Abstract
Six new lipids, trichoderols B-G (1–6), along with a known one, triharzianin B (7), were isolated from the culture of Trichoderma sp. Z43 obtained from the surface of the marine brown alga Dictyopteris divaricata. Their structures and relative configurations were identified by interpretation of 1D/2D NMR and MS data. Compounds 1–7 were assayed for inhibiting the growth of three phytopathogenic fungi (Fusarium graminearum, Gaeumannomyces graminis, and Glomerella cingulata), four marine phytoplankton species (Amphidinium carterae, Heterocapsa circularisquama, Heterosigma akashiwo, and Prorocentrum donghaiense), and one marine zooplankton (Artemia salina). Compounds 1, 4, and 7 exhibited weak antifungal activities against three phytopathogenic fungi tested with MIC ≥ 64 μg/mL. All compounds displayed moderate antimicroalgal activity with IC50 ≥ 15 μg/mL and low toxicity to the brine shrimp Artemia salina.
1. Introduction
Crop diseases caused by pathogenic fungus have seriously restricted healthy development of agriculture in the world. Fusarium head blight of wheat, take-all disease, and plant anthracnose caused by Fusarium graminearum, Gaeumannomyces graminis, and Glomerella cingulata, respectively, lead to huge economic losses in agriculture every year [1,2,3,4,5]. In addition, some marine phytoplankton species including Amphidinium carterae, Heterocapsa circularisquama, Heterosigma akashiwo, and Prorocentrum donghaiense can induce red tides that harm aquaculture industry [6,7,8,9]. Thus, it is imperative to search for natural antifungal and antimicroalgal drugs with high activity and safety. On the other hand, Trichoderma species are known for producing various metabolites with novel structures and intriguing bioactivities [10,11,12]. In recent years, natural products originated from marine-derived Trichoderma species have gained the attention of researchers [13], and a large number of new compounds with significant activities have been found, such as polyketides [14], terpenoids [15,16,17], and steroids [18]. Although Trichoderma species have proven to be a treasure-house of new natural products, only a few short-chain lipids have been isolated from this species. For example, triharzianins A-D were purified from T. harzianum obtained from mushroom Tricholoma matsutake [19], harzianumols A-H were separated from T. harzianum obtained from sponge Petrospongia nigra [20], and trichoderol A was acquired from Trichoderma sp. obtained from soil [21]. Moreover, these lipids displayed multifarious bioactivities such as feeding attractants and antimicrobial activities [19,21]. During our investigation of the chemical diversity and biological activity of marine-derived Trichoderma, the epiphytic Trichoderma sp. Z43 obtained from the marine brown alga Dictyopteris divaricata was examined. Its aerial mycelia grew rapidly. Most of them were white on the PDA medium at 25 °C, and some parts of the plate were yellow. Alternate permutation of the mycelia was observed through a common microscope. As a result, six new lipids, trichoderols B-G (1–6), along with a known one, triharzianin B (7), were isolated and identified. Herein, the details of isolation, structure elucidation, and bioactivities of these compounds are described.
2. Results and Discussion
The organic extracts of marine-derived Trichoderma sp. Z43 isolated from the marine brown alga Dictyopteris divaricata were subjected to a series of column chromatography processes to produce six new lipids, namely, trichoderols B-G (1–6), along with a known one, triharzianin B (7) (Figure 1). Compound 7 was unambiguously identified by comparing its NMR data and specific rotation value with those reported in the literature [19].
