1. Introduction
Cyanobacteria produce a variety of toxic compounds known as cyanotoxins [
1]. Human health consequences of cyanotoxins have been of increasing concern as the impacts of cyanobacterial blooms in water supplies as well as near-shore marine ecosystems have become better understood [
2]. Poisonings from cyanobacterial blooms can be catastrophic [
3], such as the death of 50 dialysis patients following exposure to inadequately treated water from the Tabocas Reservoir in Brazil or the hospitalization of 140 children and 10 adults who drank cyanobacterially-contaminated water impounded by the Solomon dam in Australia [
4,
5]. Cyanobacterial toxins have also been implicated in wildlife deaths in fisheries and terrestrial mammals, and can accumulate within the ecosystem. Potent hepatotoxins such as cyclic heptapeptides including microcystins, and cyclic pentapeptides including nodularins are produced by
Anabaena, Microcystis, Oscillatoria, Nostoc, Anabaenopsis, and
Nodularia species. Different neurotoxins are known to be produced by cyanobacteria including anatoxin-a, homoanatoxin-a, anatoxin-a(S), and saxitoxins from
Anabaena,
Aphanizomenon, Oscillatoria, Lyngbya, and
Cylindrospermopsis [
1,
3].
We found that endophytic cyanobacteria of the genus
Nostoc resident in specialized roots of the Guam cycad
Cycas micronesica Hill (Cycadaceae) produced a different neurotoxin, β-methylamino-
L-alanine (BMAA), which is found throughout cycad tissues [
8,
9]. This led to a broader examination of BMAA production by other taxa of cyanobacteria, including 30 laboratory strains of free-living cyanobacteria, representing five major microbiological taxonomic groups of Rippka
et al. [
10]. Of the thirty laboratory cultures, 29 had free or protein-bound BMAA, as did a sample of an ocean
Trichodesmium bloom [
11]. BMAA has also been found in bloom samples of cyanobacterially-contaminated water supplies in the United Kingdom, where it was found to co-occur with other cyanotoxins including microcystins, nodularin, anatoxin-a, and saxitoxins [
12].
The co-occurrence of BMAA with other cyanotoxins may be of ecological importance, since BMAA can potentiate injury to motor neurons at concentrations as low as 10 μM [
13]. Alone, BMAA has been found to selectively kill motor neurons at concentrations as low as 30 μM by AMPA/kainate receptor activation [
14].
Although BMAA is a glutamate agonist at AMPA and NMDA receptors, it is a structurally dissimilar polar amino acid with two functional groups. Also known by the synonyms α-amino-β-methylaminopropionic acid (MeDAP) and S(+)-
m-methyl-α
,β-diaminopropionic acid, BMAA has a molecular weight of 118.13. Originally isolated from the seeds of cycads, BMAA can be synthesized from α-acetamidoacrylic acid [
15,
16]. Its structure is basically that of alanine with an added methylated amino group (
Figure 1). Historically, BMAA was thought to be a possible slow toxin [
17] associated with neurodegenerative illness among the Chamorro people of Guam [
18]. The interest in BMAA has recently been renewed with evidence that BMAA is incorporated into the protein fraction of cyanobacteria, cycads, animals that forage on cycad seeds, as well as indigenous people with traditional diets rich in BMAA [
8,
19]. The incorporation and storage of BMAA in protein explains not only how biomagnification of a water-soluble molecule can occur, but also may account for the observed disease latency in ALS/PDC due to the intermittent release of the endogenous neurotoxic reservoir stored in the proteins [
19]. Some forms of progressive neurodegenerative illness are associated with protein misfolding and protein truncation—likely occurrences in the case of a foreign amino acid insertion during protein synthesis—and could conceivably trigger neurodegeneration.
The incorporation of BMAA into protein was first demonstrated by Polsky, Nunn and Bell, who found BMAA in an extract removed from cycad seeds; acid hydrolysis of the extract released BMAA in greater amounts than found in the free fraction [
20]. Subsequently, the insertion of BMAA into artificial peptides [
21] further demonstrated the potential for protein-bound BMAA in natural systems. However, association of a non-protein amino acid like BMAA with proteins is not unusual, since there are many other cases of unusual amino acids being associated with cyanobacterial proteins [
22]. For such studies, hydrochloric acid (HCl) hydrolysis is by far the most commonly used method of releasing amino acids from proteins [
23]. The conventional acidic hydrolysis uses 6N HCl for 18 to 24 hours
in vacuo and provides high recovery of all protein-bound amino acids except tryptophan.
