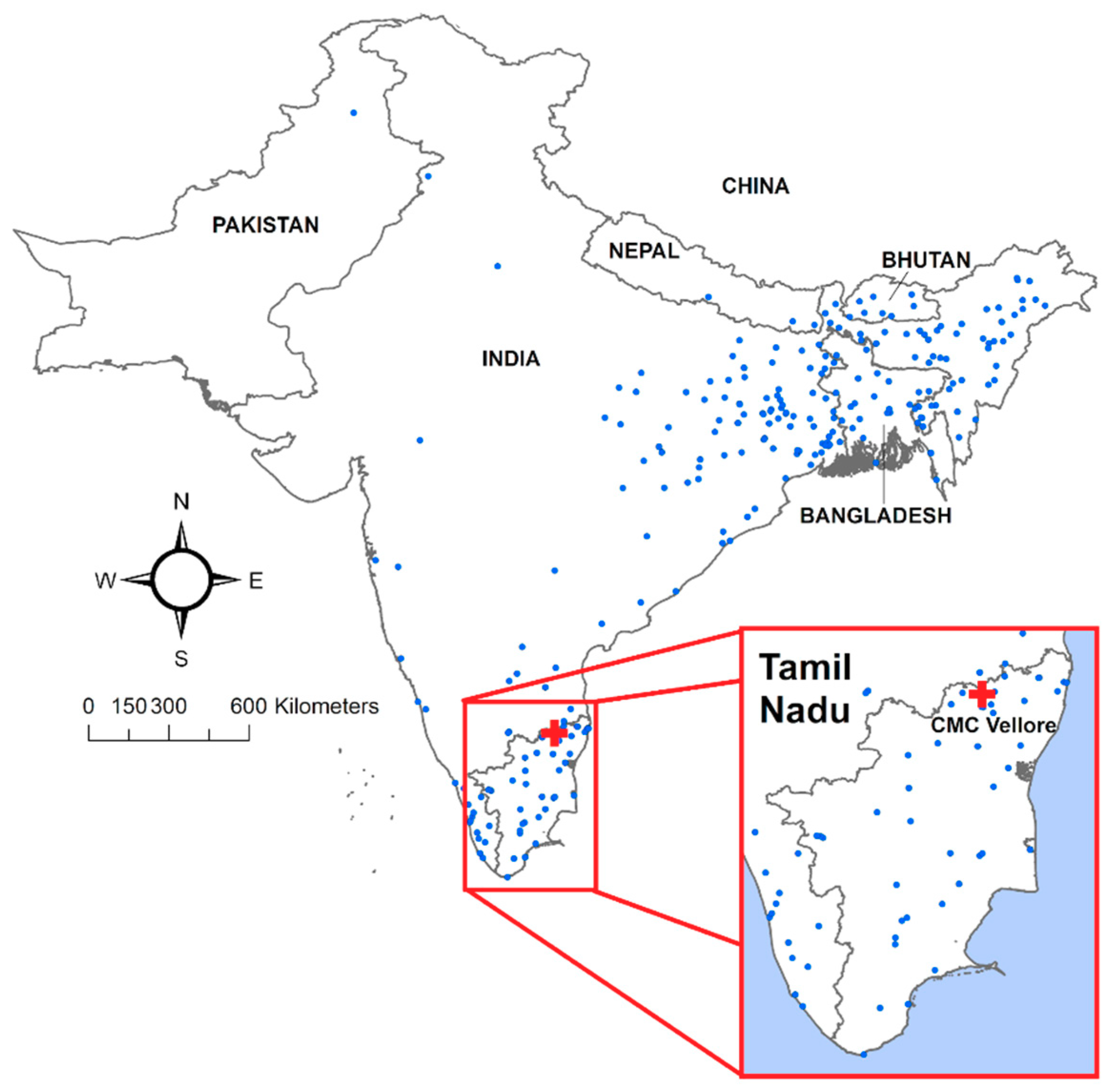
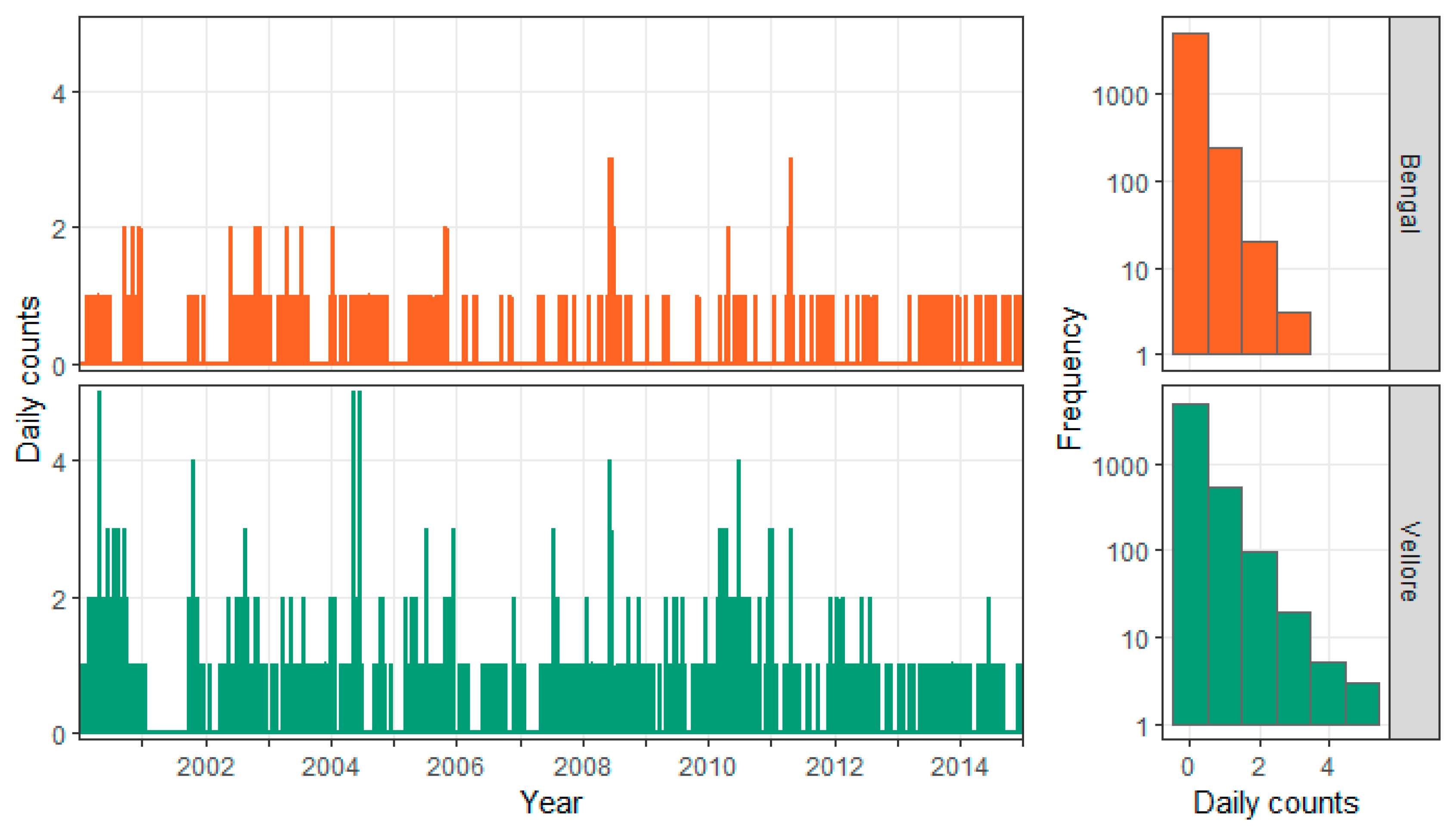
3.1. Exploratory Analysis
3.2. Point Process Intensity
3.3. Point Process Modeling
3.4. Temporal Effects and Patient Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Short, D.; Frischer, M. Supporting the evidence-based treatment of individual drug users: The case for ‘patient profile decision analysis’. Int. J. Drug Policy 2005, 16, 143–147. [Google Scholar] [CrossRef]
- Raghupathi, W.; Raghupathi, V. Big data analytics in healthcare: Promise and potential. Health Inf. Sci. Syst 2014, 2. [Google Scholar] [CrossRef]
- O’Sullivan, D.; McLoughlin, E.; Bertolotto, M.; Wilson, D.C. Mobile case-based decision support for intelligent patient knowledge management. Health Inform. J 2007, 13, 179–193. [Google Scholar] [CrossRef]
- Gil-Prieto, R.; Walter, S.; Alvar, J.; de Miguel, A.G. Epidemiology of leishmaniasis in Spain based on hospitalization records (1997–2008). Am. J. Trop. Med. Hyg. 2011, 85, 820–825. [Google Scholar] [CrossRef]
- Mor, S.M.; DeMaria, A., Jr.; Naumova, E.N. Hospitalization Records as a Tool for Evaluating Performance of Food- and Water-Borne Disease Surveillance Systems: A Massachusetts Case Study. PLoS ONE 2014, 9, e93744. [Google Scholar] [CrossRef]
- Birkhead, G.S.; Klompas, M.; Shah, N.R. Uses of electronic health records for public health surveillance to advance public health. Annu. Rev. Public Health 2015, 36, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Klompas, M.; Cocoros, N.M.; Menchaca, J.T.; Erani, D.; Hafer, E.; Herrick, B.; Josephson, M.; Lee, M.; Payne Weiss, M.D.; Zambarano, B.; et al. State and Local Chronic Disease Surveillance Using Electronic Health Record Systems. Am. J. Public Health 2017, 107, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Chui, K.K.C.; Steven, A.; Naumova, E.N. Snowbirds and infection–new phenomena in pneumonia and influenza