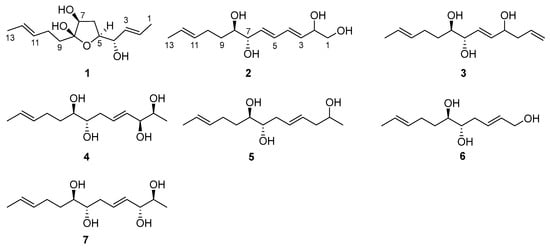
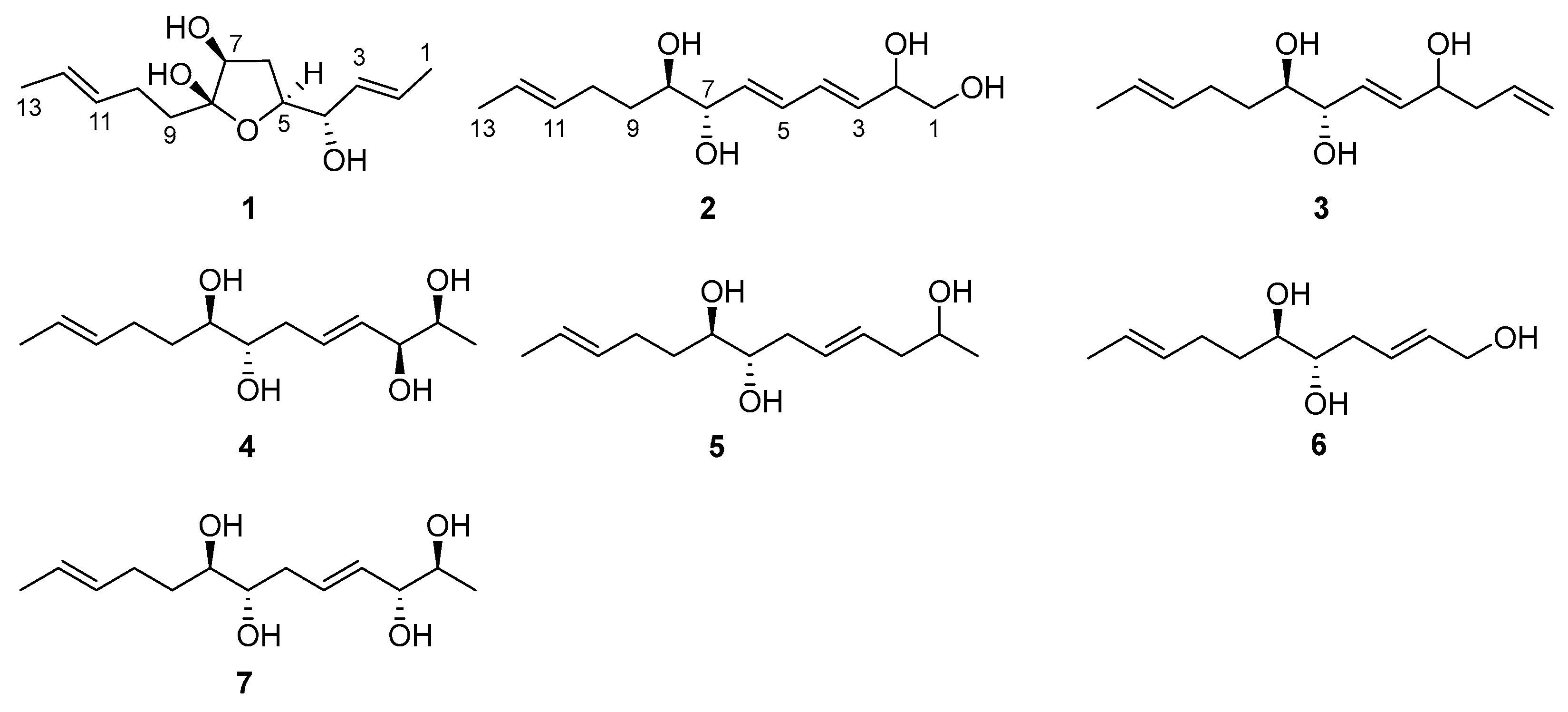
Figure 1.
Chemical structures of 1–7.
2.1. Structural Elucidation
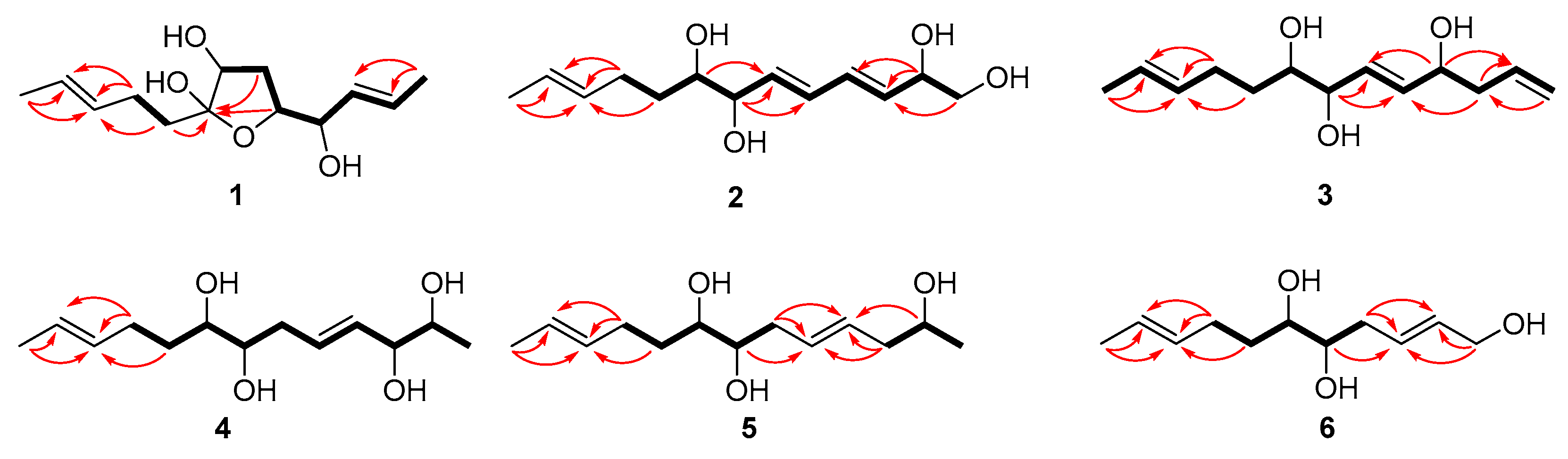
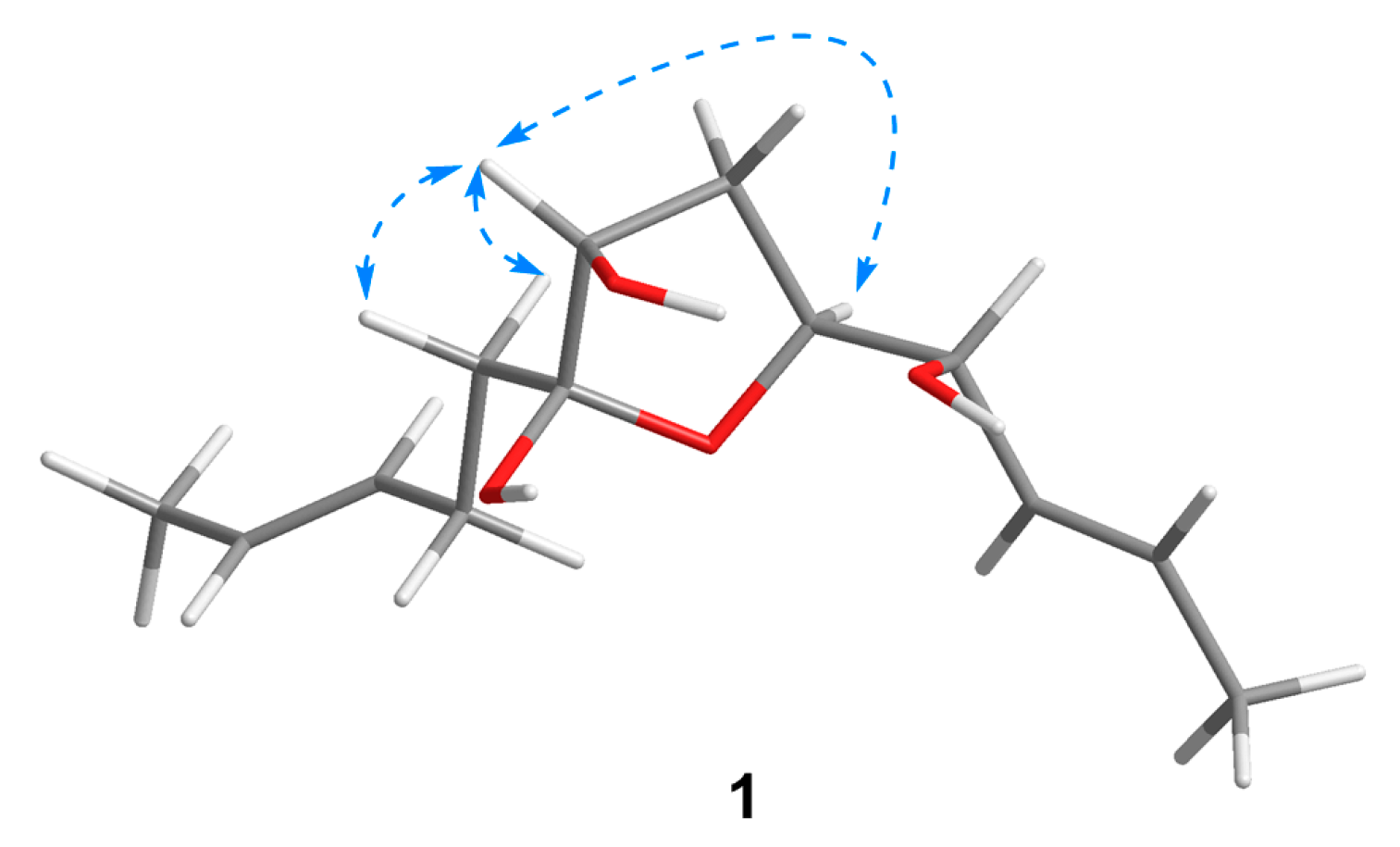
Compound 1 was isolated as a colorless oil with a molecular formula of C13H22O4 established by HRESIMS (m/z 265.1420 [M + Na]+), implying three degrees of unsaturation. The 1H NMR spectrum (Table 1) showed two methyl doublets, one double quartet, one double doublet, and two multiplets assignable to four olefinic protons and one double multiplet and two double doublets attributable to three oxymethines. The 13C NMR and DEPT spectra (Table 2) displayed the presence of two methyls, three methylenes, seven methines, and one nonprotonated carbon. COSY correlations of H-1/H-2/H-3/H-4/H-5/H-6/H-7 established the linkage of C-1 to C-7 (Figure 2). A pentenyl group was confirmed by the COSY correlation of H-9 with H-10 and the HMBC correlations from H-10/H-13 to C-11 and C-12 and from H-9 to C-11, which was then elongated to C-7 via C-8 by the HMBC correlations from H-6/H-9 to C-8. To satisfy the molecular formula, an ether linkage was situated between C-5 and C-8, which was verified by the HMBC correlation from H-5 to C-8 (Figure 2). Thus, the planar structure of 1 was validated. The double bond at C-2 was attributed to be trans by the large coupling constant (J = 15.2) between