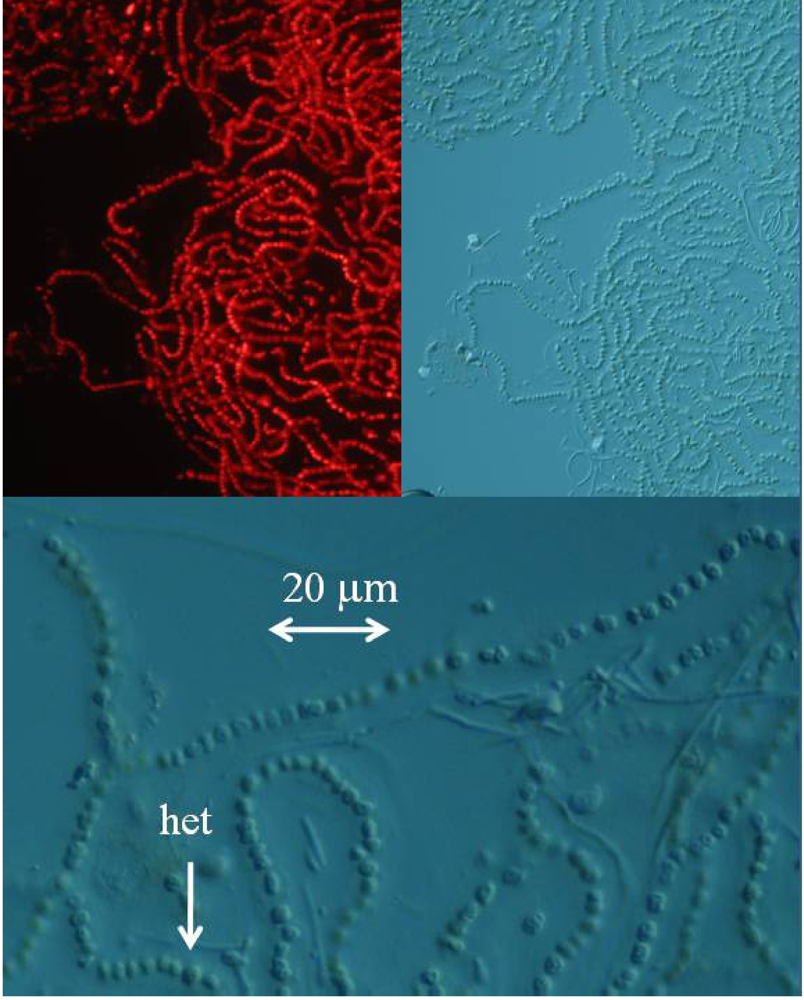
The detection and quantification of BMAA in a sample increases in difficulty as the concentration of BMAA decreases and the complexity of the physiological matrix increases. While it is relatively simple to detect the high concentration of BMAA in a cycad seed gametophyte or even a low concentration of synthetic BMAA standard with standard chromatographic techniques, detection of BMAA in low concentration in a complex physiological matrix (such as in a cyanobacterium, or a human brain sample) requires careful method development and validation. Single injection experiments, particularly those performed by investigators with little previous amino acid experience are unlikely to result in detection. Multiple techniques to confirm the presence of BMAA in a sample are preferred. To unequivocally demonstrate that a marine cyanobacterium, a laboratory strain of
Nostoc sp. known as CMMED-01 (
Figure 2), produces BMAA, we used five different techniques to assay it for the production of BMAA.
3. Conclusions
The use of three different HPLC techniques and two different LC/MS techniques conclusively demonstrates that the marine cyanobacterial species represented by
Nostoc CCMED-001 produces the neurotoxic non-protein amino acid, β-methylamino-
L-alanine. The precise fragmentation pattern and collision-induced dissociation ion ratios of the LC/MS/MS provide unequivocal evidence of BMAA in the sample. Although BMAA is present in the cyanobacterium, we detected some variation in concentration since the samples tested were taken at different times from the parent culture. Variation in concentration is not unexpected, since nitrogenase activity within marine cyanobacteria can vary dramatically within a single day as well as throughout the life cycle depending on the specific taxon, photon flux, and culture senescence [
27–
29]. Our study confirming through five different techniques the presence of BMAA in a marine
Nostoc species supports the addition of BMAA to the list of phycotoxins produced by cyanobacteria [
30].
Originally discovered in 1967 in cycad seeds from Guam [
15,
16], BMAA for 45 years was believed to occur only in cycads. As such, the putative link of BMAA to the debilitating paralytic disease ALS/PDC suffered by the Chamorro people was believed to be related only to the Chamorro consumption of flour made from the seed gametophytes of
Cycas micronesica Hill [
15–
20]. Claims that Chamorro processing procedures remove nearly all of the BMAA from cycad flour, combined with no clear etiological linkage of cycad consumption in the two other foci of ALS/PDC in West Papua and the Kii peninsula of Japan, led to BMAA being discounted as a possible cause of neurodegenerative illness among the Chamorro people [
18]. Furthermore, there was no conceivable connection of BMAA to other forms of progressive neurodegenerative illness in the parts of the world where cycads do not occur.