hospitalizations from winter migration of older adults: A spatiotemporal analysis. BMC Public Health 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Nair, G.B.; Ramamurthy, T.; Bhattacharya, S.K.; Mukhopadhyay, A.K.; Garg, S.; Bhattacharya, M.K.; Takeda, T.; Shimada, T.; Takeda, Y.; Deb, B.C. The Spread of Vibrio cholerae O139 in India. J. Infect. Dis 1994, 169, 1029–1034. [Google Scholar] [CrossRef]
- Albert, M.J.; Ansaruzzaman, M.; Bardhan, P.K.; Faruque, A.S.G.; Faruque, S.M.; Islam, M.S.; Mahalanabis, D.; Sack, R.B.; Salam, M.A.; Siddique, A.K.; et al. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae 0139 synonym Bengal. Lancet 1993, 342, 387–390. [Google Scholar]
- Nair, G.B.; Shimada, T.; Kurazono, H.; Okuda, J.; Pal, A.; Karasawa, T.; Mihara, T.; Uesaka, Y.; Shirai, H.; Garg, S. Characterization of phenotypic, serological, and toxigenic traits of Vibrio cholerae O139 bengal. J. Clin. Microbiol 1994, 32, 2775–2779. [Google Scholar] [PubMed]
- Jesudason, M.V.J.; Jacob, T. The Vellore Vibrio Watch. Lancet 1996, 347, 1493–1494. [Google Scholar] [CrossRef]
- Faruque, S.M.; Sack, D.A.; Sack, R.B.; Colwell, R.R.; Takeda, Y.; Nair, G.B. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA 2003, 100, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Bharati, K.; Bhattacharya, S.K. Cholera Outbreaks in South-East Asia. In Cholera Outbreaks; Nair, G.B., Takeda, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 87–116. [Google Scholar]
- Ministry of Tourism. India Tourism Statistics at a Glance. Available online: http://tourism.gov.in/sites/default/files/Other/India%20Tourism%20Statistics%20at%20a%20Glance%202014New.pdf (accessed on 6 August 2017).
- Nielsen India Pvt. Ltd. Tourism Survey for Tamil Nadu (January–December 2014) Final Report. Available online: http://tourism.gov.in/sites/default/files/Other/Executive%20Summary%20%20State%20Report%20-%20Tamilnadu.pdf (accessed on 8 August 2017).