H-2 and H-3. The chemical shifts of other two olefinic carbon atoms (C-11, δC 130.9; C-12, δC 125.5) were highly similar to those of triharzianin B (7) (C-11, δC 130.9; C-12, δC 124.6) [19], suggesting a trans configuration of the C-11 double bond, which was further verified by the IR absorption at 969 cm−1. The relative configurations of H-5, OH-7, and OH-8 were confirmed by NOESY correlations of H-7 with H-5 and H-9 (Figure 3), and the relationship of H-4 and H-5 was deduced to be threo due to NMR data that were similar to those of (-)-(S)-1-[(2S,5S)-5-[2-propenyl]tetrahydrofuran-2-yl]prop-2-en-1-ol [22].

Table 1.
1H NMR data for 1–6 (δ in ppm, J in Hz).

Table 2.
13C NMR data for 1–6 (δ in ppm).
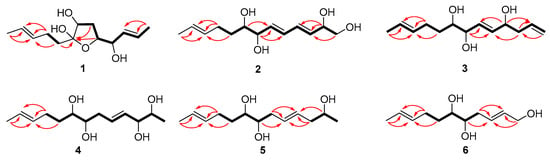
Figure 2.
Key COSY and HMBC correlations of 1–6 (bold lines for COSY and arrows for HMBC).
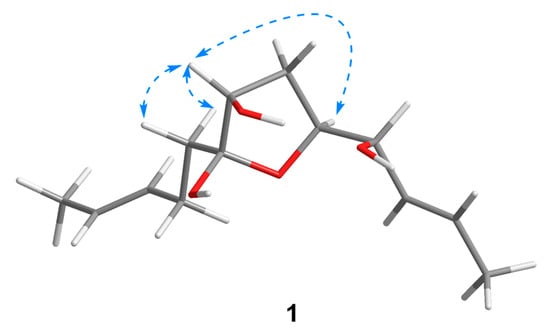
Figure 3.
Key NOESY correlations of 1.
Compound 2 was purified as a colorless oil and assigned a molecular formula of C13H22O4 by interpretation of HRESIMS (m/z 265.1420 [M + Na]+) data. In conjunction with HSQC data, the 1H NMR spectrum (Table 1) revealed notable signals including one methyl doublet, one double triplet, one double doublet, and one doublet of double doublets ascribable to three oxygenated methines, two double doublets attributable to an oxygenated methylene, and four double doublets and two multiplets