Interest in BMAA was rekindled, however, in 2002 when it was hypothesized that BMAA may accumulate within animals that eat cycads [
32]. Experimental evidence for such bioaccumulation soon followed [
33,
34]. In 2003, it was discovered that BMAA is produced by cyanobacteria of the genus
Nostoc which live as symbionts within specialized cycad roots [
8]. In 2004 it was discovered that BMAA can be bound to proteins, including the proteins of cycad flour which had not been previously investigated [
19]. Thus a new pictured emerged: BMAA was present within the traditional Chamorro diet from multiple sources in far higher concentrations than had previously been suspected [
8,
19,
34]. In 2005, it was discovered that BMAA can be produced by diverse taxa of cyanobacteria [
11]. It was thought that this broad pattern of cyanobacterial production of BMAA might provide a clue into why BMAA was found in brain tissues of patients with progressive neurodegenerative illness who lived far from Guam [
19,
35].
Confirmation through five different techniques that BMAA is produced by the marine
Nostoc species represented by the CCMED-01 samples increases our interest in finding out if this potent neurotoxin, which selectively targets motor neurons [
13,
14], can be biomagnified within marine ecosystems. We await with interest the results of ongoing studies of BMAA biomagnification at the University of Hawaii, Stockholm University, Chiba University, and our Institute designed to test this possibility. It also would be interesting to know if marine (or even fresh-water) cyanobacteria play a role in human exposure to BMAA in the ALS/PDC focus in Kii, Japan. Furthermore, the recent finding that BMAA co-occurs with other cyanotoxins in contaminated water supplies raises the possibility that low-level human exposure to BMAA may occur in many parts of the world [
12]. In contrast to ALS/PDC foci, such as Guam, where exposure to high levels of BMAA in traditional diets may constitute a broad threat to human health, exposures to low concentrations of BMAA in marine or freshwater ecosystems may trigger progressive neurodegenerative illness among genetically vulnerable individuals, potentially resulting in a new paradigm for sporadic motor neuron disease [
19,
36]. Until more is known about the possible link between BMAA and gene/environment interactions in progressive neurodegenerative illnesses, we again suggest [
11] that it is now prudent to begin to assemble baseline data on the presence of BMAA in water supplies and human diets.
4. Experimental
General
The marine Nostoc sp. culture CMMED-01 was based on a sample originally collected on June 2, 2004 in Kaneohe Bay, on the island of Oahu in Hawaii by D. Hashimoto and identified by B. Anderson. This collection was cultured and maintained by R. Bidigare, G, Tien and colleagues at The Center for Marine Microbial Ecology and Diversity at the University of Hawaii, Manoa and sent to our laboratory by S. Christensen and A. Messner.
4a. Fluorescence microscopy
The lyophilized marine Nostoc sp. culture was reconstituted in water, and fresh mounted or mounted with Permount SP15-100 Toluene solution (Fisher Scientific, Fair Lawn, NJ) on microscope slides. The cyanobacteria were imaged with a Zeiss Axioplan 2 microscope. The cyanobacteria were examined with differential interference contrast and fluorescence microscopy with excitation at 550 nm and emission at 590 nm using a Chroma filter set (Chroma Technology, Rockingham, VT).