- Rajan, S.I.; D’Sami, B.; Raj, S.A. Non-Resident Tamils and Remittances: Results from Tamil Nadu Migration Survey 2015; State Planning Commission, Government of Tamil Nadu: Chennai, India, 2015. [Google Scholar]
- Waldor, M.K.; Mekalanos, J.J. Emergence of a New Cholera Pandemic: Molecular Analysis of Virulence Determinants in Vibrio cholerae O139 and Development of a Live Vaccine Prototype. J. Infect. Dis 1994, 170, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Azman, A.S.; Rudolph, K.E.; Cummings, D.A.; Lessler, J. The incubation period of cholera: A systematic review. J. Infect 2013, 66, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Bivand, R.; Pebesma, E.J.; Gomez-Rubio, V. Applied Spatial Data Analysis with R, 2nd ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Cruz, M.S.; Alarcon-Falconi, T.M.; Hartwick, M.A.; Venkat, A.; Ehrlich, H.Y.; Anandan, S.; Ward, H.D.; Veeraraghavan, B.; Naumova, E.N. From hospitalization records to surveillance: The use of local patient profiles to characterize cholera in Vellore, India. PLoS ONE 2017, 12, e0182642. [Google Scholar] [CrossRef]
- Kahle, D.; Wickham, H. ggmap: Spatial Visualization with ggplot2. R J 2013, 5, 144–161. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Version 3.5.1.; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- RStudio Team. RStudio: Integrated Development Environment for R; Version 1.1.463.; RStudio, Inc.: Boston, MA, USA, 2018. [Google Scholar]
- Becker, R.; Wilks, A.R.; Brownrigg, R.; Minka, T.P.; Deckmyn, A. maps: Draw Geographical Maps. Version 3.2.0. Available online: https://cran.r-project.org/web/packages/maps/index.html (accessed on 2 November 2019).
- Bivand, R.; Lewin-Koh, N. maptools: Tools for Reading and Handling Spatial Objects. Version 0.9-2. 2017. Available online: https://cran.r-project.org/web/packages/maptools/index.html (accessed on 2 November 2019).