assignable to six olefinic protons. DEPT experiments displayed 13 resonances in the 13C NMR spectrum, which were assigned to one methyl, three methylenes, and nine methines. COSY correlations of H-1/H-2/H-3/H-4/H-5/H-6/H-7/H-8/H-9/H-10 established the linkage of C-1 to C-10, which was further confirmed by the HMBC correlations from H-1 to C-3, from H-2 to C-3 and C-4, from H-7 to C-5 and C-6, and from H-8 to C-6 (Figure 2). A propenyl group was located at C-10, verified by the HMBC correlations from H-9 to C-11 and from H-10/H-13 to C-11 and C-12 (Figure 2). Thus, the planar structure of 2 was affirmed. The double bonds at C-3 and C-5 were determined to be trans by the large coupling constants (J = 13.1), and the identical 13C NMR data of 2 (C-11, δC 132.2; C-12, δC 126.0) with those of triharzianin C (C-11, δC 132.2; C-12, δC 126.0) [19] suggested the double bond at C-11 to be trans. The relationship of OH-7 and OH-8 was assigned to be erythro by comparison of NMR data with those for triharzianin C [19]. Although the chemical shifts of C-1, C-2, and C-3 in 2 were the same as those of (S)-but-3-ene-1,2-diol [23], the relative configuration of OH-2 could not be confirmed due to the existence of only one chiral carbon atom (C-2) in this moiety. Thus, compound 2 was named (3E,5E,11E)-trideca-3,5,11-trien-1,2,7,8-tetraol, and its structural formula was HOCH2CH(OH)CH=CHCH=CHCH(OH)CH(OH)CH2CH2CH=CHCH3.