4 b1. Fluorescence HPLC methods for analysis of free-BMAA
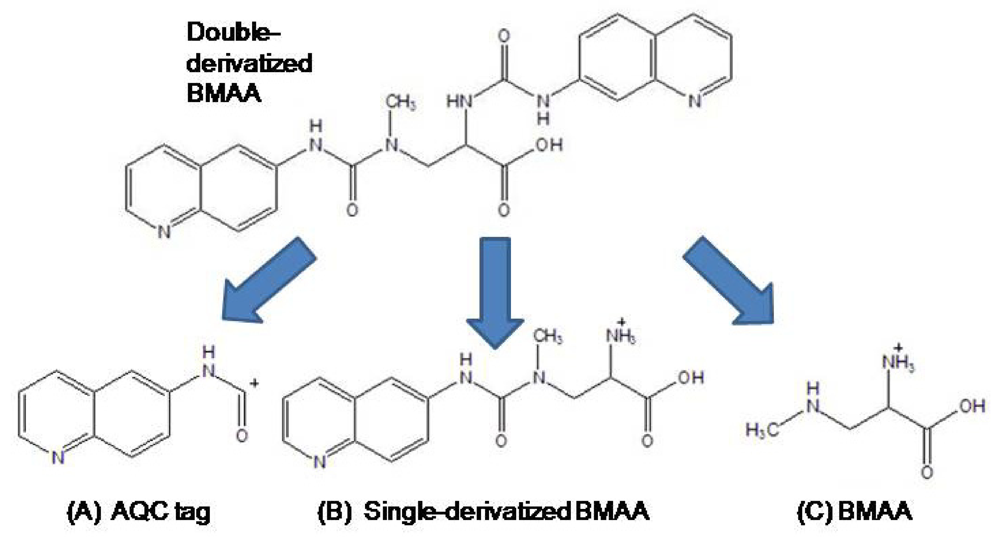
Freeze-dried cyanobacterial samples were macerated in liquid nitrogen, weighed, hand macerated in trichloroacetic acid (0.4 mg/mL 0.1N TCA), vortexed, and extracted for 12 h at 4°C. Samples were centrifuged and the supernatant decanted. The pellet was remacerated in TCA (50 μL), vortexed, and re-extracted for 1 h at room temperature. Samples were centrifuged and the TCA supernatants pooled and filtered (0.22 μm Ultrafree-MC, Millipore) at 13,800 g for 5 min (Labnet Spectrafuge 16M). Free amino acid extracts were derivatized with 6-aminoquinolyl-
N-hydroxysuccinimidyl carbamate (AQC Waters AccQTag reagent, PN WAT052880) following standardized, validated protocols: sample (20 μL) was buffered with 0.5 M borate (80 μL) and derivatized with AQC (20 μL) [
9]. Sample batches for analysis included derivatized samples, blanks [20 mM HCl (20 μL with 0.5 M borate (80 μL) and derivatized with AQC (20 μL)], derivatized 17 amino acid standard complex (Pierce #NCI0180, Rockford, IL) spiked with BMAA to assure adequate separation, derivatized BMAA standard for quantification, water blanks, and derivatized low-level spiked samples for peak identification and to check for a balanced reaction. BMAA was separated from other amino acids by reverse-phase elution (Waters Nova-Pak C18 column, 3.9 x 300 mm) on a Waters 1525 binary HPLC pump and a Waters 717 Autosampler using a gradient method with 140 mM sodium acetate, 5.6 mM triethylamine, pH 5.2 (Eluent A), and 52% acetonitrile in water (Eluent B) at 37°C, a flow rate of 1.0 mL/min, and a 10μL sample injection. The standard amino acid elution gradient was modified for BMAA separation on a 60 min gradient (0.0 min = 100% A; 2 min = 90% A curve 11; 5 min = 84% A curve 11; 10 min = 84 % A curve 6; 18 min = 75% A curve 6; 33 min = 65% A curve 10; 35 min = 40% A curve 6; 37.5 min = 100% B curve 6; 47 min = 100% B curve 6; 50 min =100% A curve 6; 60 min =100% A curve 6). BMAA concentration was quantified by detection of the fluorescent tag (Waters 2475 Multi γ-Fluorescence Detector) with excitation at 250 nm and emission at 395 nm (
Figure 2) and by comparison to an authenticated synthetic BMAA standard (P. Nunn, Univ. of Portsmouth, UK).
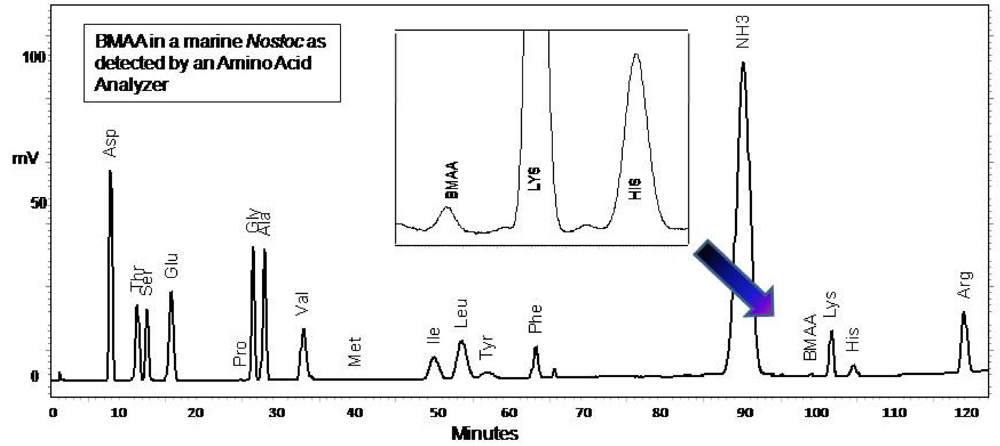
4b2. Fluorescence HPLC methods for analysis of total BMAA
