Chrysomycins, Anti-Tuberculosis C-Glycoside Polyketides from Streptomyces sp. MS751
Abstract
1. Introduction
2. Results
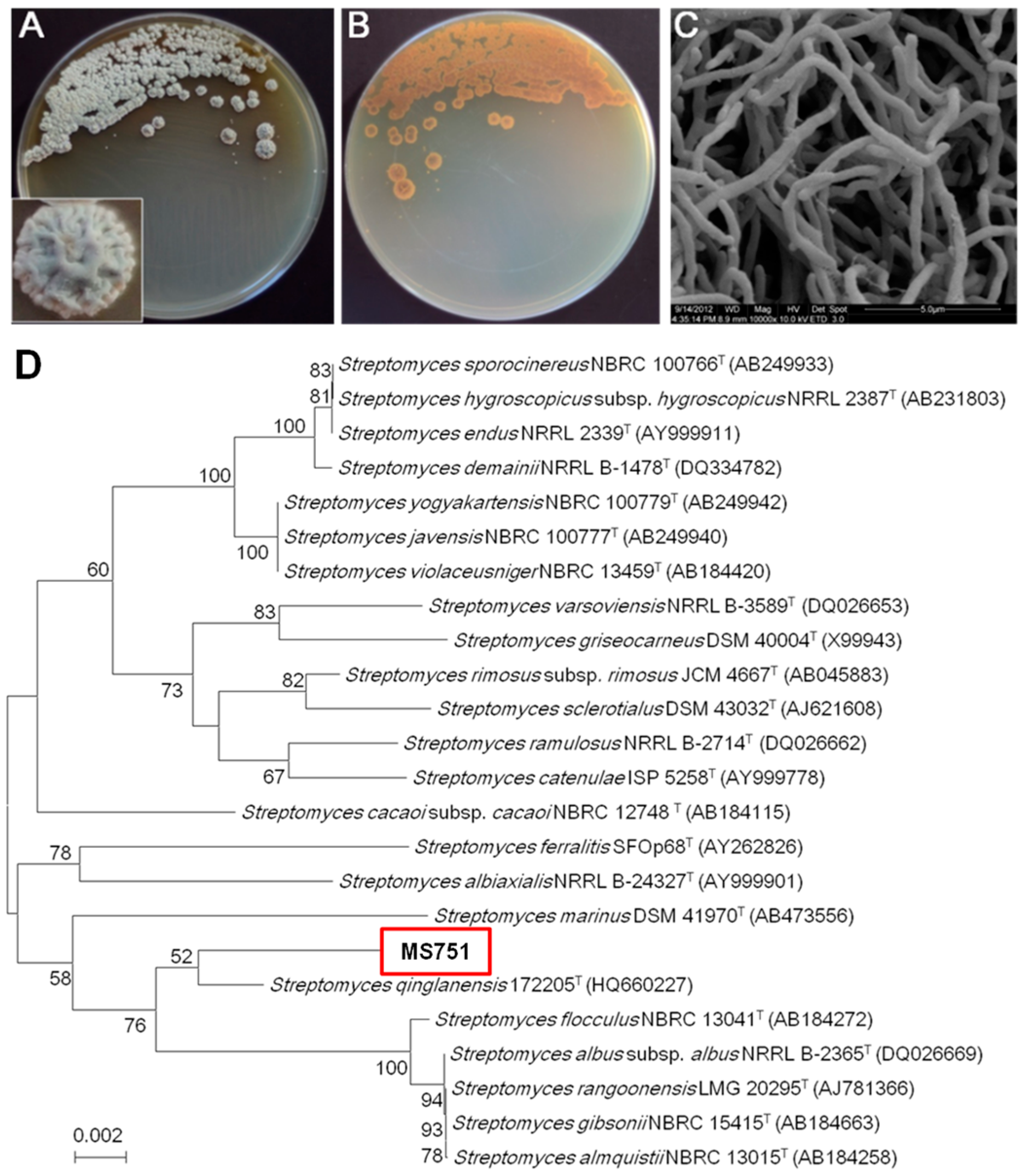
2.1. Identification, Fermentation of Strain MS751, and Purification of New Chrysomycins
2.2. Structure Elucidation of New Chrysomycins

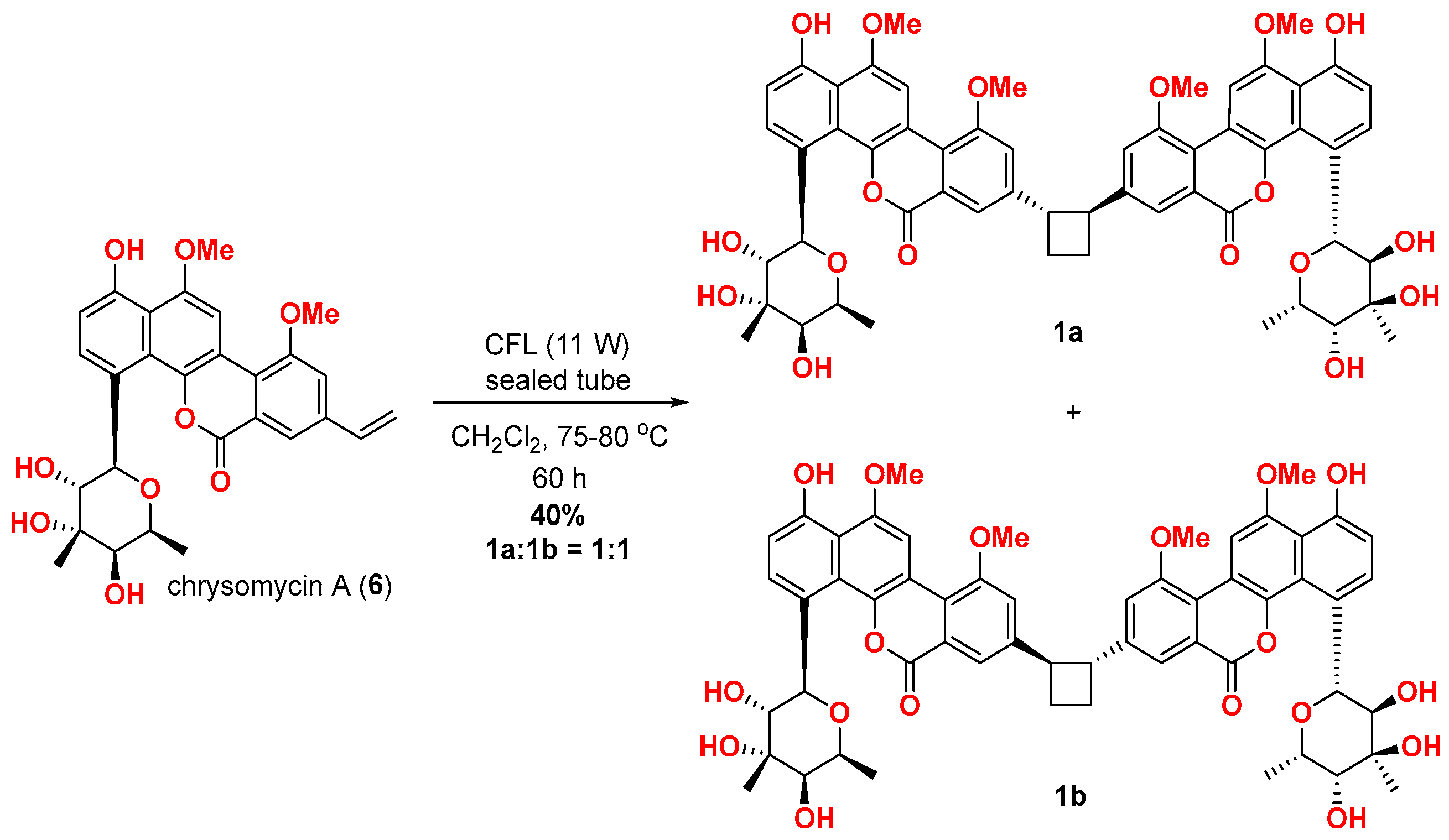
2.3. Biomimetic [2 + 2] Photodimerisation of Chrysomycin A
2.4. Anti-Tuberculosis Activity Evaluation
3. Discussion
4. Materials and Methods
4.1. General Experimental Section
4.2. Characterisation of Streptomyces sp. Strain MS751
4.3. Cultivation, Extraction, and Compounds Purification
4.4. Synthesis, Isolation, and Characterisation of trans-Dimers 1a and 1b
4.5. Bioassays for Purified Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Zumla, A.; Memish, Z.A.; Maeurer, M.; Bates, M.; Mwaba, P.; Al-Tawfiq, J.A.; Denning, D.W.; Hayden, F.G.; Hui, D.S. Emerging novel and antimicrobial-resistant respiratory tract infections: New drug development and therapeutic options. Lancet Infect. Dis. 2014, 14, 1136–1149. [Google Scholar] [CrossRef]
- Bloemberg, G.V.; Keller, P.M.; Stucki, D.; Trauner, A.; Borrell, S.; Latshang, T.; Coscolla, M.; Rothe, T.; Hömke, R.; Ritter, C.; et al. Acquired Resistance to Bedaquiline and Delamanid in Therapy for Tuberculosis. N. Engl. J. Med. 2015, 373, 1986–1988. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Wang, Q.; Song, F.; Xiao, X.; Huang, P.; Li, L.; Monte, A.; Abdel-Mageed, W.M.; Wang, J.; Guo, H.; He, W.; et al. Abyssomicins from the South China Sea deep-sea sediment Verrucosispora sp.: Natural thioether Michael addition adducts as antitubercular prodrugs. Angew. Chem. Int. Ed. 2013, 52, 1231–1234. [Google Scholar] [CrossRef]
- Song, F.; Liu, X.; Guo, H.; Ren, B.; Chen, C.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M.; et al. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Guo, H.; Hou, W.; Yang, N.; Ren, B.; Liu, M.; Dai, H.; Liu, X.; Song, F.; et al. Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 2013, 97, 3885–3892. [Google Scholar] [CrossRef]