Compound 3 was acquired as a colorless oil, and its molecular formula was determined to be C13H22O3 by HRESIMS (249.1448 [M + Na]+) data. The 1H NMR spectrum, along with HSQC data, exhibited notable signals including one methyl doublet, one double triplet, one double doublet, one doublet of double doublets assignable to three oxygenated methines, two broad singlets attributable to a terminal double bond, and three multiplets and two doublets of double doublets ascribable to five olefinic protons. The 13C NMR spectrum displayed 13 resonances, sorted into one methyl, four methylenes, and eight methines. The 1H and 13C NMR data (Table 1 and Table 2) revealed the presence of a similar skeleton to 2. COSY correlations of H-1/H-2/H-3/H-4/H-5/H-6/H-7/H-8/H-9/H-10/H-11/H-12/H-13 and HMBC correlations from H-1 to C-3, from H-3 to C-2 and C-5, from H-4 to C-2 and C-6, from H-7 to C-5 and C-6, from H-10/H-13 to C-11 and C-12, and from H-9 to C-11 (Figure 2) confirmed the planar structure of 3. The double bonds at C-5 and C-11 were deduced to be trans by the large coupling constant (J = 15.5) between H-5 and H-6 and by the identical NMR data of C-11 and C-12 with those of 2. The relative configurations of OH-7 and OH-8 were determined to be the same as those of 2 on the basis of highly similar chemical shifts and coupling constants of them. Despite the similar NMR data of C-1 to C-5 in 3 with those of 12(S)-hydroxy-5(Z),8(Z),10(E),14(E)-eicosatetraenoic acid [24], the relative configuration of OH-4 was still unsolved. Compound 3 was named (5E,11E)-trideca-1,5,11-trien-4,7,8-triol, and its structural formula was CH2=CHCH2CH(OH)CH=CHCH(OH)CH(OH)CH2CH2CH=CHCH3.
Compound 4 was obtained as a colorless oil and given a molecular formula of C13H24O4 by interpretation of HRESIMS data (m/z 243.1558 [M]-), requiring two degrees of unsaturation. Its NMR data (Table 1 and Table 2) were highly similar to those of triharzianin B (7) [19], except for chemical shifts of C-2 and C-3 (C-2, δC 70.2; C-3, δC 76.6 for triharzianin B; C-2, δC 71.6; C-3, δC 77.9 for 4), which indicated that 4 and triharzianin B possessed the same planar structure. COSY correlations of H-1/H-2/H-3/H-4/H-5/H-6/H-7/H-8/H-9/H-10 and HMBC correlations from H-9 to C-11 and from H-10/H-13 to C-11 and C-12 (Figure 2) further afforded to this structure. The geometry of two double bonds at C-2 and C-11 and the relative configurations of OH-7 and OH-8 were deduced to be the same as those of triharzianin B (7) [19] on the basis of their identical NMR data. The relationship of H-2 and H-3 was threo by analysis of NMR data with those for separacenes A and B [25]. Therefore, compound 4 was named (4E,11E)-trideca-4,11-diene-2,3,7,8-tetraol, and its structural formula was CH3CH(OH)CH(OH)CH=CHCH2CH(OH)CH(OH)CH2CH2CH=CHCH3.
Compound 5 was isolated as a colorless oil. HRESIMS analysis gave the molecular formula of C13H24O3, consistent with two degrees of unsaturation. Its NMR data (Table 1 and Table 2) resembled those of 4, except for the presence of signals for a methylene and the lack of signals for a hydroxymethine group. COSY correlations of H-1/H-2/H-3 confirmed the kinkage from C-1 to C-3, which was then elongated to C-10 by the HMBC correlations from H-2 to C-4, from H-3/H-6 to C-4 and C-5, and from H-7 to C-5 and the COSY correlations of H-6/H-7/H-8/H-9/H-10. A propenyl group was situated at C-10 by the HMBC correlations from H-9 to C-11 and from H-10/H-13 to C-11 and C-12 (Figure 2). Thus, the planar structure of 5 was confirmed. The configurations of OH-7, OH-8, and the double bond at C-11 were the same as those of 4 due to their similar NMR data, and the geometry of double bond at C-4 was deduced to be trans by the large coupling constant (J = 15.4) between H-4 and H-5. The relative configuration of OH-2 was uncertain in spite of comparing the NMR data of 5 with those of (S)-2-hexanol and (R)-octan-2-ol carefully [26]. Thus, compound 5 was named (4E,11E)-trideca-4,11-dien-2,7,8-triol, and its structural formula was CH3CH(OH)CH2CH=CHCH2CH(OH)CH(OH)CH2CH2CH=CHCH3.