CMMED-01 was hydrolyzed (63 mg/mL wet weight) in 6 N HCl (400 μL) for 16 hours at 110EC. The hydrolysate (200 μL) was filtered at 13,800 g for 3 min (0.22 μm Ultrafree-MC, Millipore). The residue was dried down using a speed vacuum (Thermo-Savant SC250DDA Speed Vac Plus, Waltham, MA) and resuspended in 20 mM HCl (100 μL). The sample was diluted 1:10 using 20 mM HCl before derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate [sample (20 μL) was buffered with 0.5 M borate (80 μL) and derivatized with AQC (20 μL)]. Sample batch included blanks [20 mM HCl (20 μL) with 0.5 M borate (80 μL) and derivatized with AQC (20 μL)], derivatized 17 amino acid standard complex (Pierce #NCI0180, Rockford, IL) with a BMAA spike to assure adequate separation, derivatized BMAA standard for quantification, HPLC grade water blanks, derivatized low-level spiked samples for peak identification and to check for a balanced reaction, a matrix blank of hydrolyzed, derivatized Lycopersicon esculentum Mill. (Solanaceae) for a negative control, a matrix blank of hydrolyzed, derivatized L. esculentum spiked with low-levels of BMAA and a blank of derivatized L-2,4-diamino-n-butyric acid which has a molecular weight of 118.13, equivalent to BMAA but with a different structure.
BMAA was separated from other amino acids by reverse-phase elution (Waters Nova-Pak C18 column, 3.9 x 300 mm) on a Waters 1525 binary HPLC pump and a Waters 717 Autosampler using a gradient method with 140 mM sodium acetate, 5.6 mM triethylamine, pH 5.7 (Eluent A), and 52% acetonitrile in water (Eluent B) at 37°C, a flow rate of 1.0 mL/min, and a 20 μL sample injection. The standard amino acid elution gradient was modified for BMAA separation on a 30 min gradient (0.0 min = 75% A; 2.0 min = 75% A curve 6; 17.0 min = 63% A curve 7; 18.5 min = 100% B curve 6; 23.5 min = 100% B curve 6; 25.0 min = 75% A curve 6; 30.0 min = 75% A curve 6). BMAA concentration was quantified by detection of the fluorescent tag (Waters 2475 Multi γ-Fluorescence Detector) with excitation at 250 nm and emission at 395 nm (
fig. 3) and by comparison to an authenticated synthetic BMAA standard (P. Nunn).
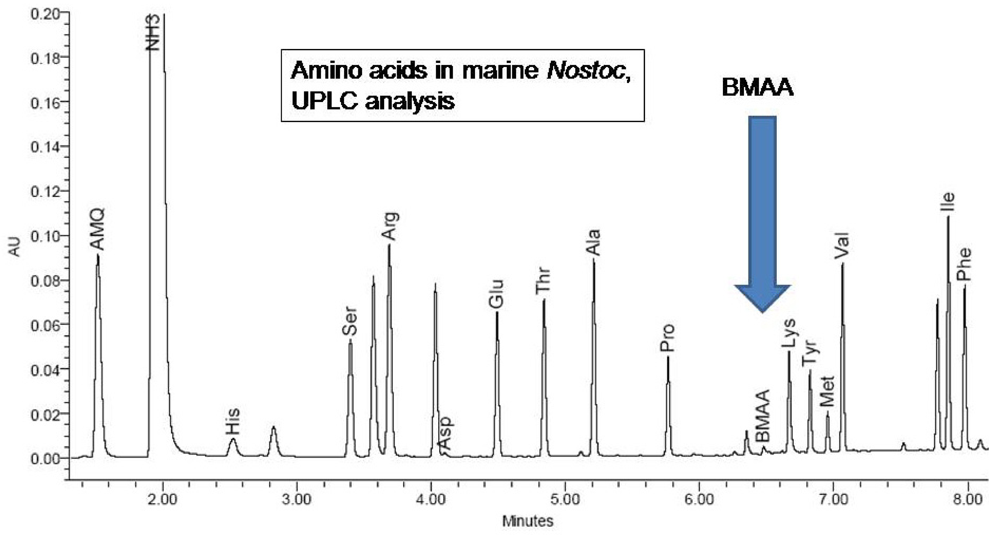
4c. UPLC-UV Methods for quantification of total BMAA in CMMED-01
Methods for quantification of BMAA were adapted from previously validated High Performance Liquid Chromatography (HPLC) methods [
11] which were transferred and optimized to Ultra Performance Liquid Chromatography (UPLC7). Waters Acquity UPLC methods for amino acid analysis with ultraviolet (UV) detection were used with a modified solvent gradient and validated in compliance with United States Pharmacopeia General Chapter 1225 [
31]. Eluents were purchased from Waters (Eluent A: part #186003838, Eluent B: part # 186003839); composition is proprietary. Separation was by reverse phase over 9.5 minutes on a Waters AccQTag Ultra column (part# 186003837, 2.1x100 mm) at 55°C with a flow rate of 0.7 mL/min (0.0 min = 0.1% B; 0.54 min = 0.1% B curve 6; 6.24 min = 9.1% B curve 7; 7.74 min = 21.2% B curve 6; 8.04 min = 59.6% B curve 6; 8.73 min= 0.1% B curve 6; 9.5 min = 0.1% B curve 6).