- Liu, M.; Abdel-Mageed, W.M.; Ren, B.; He, W.; Huang, P.; Li, X.; Bolla, K.; Guo, H.; Chen, C.; Song, F.; et al. Endophytic Streptomyces sp. Y3111 from traditional Chinese medicine produced antitubercular pluramycins. Appl. Microbiol. Biotechnol. 2014, 98, 1077–1085. [Google Scholar] [CrossRef]
- Jain, S.K.; Pathania, A.S.; Parshad, R.; Raina, C.; Ali, A.; Gupta, A.P.; Kushwaha, M.; Aravinda, S.; Bhushan, S.; Bharate, S.B.; et al. Chrysomycins A–C, antileukemic naphthocoumarins from Streptomyces sporoverrucosus. RSC Adv. 2013, 3, 21046–21053. [Google Scholar] [CrossRef]
- Strelitz, F.; Flon, H.; Asheshov, I.N. Chrysomycin: A new antibiotic substance for bacterial viruses. J. Bacteriol. 1955, 69, 280–283. [Google Scholar] [CrossRef]
- Crimmins, M.T. Synthetic applications of intramolecular enone-olefin photocycloadditions. Chem. Rev. 1988, 88, 1453–1473. [Google Scholar] [CrossRef]
- Lee-Ruff, E.; Mladenova, G. Enantiomerically pure cyclobutane derivatives and their use in organic synthesis. Chem. Rev. 2003, 103, 1449–1483. [Google Scholar] [CrossRef]
- Poplata, S.; Tröster, A.; Zou, Y.-Q.; Bach, T. Recent advances in the synthesis of cyclobutanes by olefin [2 + 2] photocycloaddition reactions. Chem. Rev. 2016, 116, 9748–9815. [Google Scholar] [CrossRef]
- Iriondo-Alberdi, J.; Greaney, M.F. Photocycloaddition in natural product synthesis. Eur. J. Org. Chem. 2007, 2007, 4801–4815. [Google Scholar] [CrossRef]
- Bach, T.; Hehn, J.P. Photochemical reactions as key steps in natural product synthesis. Angew. Chem. Int. Ed. 2011, 50, 1000–1045. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Song, F.; Wang, S.; Guo, H.; Wei, Q.; Dai, H.; Chen, X.; Xia, X.; Liu, X.; et al. Chrysomycin A Derivatives for the Treatment of Multi-Drug-Resistant Tuberculosis. ACS Cent. Sci. 2020, 6, 928–938. [Google Scholar] [CrossRef]
- Fischer, C.; Lipata, F.; Rohr, J. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J. Am. Chem. Soc. 2003, 125, 7818–7819. [Google Scholar] [CrossRef]