Compound 6 was obtained as a colorless oil and was given a molecular formula of C11H20O3 by analysis of HRESIMS data, requiring two degrees of unsaturation. Its NMR data exhibited high similarities to those of 4 except for the presence of signals for a hydroxymethylene group and lack of signals for a methyl and two hydroxymethine groups. COSY correlations of H-1/H-2/H-3/H-4/H-5/H-6/H-7/H-8 determined the linkage of C-1 to C-8, and a propenyl group was located at C-8, confirmed by the HMBC correlations from H-7 to C-9 and from H-8/H-11 to C-9 and C-10 (Figure 2). Other HMBC correlations further verified the structure of 6 (Figure 2). The large coupling constant (J = 15.4) between H-2 and H-3 demonstrated that the double bond at C-2 was trans. The identical NMR data of the spin system in 6 (from C-2 to C-11) and 4 (from C-4 to C-13) (Table 1 and Table 2) ascertained the trans configuration of a double bond at C-9 and the erythro relationship of the vicinal diol (OH-5 and OH-6). Compound 6 was named (2E,9E)-undeca-2,9-dien-1,5,6-triol, and its structural formula was HOCH2CH=CHCH2CH(OH)CH(OH)CH2CH2CH=CHCH3.
2.2. Bioactivity of Isolated Compounds
Compounds 1–7 were assayed for antifungal activity against Fusarium graminearum, Gaeumannomyces graminis, and Glomerella cingulata. The result showed that compounds 1, 4, and 7 displayed weak antifungal activity (Table 3). Compound 1 could inhibit the three phytopathogenic fungi tested with MIC values ranging from 64 to 256 μg/mL. Compounds 4 and 7 possessed identical inhibition against Glomerella cingulata and Gaeumannomyces graminis, with MIC values of 128 and 256 μg/mL, respectively. In addition, all the isolates were evaluated for antimicroalgal activity against Amphidinium carterae, Heterocapsa circularisquama, Heterosigma akashiwo, and Prorocentrum donghaiense (Table 4). It was worth noting that only compound 1 was active against all the phytoplankton species tested with IC50 values ranging from 15 to 28 μg/mL. Moreover, the antimicroalgal activity of compounds 2–7 (IC50 ≥ 30 μg/mL) was weaker than that of compound 1. The above results suggested that the tetrahydrofuran ring could improve antifungal and antimicroalgal activity of these lipids by analysis of their structure–activity relationships. In addition, the brine shrimp lethality of 1–7 was also estimated, with the lethal rates against Artemia salina of these compounds being less than 10% at 100 μg/mL (Table 4). All isolates showed low toxicity to the brine shrimp Artermia salina, which demonstrated the safety of their further exploitation. An in-depth study, such as chemical modification, should be conducted to promote the bioactivity of these compounds, increasing their prospective use in the development of antifungal and antimicroalgal agents.

Table 3.
Inhibition of three phytopathogenic fungi by 1–7.

Table 4.
Inhibition of phytoplankton species by 1–7.
3. Materials and Methods
3.1. General Experimental Producers
NMR spectra were obtained on a Bruker Avance III 500 NMR spectrometer (500 and 125 MHz for 1H and 13C, respectively) using tetramethylsilane (TMS) as an internal standard (Bruker Corp., Billerica, MA, USA). Chemical shifts are reported in parts per million (δ) in CDCl3/CD3OD (δH reported referred to CDCl3/CD3OD at 7.26/3.31 ppm; δC reported referred to CDCl3/CD3OD at 77.16/49.00 ppm) and coupling constants (J) in Hz. Optical rotations were measured on an SGW-3 polarimeter (Shanghai Shenguang Instrument Co., Ltd., Shanghai, China). IR spectra were recorded on a Nicolet iS50 FT-IR spectrometer (Thermo Fisher Scientific, Waltham, MA, USA); peaks are reported in cm−1. High-resolution ESI mass spectra were determined on a Xevo G2-XS QTof mass spectrometer (Water Crop., Milford, MA, USA). Column chromatography (CC) was carried out with silica gel (200–300 mesh, Qingdao Haiyang Chemical Co., Qingdao, China), RP-18 (AAG12S50, YMC Co., Ltd., Kyoto, Japan), and Sephadex LH-20 (GE Healthcare, Uppsala, Sweden). Thin-layer chromatography (TLC) was performed with precoated silica gel plates (GF-254, Qingdao Haiyang Chemical Co., Qingdao, China).