Validation was completed using a BMAA standard (synthesized standard P. Nunn) derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Ruggedness was evaluated by calculating the precision of biweekly triplicate injections at one concentration over 32 weeks (RSD 3.6%). Linearity was evaluated by plotting peak area as a function of analyte concentration based on 9 injections from each of 21 different preparations and regression analysis was performed: slope = 5589; intercept = −3198; correlation coefficient = 0.9998; residual sum of squares = 1291469639. The limit of detection (LOD) and limit of quantification (LOQ) were determined as 0.1 pmol and 1.1 pmol respectively, with a linear range to 539 pmol per injection. Accuracy (recovery = 98%) was calculated by spiking negative control matrix, Lycopersicon esculentum, with known amounts of BMAA in triplicate at five concentrations. Precision was evaluated with two measures, repeatability and intermediate precision. Repeatability was assessed with triplicate injections at five concentrations on two consecutive days (RSD 1.2%). Intermediate precision was calculated biweekly over 32 weeks with triplicate injections at three concentrations (RSD 3.9%). Selectivity was verified with in-line LC/MS as noted below.
Methods for sample preparation were adapted from validated HPLC methods [
11]. Twenty mg of freeze dried cells from cultured CMMED-01 were weighed in triplicate and hydrolyzed in 6 M HCl (400 μL) at 110°C for 16 hours. Hydrolysate (200 μL) was filtered with a 0.22 μm centrifugal filter device (Millipore Ultrafree-MC), dried to completeness, then suspended in 20 mM HCl (100 μL) and diluted 1:10. Sample (20 μL) was buffered with 0.5 M borate (80 μL) and derivatized with Waters AccQTag reagent (20 μL); 2 μL were injected. BMAA in the cyanobacterial samples was quantified using the validated UPLC method above, with UV detection at 260 nm. Sample batch included blanks [20 mM HCl (20 μL) with 0.5 M borate (80 μL) and derivatized with AQC (20 μL)], derivatized 17 amino acid standard complex (Pierce #NCI0180, Rockford, IL) spiked and unspiked with BMAA to assure adequate separation, derivatized BMAA standard for quantification, derivatized low-level spiked samples for peak identification and to check for a balanced reaction, a negative matrix blank of hyrdolyzed
Homolanthus nutans Muell. Arg. (Euphorbiaceace), and a matrix blank of hydrolyzed
H. nutans spiked with low-levels of BMAA. The presence of the non-protein amino acid BMAA as well as some of the 20 essential protein amino acids were determined by reference to synthetic BMAA and a multiple amino acid standard (Pierce #NCI0180, Rockford, IL.;
Figures 4,
5).