- Shepherd, M.D.; Liu, T.; Méndez, C.; Salas, J.A.; Rohr, J. Engineered biosynthesis of gilvocarcin analogues with altered deoxyhexopyranose moieties. Appl. Environ. Microbiol. 2011, 77, 435–441. [Google Scholar] [CrossRef]
- Pahari, P.; Kharel, M.K.; Shepherd, M.D.; van Lanen, S.G.; Rohr, J. Enzymatic total synthesis of defucogilvocarcin M and its implications for gilvocarcin biosynthesis. Angew. Chem. Int. Ed. 2012, 51, 1216–1220. [Google Scholar] [CrossRef]
- Weiss, U.; Yoshihira, K.; Highet, R.J.; White, R.J.; Wei, T.T. The chemistry of the antibiotics chrysomycin A and B. Antitumor activity of chrysomycin A. J. Antibiot. 1982, 35, 1194–1201. [Google Scholar] [CrossRef]
- Liu, D.-N.; Liu, M.; Zhang, S.-S.; Shang, Y.-F.; Song, F.-H.; Zhang, H.-W.; Du, G.-H.; Wang, Y.-H. Chrysomycin A inhibits the proliferation, migration and invasion of U251 and U87-MG glioblastoma cells to exert its anti-cancer effects. Molecules 2022, 27, 6148. [Google Scholar] [CrossRef]
- Xu, Z.; Zheng, S.; Gao, X.; Hong, Y.; Cai, Y.; Zhang, Q.; Xiang, J.; Xie, D.; Song, F.; Zhang, H.; et al. Mechanochemical preparation of chrysomycin A self-micelle solid dispersion with improved solubility and enhanced oral bioavailability. J. Nanobiotechnology 2021, 19, 164. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, S.-S.; Liu, D.-N.; Yang, Y.-L.; Wang, Y.-H.; Du, G.-H. Chrysomycin A attenuates neuroinflammation by down-regulating NLRP3/cleaved caspase-1 signaling pathway in LPS-stimulated mice and BV2 cells. Int. J. Mol. Sci. 2021, 22, 6799. [Google Scholar] [CrossRef]
- Liu, H.; Cai, Y.; Chu, Y.; Yu, X.; Song, F.; Wang, H.; Zhang, H.; Sun, X. Formulation of chrysomycin A cream for the treatment of skin infections. Molecules 2022, 27, 4613. [Google Scholar] [CrossRef]
- Muralikrishnan, B.; Dan, V.M.; Vinodh, J.; Jamsheena, V.; Ramachandran, R.; Thomas, S.; Dastager, S.G.; Kumar, K.S.; Lankalapalli, R.S.; Kumar, R.A. Anti-microbial activity of chrysomycin A produced by Streptomyces sp. against Mycobacterium tuberculosis. RSC Adv. 2017, 7, 36335–36339. [Google Scholar] [CrossRef]
- Hui, C.; Wang, Z.; Xie, Y.; Liu, J. Contemporary synthesis of bioactive cyclobutane natural products. Green Synth. Catal. 2023, 4, 1–6. [Google Scholar] [CrossRef]