- Bivand, R.; Rundel, C. rgeos: Interface to Geometry Engine-Open Source (GEOS). Version 0.3-23. 2017. Available online: https://cran.r-project.org/web/packages/rgeos/index.html (accessed on 2 November 2019).
- Pebesma, E.; Bivand, R.S. sp: Classes and Methods for Spatial Data in R. Version 1.3-1. 2018. Available online: https://cran.r-project.org/web/packages/sp/index.html (accessed on 2 November 2019).
- Baddeley, A.; Rubak, E.; Turner, R. Spatial Point Patterns: Methodology and Applications with R; Chapman and Hall/CRC Press: London, UK, 2015. [Google Scholar]
- Diggle, P. Statistical Analysis of Spatial and Spatio-Temporal Point Patterns; CRC Press: London, UK, 2013. [Google Scholar]
- Loader, C. Local Regression and Likelihood; Springer: New York, USA, 2006. [Google Scholar]
- Diggle, P. A kernel method for smoothing point process data. J. R. Stat. Soc. Ser. C 1985, 34, 138–147. [Google Scholar] [CrossRef]
- National Informatics Centre. National Portal of India. Available online: https://india.gov.in/calendar (accessed on 24 January 2016).
- National Informatics Centre. Public Holidays for the year 2016: Tamil Nadu Government Portal. Available online: http://www.tn.gov.in/holiday/2016 (accessed on 24 January 2016).