4d. Amino Acid Analyzer methods for CCMED-01
Freeze dried CMMED-01
Nostoc was hydrolyzed (63 mg/mL) in 6 N HCl (400 mL) for 16 hours at 110°C. The hydrolysate (200 μL) was filtered at 13,800 g for 3 min (0.22 μm Ultrafree-MC, Millipore). The residue was dried down using a speed vacuum (Thermo-Savant SC250DDA Speed Vac Plus, Waltham, MA) and resuspended in 20 mM HCL (100 μL). The sample was diluted 1:10 using 20 mM HCl. Underivatized sample (20 μL) was injected into a Hitachi Amino Acid Analyzer L8900 equipped with a Hitachi Reaction column (PN 855-3533) at 135°C, a high speed physiological fluid analysis analytical column Li-form resin #2622SC 6 mm ID x 40 L 060928C (PN855-4515), precolumn (PN855-3643), guard column #2619 4 mm ID x 5 L 2007.07.19 070639 (PN855-5268), and AmmoniaFilter column (Ion exchange 4.6 x 40 Column #2650L, PN 855-3523). Hitachi pre-made buffers were used as follows: (B1) PF-1/AN0-8711, (B2) PF-2 /AN0-8712, (B3) PF-3/An0-8713, (B4) PF-4/An0-8714, (B5) 5% methanol, (B6) PF-RG/An0-8715, (R1) ninhydrin solution, (R2) ninhydrin-buffer of lithium acetate dihydrate, (R3) 5% ethanol. Separation was made with a flow rate for pump 1 of 0.53 mL/min and 0.45 mL/min for pump 2 and a 152 min gradient elution (0.0 min = 100% B1, separation column and guard column temp 35°C, 50% R1, 50% R2; 1.5 min = column 32°C; 15.6 min = 81% B1, 19% B2, column 57°C; 36 min = 60°C; 45 min = 32°C; 57 min = 70°C; 68 min = 15% B1, 75% B2, 10% B3; 69 min = 58°C; 76.1 min = 60% B2, 40% B3; 89 min = 65°C; 95 min = 20% B2, 80% B4; 98.1 min= 25% B2, 75% B4; 112.1 min = 100% B4; 125 min = 70°C; 125.1 min = 100% R3; 129.1 min = 100% B6; 132.1 = 100%B1; 137.1 min = 50% R1, 50% R2, 35°C). The presence of BMAA was verified by reference to an authenticated synthetic standard (P. Nunn), and a multiple amino acid standard (Pierce #NCI0180, Rockford, IL,
fig. 6). Sample batches included the following underivatized samples: amino acid standards with and without BMAA spikes, BMAA,
L-2,4-diamino-
n-butyric acid, which has a molecular weight of 118.13 equivalent to BMAA, hydrolyzed
Lycopersicon esculentum, o-methyl-L-tyrosine,
L-
o-methylthreonine,
s-methyl-
L-cysteine,
n-ε-methyllysine HCl,
L-2,3,diaminoproprionic acid HCl,
Nt-methyl-
L-histidine, methylamine HcC, 3 methyl-
L-histidine,
NG-methyl-l-arginine and amino acid standard physiological basic A6282 (Sigma).
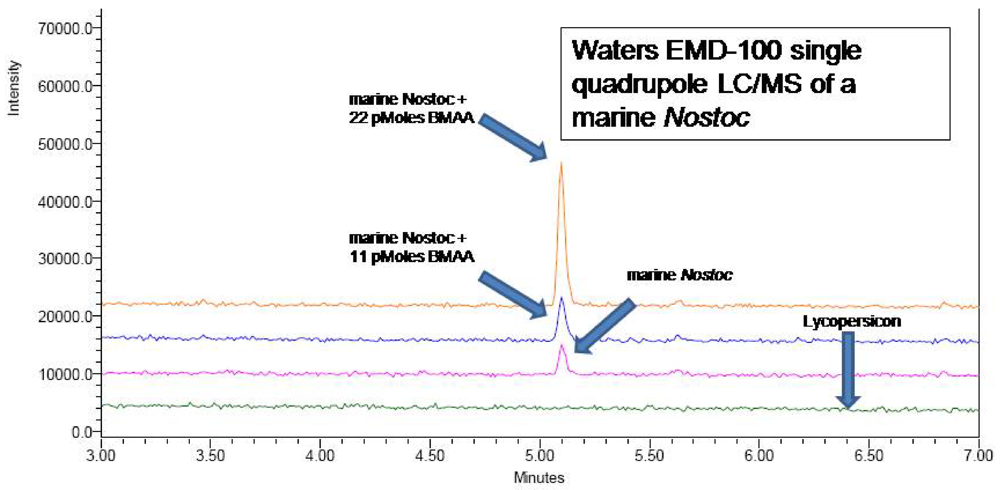
4e. LC/MS methods for verification of presence of BMAA in CMMED-01
A CMMED-01
Nostoc sample prepared as in 2a was injected (5 μL) into a Waters UPLC on a Waters AccQTag Ultra column (part# 186003837, 2.1x100 mm) at 55°C with a flow rate of 0.7 mL/min, using the UPLC method previously noted. A post-column split of the flow delivered a flow rate of 0.3 mL/min to a Waters EMD 1000 single quadrupole mass spectrometer. Nitrogen gas was purified to 99.2% using a NitroFlow nitrogen generator (Parker Balston, Haverfill, MA) and supplied to the electrospray ionization interface at 116 psi with desolvation at 500 L/hr. Positive ion mode was used to detect BMAA derivatized with 6-aminoquinolyl-
N-hydroxysuccinimidyl carbamate (AQC) (Waters AccQTag reagent, part # WAT052880), using single ion monitoring at 459.20 Daltons with a span of 0.3 Daltons and a dwell time of 0.5 seconds. The source temperature and desolvation temperature were 130°C and 300°C respectively. Voltages were as follows: capillary 3.05 kV, cone 27 V, extractor 1 V, RF lens 0.4 V. LC/MS verification of the UPLC peak (
Figure 5) was obtained (
Figure 6), as were LC/MS verifications of AQC derivatized CMMED-01 samples spiked with 11 picomoles and 22 picomoles BMAA respectively (
Figure 7). Sample batch included blanks [20 mM HCl (20 μL) with 0.5 M borate (80 μL) and derivatized with AQC (20 μL)], derivatized 17 amino acid standard complex (Pierce #NCI0180, Rockford, IL) spiked with BMAA to assure adequate separation, derivatized BMAA standard for quantification, and derivatized low-level spiked samples for peak identification and to check for a balanced reaction and a negative matrix blank of hyrdolyzed
Lycopersicon esculentum Mill. (Solanaceae), a matrix blank of hydrolyzed
L. esculentum spiked with low-levels of BMAA, and a blank of
L-2,4-diamino-
n-butyric acid which has a molecular weight of 118.13 equivalent to BMAA but with a different structure.