- Yang, P.; Jia, Q.; Song, S.; Huang, X. [2 + 2]-Cycloaddition-derived cyclobutane natural products: Structural diversity, sources, bioactivities, and biomimetic syntheses. Nat. Prod. Rep. 2023, 40, 1094–1129. [Google Scholar] [CrossRef]
- Hong, Y.J.; Tantillo, D.J. How cyclobutanes are assembled in nature--insights from quantum chemistry. Chem. Soc. Rev. 2014, 43, 5042–5050. [Google Scholar] [CrossRef]
- Ma, R.; Cheng, S.; Sun, J.; Zhu, W.; Fu, P. Antibacterial Gilvocarcin-Type Aryl-C-Glycosides from a Soil-Derived Streptomyces Species. J. Nat. Prod. 2022, 85, 2282–2289. [Google Scholar] [CrossRef]
- Bayma, J.D.; Arruda, M.S.; Müller, A.H.; Arruda, A.C.; Canto, W.C. A dimeric ArC2 compound from Peperomia pellucida. Phytochemistry 2000, 55, 779–782. [Google Scholar] [CrossRef]
- Fan, Y.-Y.; Gao, X.-H.; Yue, J.-M. Attractive natural products with strained cyclopropane and/or cyclobutane ring systems. Sci. China Chem. 2016, 59, 1126–1141. [Google Scholar] [CrossRef]
- Woods, G.L.; Brown-Elliott, B.A.; Conville, P.S.; Desmond, E.P.; Hall, G.S.; Lin, G.; Pfyffer, G.E.; Ridderhof, J.C.; Siddiqi, S.H.; Wallace, R.J.; et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011. [Google Scholar]
- Pfaller, M.A.; Chaturvedi, V.; Espinel-Ingroff, A.; Ghannoum, M.A.; Odds, F.C. Reference method for broth dilution antifungal susceptibility testing of yeasts: Approved standard-second edition. CLSI document M27-A2 (ISBN 1-56238-469-4). Clin. Lab. Stand. Inst. 2008, 22, 1–51. [Google Scholar]
| Position | δH,a mult (J in Hz) | δC b | HMBC (1H→13C) |
|---|---|---|---|
| 1/1’ | 153.2, C | ||
| 2/2’ | 6.96, d (8.4) | 112.0, CH | 1/1’, 4/4’, 12a/12a’ |
| 3/3’ | 7.83, d (8.4) | 129.3, CH | 1/1’, 4a/4a’, 13/13’ |
| 4/4’ | 128.1, C | ||
| 4a/4a’ | 125.2, C | ||
| 4b/4b’ | 142.3, C | ||
| 6/6’ | 159.9, C | ||
| 6a/6a’ | 121.9, C | ||
| 7/7’ | 7.88/7.89, d (1.2) | 119.0/118.9, CH | 6/6’, 9/9’, 10a/10a’, 18/18’ |
| 8/8’ | 146.2, C | ||
| 9/9’ | 7.58/7.61, d (1.2) | 116.5, CH | 7/7’, 10/10’, 10a/10a’, 18/18’ |