- Naumova, E.N.; MacNeill, I.B. Seasonality assessment for biosurveillance systems. In Advances in Statistical Methods for the Health Sciences; Auget, J.L., Balakrishnan, N., Mesbah, M., Molenberghs, G., Eds.; Birkhäuser: Boston, MA, USA, 2007; pp. 437–450. [Google Scholar]
- Stashevsky, P.S.; Yakovina, I.N.; Alarcon Falconi, T.M.; Naumova, E.N. Agglomerative Clustering of Enteric Infections and Weather Parameters to Identify Seasonal Outbreaks in Cold Climates. Int. J. Environ. Res. Public Health 2019, 16, 2083. [Google Scholar] [CrossRef]
- Office of the Registrar General and Census Commissioner. Table D-01: Population Classified by Place of Birth and Sex; Ministry of Home Affairs, Ed.; Government of India: New Delhi, India, 2011.
- Colombara, D.V.; Cowgill, K.D.; Faruque, A.S. Risk factors for severe cholera among children under five in rural and urban Bangladesh, 2000–2008: A hospital-based surveillance study. PLoS ONE 2013, 8, e54395. [Google Scholar] [CrossRef]
- Root, E.D.; Rodd, J.; Yunus, M.; Emch, M. The role of socioeconomic status in longitudinal trends of cholera in Matlab, Bangladesh, 1993–2007. PLoS Negl. Trop Dis 2013, 7, e1997. [Google Scholar] [CrossRef] [PubMed]
- Nazia, N.; Ali, M.; Jakariya, M.; Nahar, Q.; Yunus, M.; Emch, M. Spatial and population drivers of persistent cholera transmission in rural Bangladesh: Implications for vaccine and intervention targeting. Spat Spatio-temporal