4f. Triple Quadrupole LC/MS/MS
Identification of a BMAA peak in a marine Nostoc CMMED-001 sample was verified by liquid chromatography/mass spectroscopy/mass spectroscopy (LC/MS/MS) using product ion mode in a triple quadrupole system. The freeze-dried cyanobacteria sample was hydrolyzed for 18 hours in 6 N HCl at 110°C and then dried to remove HCl in a Thermo-Savant SC250DDA Speed Vac Plus (Waltham, MA). The sample was reconstituted in dilute HCl (20 mM) and derivatized with AQC, which increased the molecular weight of the BMAA analyte from 118 to 458. To increase the quantity of analyte injected into the LC/MS/MS, the BMAA peak was collected three times from a Waters 1525 binary HPLC pump and a Waters 717 Autosampler, with separation through a Waters 3.9 x 300 mm Nova-Pak C18, 4 μm column (Lot No.: 11143603410254) at 37°C. Individual compounds were eluted from the column with a gradient elution of 140 mM sodium acetate buffer, 5.6 mM triethylamine, pH 5.7 (mobile phase A) and 52% acetonitrile (mobile phase B) with a flow rate of 1.0 mL/min. The collected peaks were dried to reduce the volume in a Thermo-Savant SC250DDA Speed Vac Plus with a Savant refrigerator trap RVT 4104. The first collected peak was then reconstituted in HPLC grade water (50 mL), heated to 55°C, vortexed, and then added to the second and third peak iteratively which were likewise heated and vortexed in a similar fashion. The concentrated filtrate was injected into a triple quadrupole system (Thermo model TSQ Quantum Discovery Max, San Jose, CA). Separation was achieved by liquid chromatography (Thermo model Surveyor LC, San Jose, CA) utilizing a Thermo Hypersil GOLD 100 x 2.1 mm, 3 μm column at 0.28 mL/min, 0.1% formic acid in water (Eluent A), 0.1% formic acid in acetonitrile (Eluent B) gradient elution (0.0 min = 100% A; 2 min = 100% A; 16 min = 80% A; 20 min = 2% A; 22 min = 2% A; 24 min = 81% B; 26 min = 81% A; 27 min = 100% A; 31 min =100% A). Nitrogen gas was supplied to the H-ESI probe (heated electrospray ionization) with a nebulizing pressure of 25 psi and a vaporizing temperature of 300°C. The mass spectrometer was operated under the following conditions: the capillary temperature was set at 280°C, capillary offset of 35, tube lens offset of 94, source collision energy of 5, and multiplier voltage of −1194. The second quadrupole was pressurized to 1.5 mTorr with 100% Argon.
The derivatized molecule after ionization (MW 459) via HESI was selected as the precursor ion for analysis. Collision induced dissociation (CID) was achieved in the second quadrupole using the following parameters: 459 → 119 CE 19; 459 → 171 CE 32; and 459 → 289 CE 16. The resultant three product ions of the derivatized BMAA (MW 119, 289, 171) were scanned by the third quadrupole, subsequently detected, and their relative abundances quantified. The ratios of these three product ions were compared to the ratios of the product ions created by injection of AQC-derivatized ultra pure, triple crystallized BMAA standard (Peter Nunn) into the mass spectrometer under the same conditions (
Figure 9). Sample batch included blanks (20 μL of 20 mM HCl with 80 μL 0.5 M borate and derivatized with 20 μL AQC), derivatized 17 amino acid standard complex (Pierce #NCI0180, Rockford, IL) to assure adequate separation, derivatized BMAA standard for quantification, HPLC grade water blanks, derivatized norleucine blanks, and a blank of
L-2,4-diamino-
n-butyric acid which has a molecular weight of 118.13, equivalent to BMAA but with a different structure, retention time, and product ions.