| 10/10’ | 157.4/157.3, C | ||
| 10a/10a’ | 121.8, C | ||
| 10b/10b’ | 113.3, C | ||
| 11/11’ | 8.49, s | 101.6, CH | 4b/4b’, 10a/10a’, 10b/10b’, 12/12’, 12a/12a |
| 12/12’ | 151.9, C | ||
| 12a/12a’ | 115.1, C | ||
| 13/13’ | 6.01, d (9.6) | 74.6, CH | 3/3’, 4/4’, 4a/4a’, 14/14’, 17/17’ |
| 14/14’ | 3.66, dd (9.6, 8.4) | 72.6, CH | 13/13’ |
| 15/15’ | 73.1, C | ||
| 16/16’ | 3.13, d (7.8) | 75.8, CH | 15/15’ |
| 17/17’ | 4.50, brq (6.0) | 70.7, CH | 13/13’, 16/16’, 23/23’ |
| 18/18’ | 3.96, m | 47.0/46.9, CH | 8/8’, 19/19’ |
| 19a/19’a | 2.33, m | 25.4/25.5, CH2 | 18/18’ |
| 19b/19’b | 2.45, m | 18/18’ | |
| 20/20’ | 4.15/4.14, s | 56.7/56.7, CH3 | 10/10’ |
| 21/21’ | 4.11, s | 56.3, CH3 | 12/12’ |
| 22/22’ | 1.24, s | 23.9, CH3 | 14/14’, 15/15’, 16/16’ |
| 23/23’ | 1.00/1.01, d (6.6) | 17.1, CH3 | 16/16’, 17/17’ |
| 1/1’-OH | 9.81, s | 1/1’, 2/2’, 12a/12a’ | |
| 14/14’-OH | 4.16, overlap (8.4) | ||
| 15/15’-OH | 4.18, s | 14/14’, 16/16’ | |
| 16/16’-OH | 4.57, d (7.8) | 16/16’ |
| Position | Chrysomycin G (2) | Chrysomycin H (3) | Chrysomycin I (4) | Chrysomycin J (5) | ||||
|---|---|---|---|---|---|---|---|---|
| δH,a mult (J in Hz) | δC b | δH,a mult (J in Hz) | δC b | δH,a mult (J in Hz) | δC b | δH,a mult (J in Hz) | δC b | |
| 1 | 153.2, C | 153.2, C | 153.3, C | 153.2, C | ||||
| 2 | 6.97, d (8.4) | 112.0, CH | 6.96, d (8.4) | 111.9, CH | 6.96, d (8.4) | 112.1, CH | 6.97, d (8.4) | 112.0, CH |
| 3 | 7.84, d (8.4) | 129.3, CH | 7.83, d (8.4) | 129.3, CH | 7.83, d (8.4) | 129.4, CH | 7.84, d (8.4) | 129.3, CH |
| 4 | 128.0, C | 128.0, C | 128.1, C | 128.1, C | ||||
| 4a | 125.2, C | 125.2, C | 125.3, C | 125.2, C | ||||
| 4b | 142.3, C | 142.2, C | 142.4, C | 142.3, C | ||||
| 6 | 160.0, C | 160.0, C | 160.2, C | 159.9, C | ||||
| 6a | 121.7, C | 121.5, C | 121.7, C | 121.6, C | ||||
| 7 | 7.96, s | 118.4, CH | 7.84, s | 121.4 | 7.95, d (1.8) | 117.7, CH | 7.80, d (1.2) | 122.4, CH |
| 8 | 145.4, C | 142.7, C | 150.1, C | 137.6, C | ||||
| 9 | 7.60, s | 115.8, CH | 7.55, d (1.2) | 119.1 | 7.61, d, (1.8) | 115.4, CH | 7.49, d (1.2) | 119.7, CH |