Epidemiol 2018, 24, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, B.; Salje, H.; Sturm-Ramirez, K.; Azziz-Baumgartner, E.; Homaira, N.; Ahmed, M.; Iuliano, A.D.; Paul, R.C.; Rahman, M.; Hossain, M.J.; et al. Evaluating Hospital-Based Surveillance for Outbreak Detection in Bangladesh: Analysis of Healthcare Utilization Data. PLoS Med 2017, 14, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Nair, G.B.; Takeda, Y. Cholera Outbreaks; Springer: Berlin/Heidelberg, Germany, 2014; Volume 379. [Google Scholar]
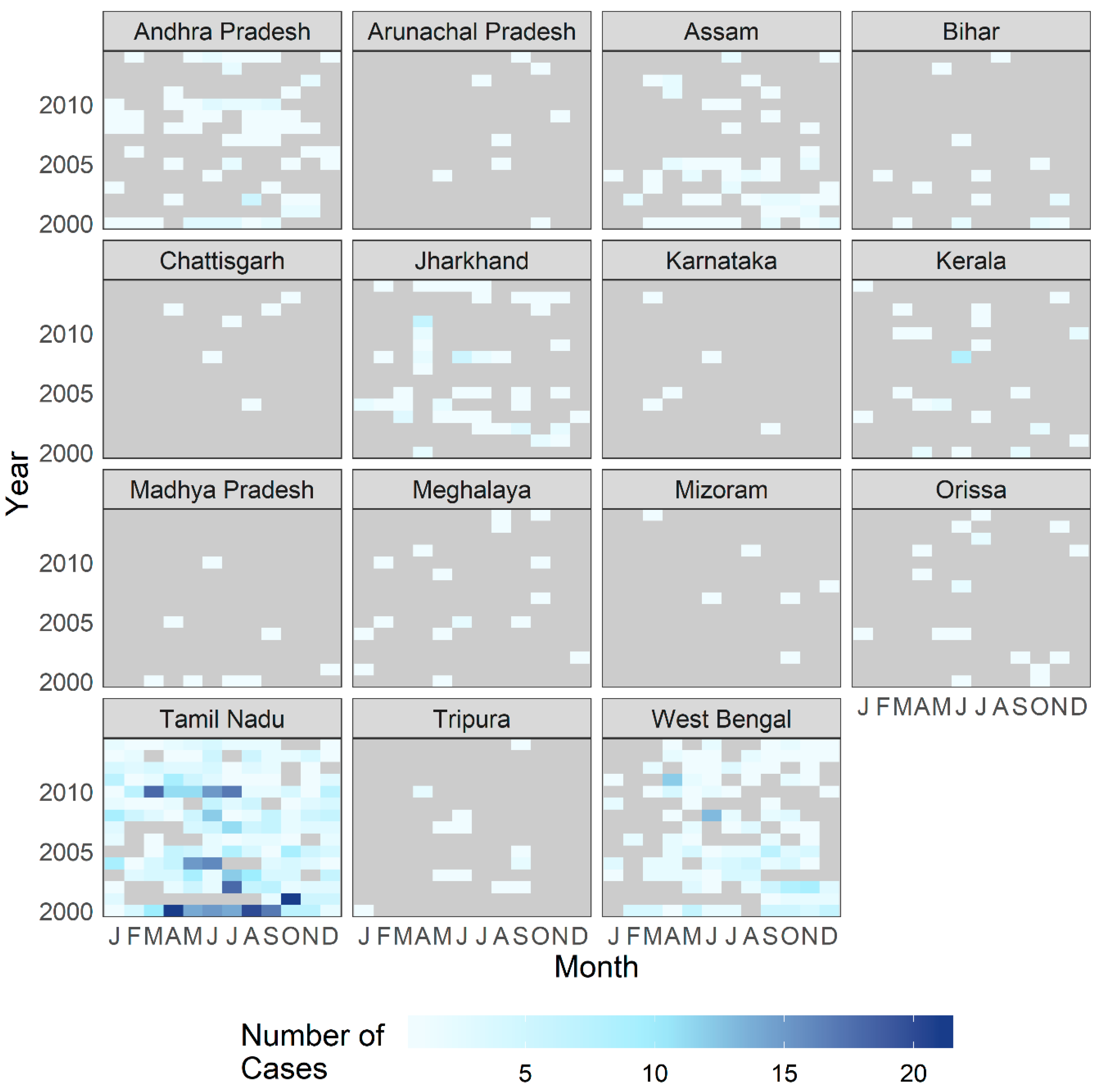
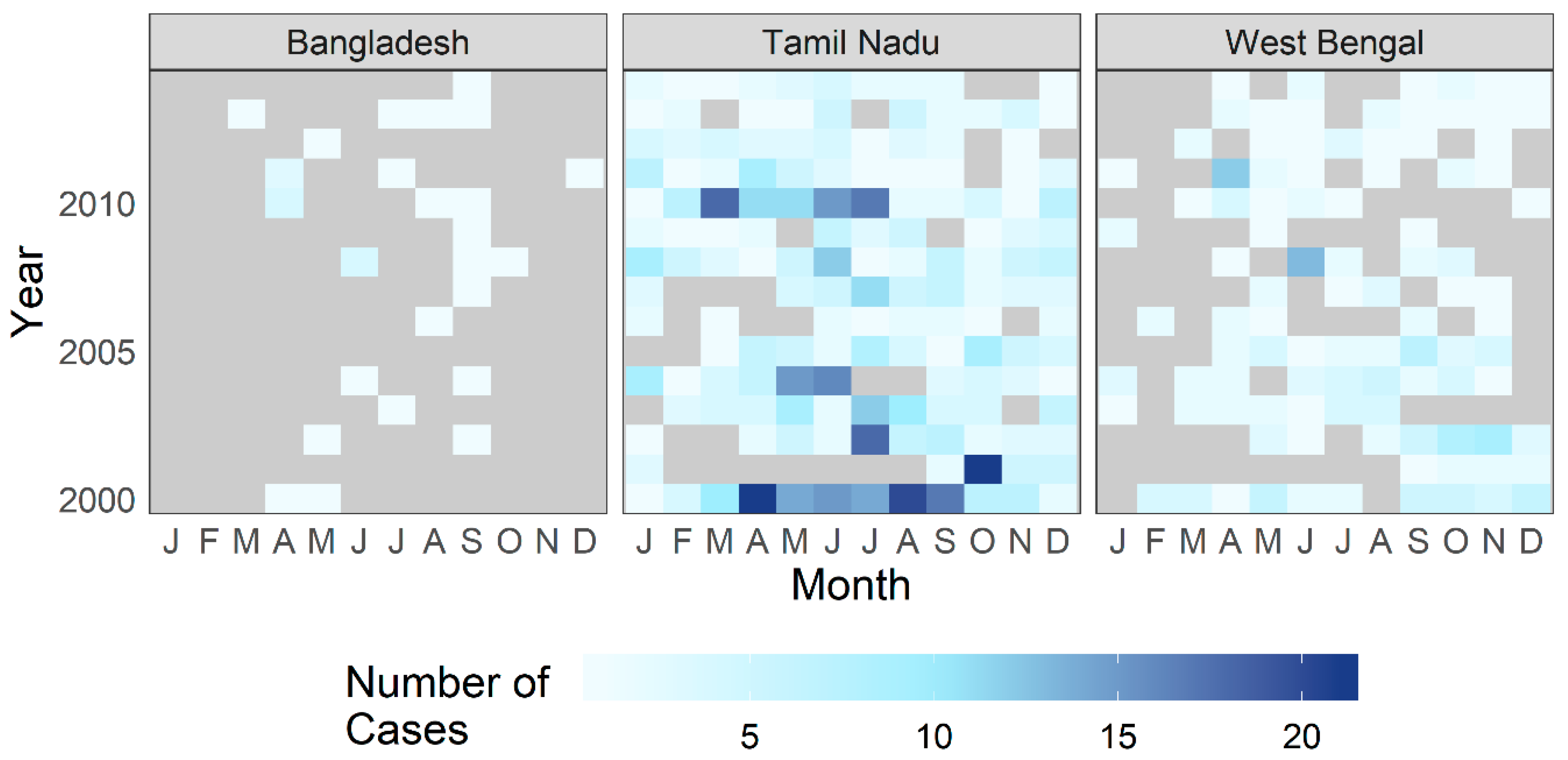
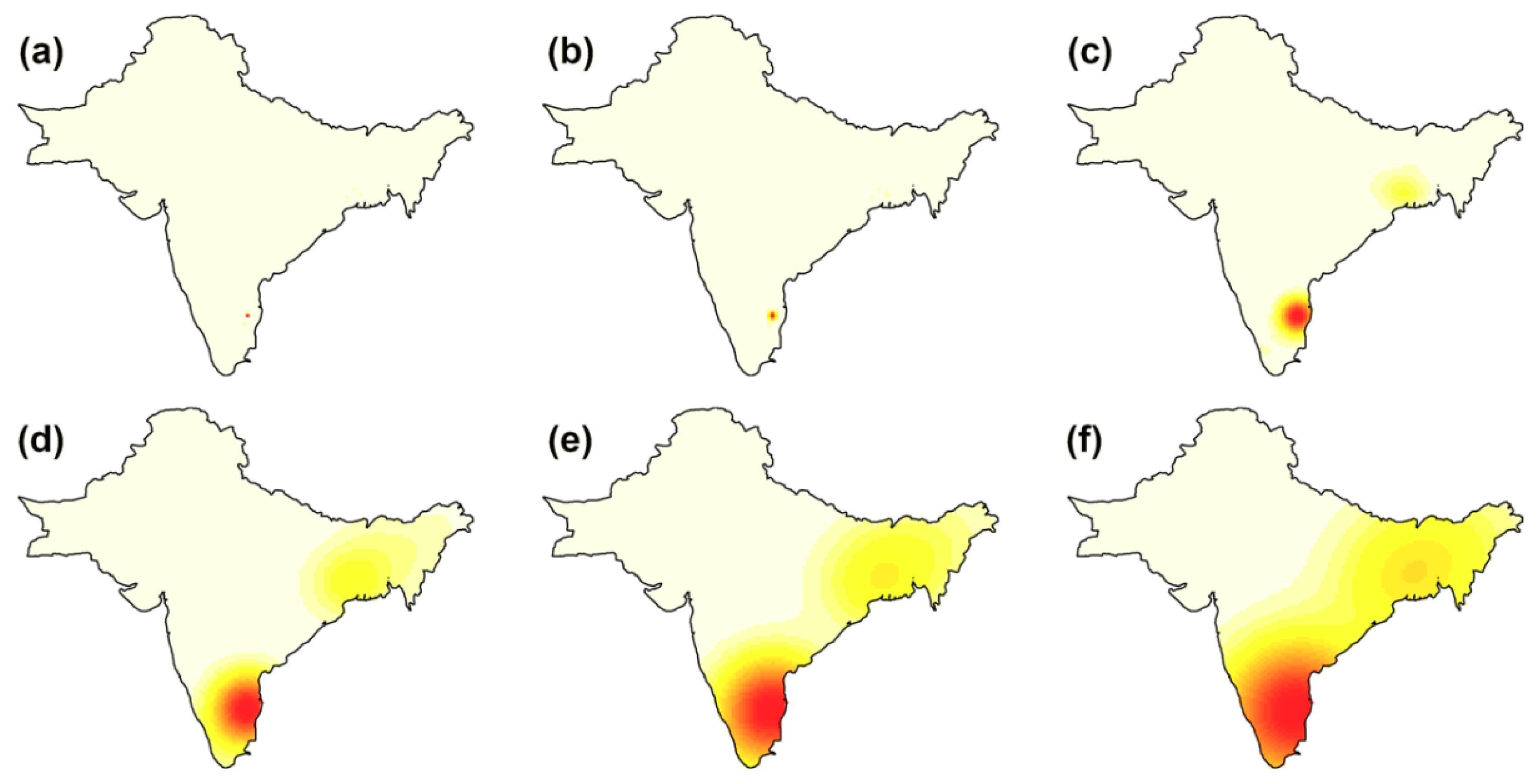
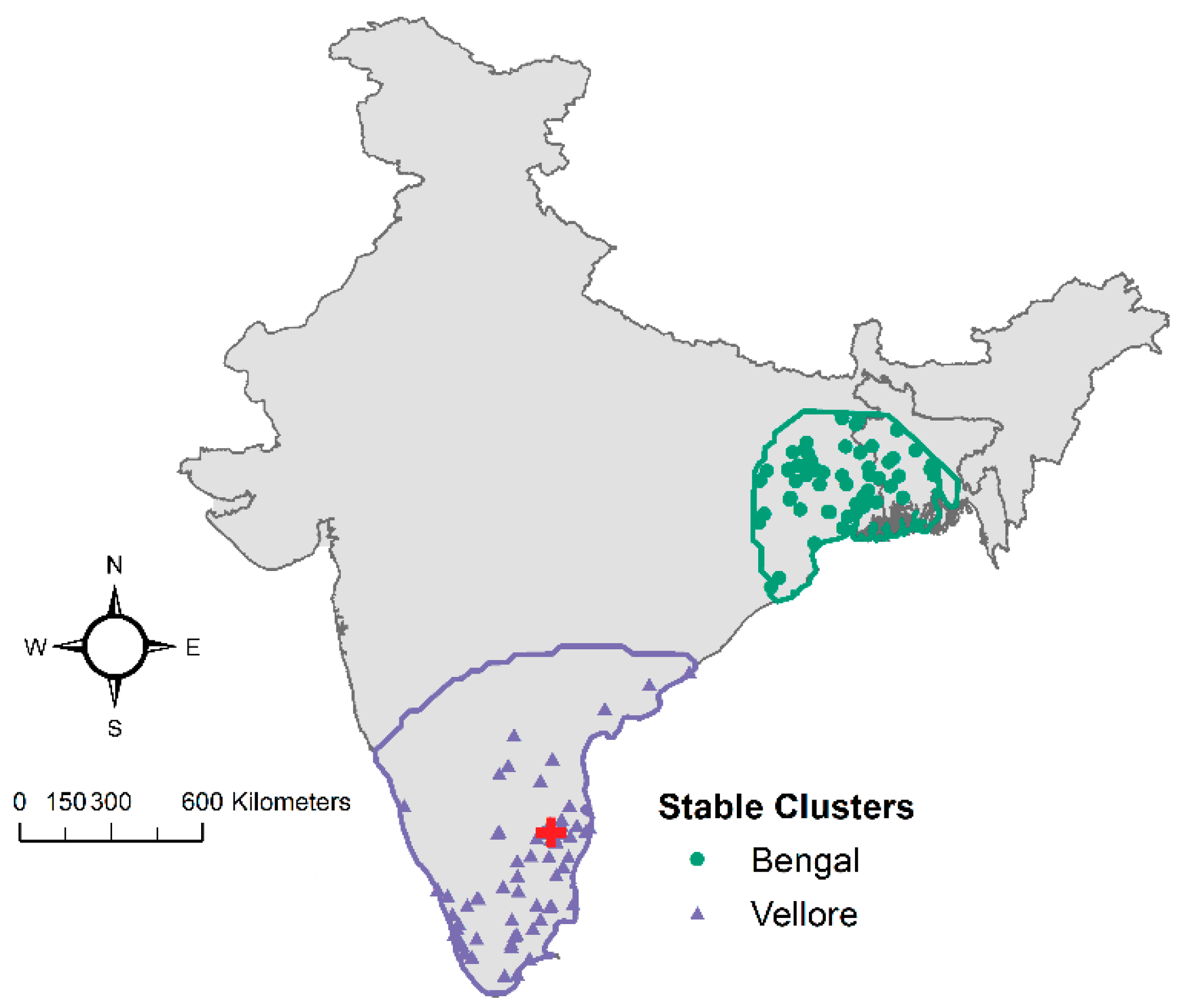
| Model | Model Formulation * |
|---|---|
| I: Trend | |
| II: Trend + Seasonality | |
| III: Trend + Seasonality + Weekends + Holidays |
| Model No. | Trend | AIC * | Cases in Vellore Cluster | Cases in Bengal Cluster | Cases in Joint Cluster |
|---|---|---|---|---|---|
| 15 | sin(x) + cos(y) | 58,132 | 836 | 493 | - |
| 14 | cos(y) + x | 58,351 | 841 | 294 | - |
| 8 | cos(y) | 58,475 | 836 | 470 | - |
| 11 | sin(x) + y | 60,160 | 800 | 500 | - |
| 13 | cos(x) + y | 60,244 | 798 | 500 | - |
| 3 | y | 60,536 | 800 | 500 | - |
| 12 | sin(y) + x | 61,123 | - | - | 1135 |
| 7 | sin(y) | 61,511 | - | - | 1327 |
| 9 | sin(x + y) | 62,028 | - | - | 1339 |