3.2. Fungal Material and Fermentation
The fungal strain Trichoderma sp. Z43 was isolated from the surface of marine brown alga Dictyopteris divaricata collected from Zhoushan, China, in July 2018. The species was identified according to morphological characteristics and analysis of ITS regions of its rDNA, deposited at GenBank (OR196112). Mass fermentation was performed statically at room temperature for 30 days in 200 × 1 L Erlenmeyer flasks, each containing 300 mL of media, by adding 40.0 g glucose, 10.0 g peptone, and 7.0 g yeast extract powder into 1000 mL purified water.
3.3. Extraction and Isolation
At the end of fermentation, the mycelia of cultures were obtained by filtration, which were then dried at room temperature, smashed, and extracted with CH2Cl2 and MeOH (1:1, v/v). After removing organic solvents under reduced pressure, the residue was partitioned between ethyl acetate (EtOAc) and H2O to afford an EtOAc-soluble extract (25.6 g). The filtrate was extracted with EtOAc and then dried to give an extract (10.0 g). The two parts were merged based on the similarity of TLC profiles and subjected to silica gel CC with step-gradient solvent systems of petroleum ether (PE)/EtOAc and CH2Cl2/MeOH to afford 11 fractions (Frs. 1–11). Fr. 8 eluted with CH2Cl2/MeOH (20:1) and was further purified by RP-18 CC to afford Fr. 8-1 (MeOH/H2O, 3:7), Fr. 8-2 (MeOH/H2O, 2:3), and Fr. 8-3 (MeOH/H2O, 9:11). Fr. 8-1 was further purified by preparative TLC (CH2Cl2/MeOH, 20:1) and Sephadex LH-20 CC (MeOH) to give compound 7 (2.6 mg). Compound 1 (4.2 mg) was isolated from Fr. 8-2 by preparative TLC (CH2Cl2/MeOH, 20:1) and Sephadex LH-20 CC (MeOH). Compounds 3 (4.0 mg), 5 (4.5 mg), and 6 (2.7 mg) were purified from Fr. 8-3 by TLC (CH2Cl2/MeOH, 10:1) and Sephadex LH-20 CC (MeOH). Fr. 9 eluted with CH2Cl2/MeOH (10:1) and was further purified by RP-18 CC (MeOH/H2O, 1:1) and preparative TLC (CH2Cl2/MeOH, 10:1) as well as Sephadex LH-20 CC (MeOH) to afford compounds 2 (3.1 mg) and 4 (3.8 mg).
3.4. Spectral and Physical Data of Compounds 1–6
Trichoderol B (1): colorless oil; [α]20D − 10.2 (c 0.14, CH3OH); IR (KBr) vmax 3443, 2923, 2854, 1633, 1453, 1384, 969 cm−1; 1H and 13C NMR data, Table 1 and Table 2; HRESIMS m/z 265.1420 [M + Na]+ (calcd for C13H22NaO4, 265.1416).
Trichoderol C (2): colorless oil; [α]20D − 4.2 (c 0.10, CH3OH); IR (KBr) vmax 3430, 2926, 2858, 1633, 1554, 1394, 1030 cm−1; 1H and 13C NMR data, Table 1 and Table 2; HRESIMS m/z 265.1420 [M + Na]+ (calcd for C13H22NaO4, 265.1416).
Trichoderol D (3): colorless oil; [α]20D + 7.9 (c 0.13, CH3OH); IR (KBr) vmax 3422, 2924, 2856, 1661, 1633, 1538, 1394, 1025 cm−1; 1H and 13C NMR data, Table 1 and Table 2; HRESIMS m/z 249.1448 [M + Na]+ (calcd for C13H22NaO3, 249.1467).
Trichoderol E (4): colorless oil; [α]20D − 12.2 (c 0.13, CH3OH); IR (KBr) vmax 3415, 2924, 1634, 1538, 1385, 1023 cm−1; 1H and 13C NMR data, Table 1 and Table 2; HRESIMS m/z 243.1588 [M]− (calcd for C13H23O4, 243.1598).
3.5. Assay for Antifungal Activity
Antifungal activity against Fusarium graminearum, Gaeumannomyces graminis, and Glomerella cingulata was performed using the microdilution method in a 96-well plate, as described previously [17]. Briefly, a stock solution of each fungus tested was diluted in potato dextrose broth (PDB) to 5 × 105 cfu/mL. Each sample was prepared in dimethyl sulfoxide (DMSO) and was diluted to final concentrations of 5120, 2560, 1280, 640, 320, 160, 80, 40, 20, 10, and 5 µg/mL in a set of capped test tubes by two-fold serial dilution. An amount of 5 µL diluent was added into each well of a 96-well flat-bottom microtiter plate containing 95 µL fungal suspension (the final sample concentrations were 256 to 0.25 µg/mL), and the fungi were cultivated at 28 °C for 48 h. The MIC value for each sample was defined as the minimum concentration of the compound with invisible microbial growth. Amphotericin B and DMSO were chosen as positive and negative controls, respectively.