| 10 | 157.1, C | 156.9, C | 157.2, C | 156.9, C | ||||
| 10a | 121.9, C | 121.4, C | 122.0, C | 121.9, C | ||||
| 10b | 113.4, C | 113.4, C | 113.5, C | 113.2, C | ||||
| 11 | 8.52, s | 101.7, CH | 8.50, s | 101.7, CH | 8.49, s | 101.8, CH | 8.48, s | 101.6, CH |
| 12 | 151.9, C | 151.9, C | 152.0, C | 151.8, C | ||||
| 12a | 115.1, C | 115.1, C | 115.2, C | 115.1, C | ||||
| 13 | 6.03, d (9.6) | 74.6, CH | 6.03, d (9.6) | 74.6, CH | 6.02, d (9.6) | 74.7, CH | 6.03, d (9.6) | 74.6, CH |
| 14 | 3.69, dd (9.6, 8.4) | 72.5, CH | 3.67, dd (9.6, 8.4) | 72.6, CH | 3.68, dd (9.6, 8.4) | 72.7, CH | 3.67, dd (9.6, 8.4) | 72.6, CH |
| 15 | 73.1, C | 73.1, C | 73.3, C | 73.1, C | ||||
| 16 | 3.15, d (7.8) | 75.8, CH | 3.14, d (7.8) | 75.8, CH | 3.15, d (7.8) | 75.9, CH | 3.14, d (7.8) | 75.8, CH |
| 17 | 4.52, q (6.6) | 70.7, CH | 4.51, q (6.6) | 70.7, CH | 4.52, q (6.6) | 70.8, CH | 4.51, q (6.6) | 70.7, CH |
| 18 | 4.70, d (5.4) | 62.2, CH2 | 2.93, t (6.6) | 38.7, CH2 | 4.92, qd (6.6, 4.8) | 67.7, CH | 4.04, s | 49.0, CH2 |
| 19 | 3.75, td (6.6, 5.4) | 61.5, CH2 | 1.43, d (6.6) | 25.7, CH3 | 205.3, C | |||
| 19’ | 2.23, s | |||||||
| 20 | 4.13, s | 56.6, CH3 | 4.13, s | 56.7, CH3 | 4.13, s | 56.8, CH3 | 4.10, s | 29.8, CH3 |
| 21 | 4.13, s | 56.4, CH3 | 4.13, s | 56.4, CH3 | 4.11, s | 56.5, CH3 | 4.12, s | 56.7, CH3 |
| 22 | 1.26, s | 23.9, CH3 | 1.25, s | 23.9, CH3 | 1.25, s | 24.0, CH3 | 1.26, s | 56.3, CH3 |
| 23 | 1.01, d (6.6) | 17.1, CH3 | 1.01, d (6.6) | 17.1, CH3 | 1.01, d (6.6) | 17.2, CH3 | 1.02, d (6.6) | 23.9, CH3 |
| 1-OH | 9.83, s | 9.82, s | 9.82, s | 9.82, s | 17.1, CH3 | |||
| 14-OH | 4.17, d (8.4) | 4.17, d (8.4) | 4.19, d (8.4) | 4.18, d (8.4) | ||||
| 15-OH | 4.20, s | 4.19, s | 4.22, s | 4.20, s | ||||
| 16-OH | 4.57, d (7.8) | 4.57, d (7.8) | 4.63, d (7.8) | 4.58, d (7.8) | ||||
| 18-OH | 5.54, t (5.4) | 5.55, d (4.8) | ||||||
| 19-OH | 4.75, t (5.4) | |||||||
| Microorganism (Strain) | Minimum Inhibitory Concentration (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 7 | 8 | Control | |
| M. bovis BCG | 50.0 | 12.5 | >100 | >100 | >100 | 6.25 | 3.13 | 0.05 [a] |