| 2 | x | 62,060 | 843 | 291 | - |
| 5 | sin(x) | 62,193 | - | - | 1339 |
| 6 | cos(x) | 62,214 | - | - | 1339 |
| 10 | cos(x + y) | 62,419 | - | - | 1339 |
| 1 | 1 | 62,558 | - | - | 1339 |
| Variable | Measure | Vellore | Bengal |
|---|---|---|---|
| Total Cases | Count | 836 | 294 |
| Holiday | Count (%) | 116 (13.88) | 45 (15.31) |
| Weekend | Count (%) | 181 (21.65) | 72 (24.49) |
| Male | Count (%) | 468 (56.05) | 185 (62.93) |
| Age | Mean (Sd) | 25.73 (23.14) | 33.92 (18.81) |
| O1 serotype | Count (%) | 574 (68.66) | 180 (61.22) |
| O139 serotype | Count (%) | 49 (5.86) | 12 (4.08) |
| Model | Vellore | Bengal | ||
|---|---|---|---|---|
| AIC * | VE (%) ** | AIC | VE (%) | |
| I: Trend | 4865 | 1.93 | 2308 | 1.02 |
| II: Trend + Seasonality | 4815 | 4.05 | 2288 | 2.81 |
| III: Trend + Seasonality + Weekends + Holidays | 2290 | 4.72 | 2290 | 2.96 |
| Variable | Vellore | Bengal |
|---|---|---|
| Trend as % change in disease counts per 30 days | −0.519 (−0.667, −0.371) | −0.455 (−0.690, −0.222) |
| Weekend effect as % difference | −31.27 (−42.96, −17.52) | −18.67 (−39.00, 7.31) |
| Holiday effect as % difference | −9.47 (−27.78, 12.73) | 2.18 (−28.08, 42.33) |
| Peak timing in days | 186.2 (170.5, 202.0) | 194.6 (171.5, 217.8) |
| Peak timing | July 7 ± 16 days | July 14 ± 23 days |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkat, A.; Falconi, T.M.A.; Cruz, M.; Hartwick, M.A.; Anandan, S.; Kumar, N.; Ward, H.; Veeraraghavan, B.; Naumova, E.N. Spatiotemporal Patterns of Cholera Hospitalization in Vellore, India. Int. J. Environ. Res. Public Health 2019, 16, 4257. https://doi.org/10.3390/ijerph16214257
Venkat A, Falconi TMA, Cruz M, Hartwick MA, Anandan S, Kumar N, Ward H, Veeraraghavan B, Naumova EN. Spatiotemporal Patterns of Cholera Hospitalization in Vellore, India. International Journal of Environmental Research and Public Health. 2019; 16(21):4257. https://doi.org/10.3390/ijerph16214257
Chicago/Turabian StyleVenkat, Aishwarya, Tania M. Alarcon Falconi, Melissa Cruz, Meghan A. Hartwick, Shalini Anandan, Naveen Kumar, Honorine Ward, Balaji Veeraraghavan, and Elena N. Naumova. 2019. "Spatiotemporal Patterns of Cholera Hospitalization in Vellore, India" International Journal of Environmental Research and Public Health 16, no. 21: 4257. https://doi.org/10.3390/ijerph16214257
APA StyleVenkat, A., Falconi, T. M. A., Cruz, M., Hartwick, M. A., Anandan, S., Kumar, N., Ward, H., Veeraraghavan, B., & Naumova, E. N. (2019). Spatiotemporal Patterns of Cholera Hospitalization in Vellore, India. International Journal of Environmental Research and Public Health, 16(21), 4257. https://doi.org/10.3390/ijerph16214257