| M. tuberculosis H37Rv | >100 | 50.0 | >100 | >100 | >100 | 1.56 | 1.56 | 0.02 [b] |
| M. tuberculosis (Hr1) | >100 | >100 | >100 | >100 | >100 | 1.56 | 1.56 | 1.00 [b] |
| M. tuberculosis (Hr2) | >100 | >100 | >100 | >100 | >100 | 1.56 | 1.56 | 2.00 [b] |
| M. tuberculosis (Hr3) | >100 | >100 | >100 | >100 | >100 | 3.12 | 1.56 | 0.50 [b] |
| M. tuberculosis (Hr4) | >100 | >100 | >100 | >100 | >100 | 1.56 | 3.12 | 1.00 [b] |
| M. tuberculosis (Hr5) | >100 | >100 | >100 | >100 | >100 | 3.12 | 1.56 | 2.00 [b] |
| M. smegmatis mc2155 | 100 | >100 | >100 | >100 | >100 | 1.56 | 3.13 | 3.13 [a] |
| MRSA | >25.0 | >100 | >100 | >100 | >100 | 6.25 | 6.25 | 1.00 [c] |
| S. aureus ATCC 6538 | >25.0 | >100 | >100 | >100 | >100 | 3.13 | 6.25 | 1.00 [c] |
| S. pneumoniae ATCC 49619 | >100 | 50.0 | >100 | >100 | >100 | 3.13 | 25.0 | 5.00 [d] |
| C. albicans | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 0.02 [e] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Guo, H.; Zhang, J.; Hu, J.; He, H.; Chen, C.; Yang, N.; Yang, F.; Lin, Z.; Dai, H.; et al. Chrysomycins, Anti-Tuberculosis C-Glycoside Polyketides from Streptomyces sp. MS751. Mar. Drugs 2024, 22, 259. https://doi.org/10.3390/md22060259
Yu J, Guo H, Zhang J, Hu J, He H, Chen C, Yang N, Yang F, Lin Z, Dai H, et al. Chrysomycins, Anti-Tuberculosis C-Glycoside Polyketides from Streptomyces sp. MS751. Marine Drugs. 2024; 22(6):259. https://doi.org/10.3390/md22060259
Chicago/Turabian StyleYu, Jiaming, Hui Guo, Jing Zhang, Jiansen Hu, Hongtao He, Caixia Chen, Na Yang, Fan Yang, Zexu Lin, Huanqin Dai, and et al. 2024. "Chrysomycins, Anti-Tuberculosis C-Glycoside Polyketides from Streptomyces sp. MS751" Marine Drugs 22, no. 6: 259. https://doi.org/10.3390/md22060259
APA StyleYu, J., Guo, H., Zhang, J., Hu, J., He, H., Chen, C., Yang, N., Yang, F., Lin, Z., Dai, H., Ouyang, L., Liu, C., Lei, X., Zhang, L., Zhu, G., & Song, F. (2024). Chrysomycins, Anti-Tuberculosis C-Glycoside Polyketides from Streptomyces sp. MS751. Marine Drugs, 22(6), 259. https://doi.org/10.3390/md22060259