3.6. Assay for Antimicroalgal Activity
The inhibition of four marine phytoplankton species (Amphidinium carterae, Heterocapsa circularisquama, Heterosigma akashiwo, and Prorocentrum donghaiense) was assayed using our previously reported method [17]. In brief, each microalga inoculum was cultured for 5 days using the sterilized f/2 medium in an incubator (20 °C, 14:10 light–dark cycle, 2000 lx light) and reached the exponential growth phase. The microalga suspension was diluted to 4–5 × 104 cells/mL and then added into a 96-well flat-bottom microtiter plate with 195 µL in each well. An amount of 5 µL sample solution (in DMSO) was pipetted into each well (the final sample concentrations were 100 to 0.125 µg/mL) and mixed uniformly. After 24 h inoculation, the live cells were counted using hemocytometer, and the inhibition rate was calculated as follows. Inhibition rate (IR) = (NCK − NT)/NCK × 100% (NCK: the number of live algal cells under negative control, NT: the number of live algal cells under treatment). DMSO and K2Cr2O7 were taken as negative and positive controls, respectively.
3.7. Assay for Brine Shrimp Lethal Activity
The inhibition of the brine shrimp Artermia salina was assayed according to the procedures described in our previous report [15]. In brief, brine shrimp eggs were left to hatch in sea water for 48 h at 25 °C under natural light. About 10 brine shrimp were placed in a 96-well flat-bottom microtiter plate with a volume of 195 µL sea water in each well. An amount of 5 µL sample solution (in DMSO) was added into each well (the final sample concentrations were 100 to 0.125 µg/mL) and mixed uniformly. The lethality was observed after 24 h of cultivation. DMSO and K2Cr2O7 served as negative and positive controls, respectively.
4. Conclusions
Chemical investigation towards the marine algicolous fungus Trichoderma sp. Z43 resulted in the isolation of seven lipids, including six new ones (trichoderols B-G (1–6)) and a known one, triharzianin B (7). The C13 and C11 lipids are rarely found in nature, especially in Trichoderma species, and these new compounds greatly enrich the chemical diversity of marine-derived natural products. Finding and stimulating silent biosynthetic gene clusters may be an effective means to excavate this kind of metabolite. These isolations were evaluated for inhibition against three phytopathogenic fungi and four marine phytoplankton species. Several of them exhibited inhibition of one or more fungi/plankton species tested, and the tetrahydrofuran ring could improve antifungal and antimicroalgal activity of these lipids by analysis of their structure–activity relationships. Moreover, all isolates exhibited low toxicity to the brine shrimp Artermia salina, suggesting the security for their further exploitation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21080453/s1, Figures S1–S42: 1D/2D NMR, HRESIMS spectra of 1–6.
Author Contributions
Conceptualization, N.-Y.J.; data curation, Z.-Z.S.; formal analysis, Z.-Z.S.; funding acquisition, Z.-Z.S. and N.-Y.J.; investigation, Z.-Z.S.; methodology, X.-L.Y.; project administration, N.-Y.J.; supervision, N.-Y.J.; writing—original draft, Z.-Z.S.; writing—review and editing, N.-Y.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (42206130 and 42076096), the Natural Science Foundation of Shandong Province (ZR2020QD102), the Taishan Scholar Project Special Funding (tsqn201909164), and the Science & Technology Specific Projects in Agricultural High-Tech Industrial Demonstration Area of the Yellow River Delta (2022SZX01).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data of the compounds are available in Supplementary Materials.
Acknowledgments
The authors acknowledge Ke Li (Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences) for her technical support of HRESIMS.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Phoulivong, S.; Cai, L.; Chen, H.; McKenzie, E.H.C.; Abdelsalam, K.; Chukeatirote, E.; Hyde, K.D. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 2010, 44, 33–43. [Google Scholar] [CrossRef]
- Wang, B.; Li, B.H.; Wang, C.X.; Zhang, Z.F. Effects of temperature, wetness duration, and moisture on the conidial germination, infection, and disease incubation period of Glomerella cingulate. Plant Dis. 2015, 99, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases-a field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]
- Freeman, J.; Ward, E. Gaeumannomyces graminis, the take-all fungus and its relatives. Mol. Plant Pathol. 2004, 5, 235–252. [Google Scholar] [CrossRef]
- Murray, S.A.; Kohli, G.S.; Farrell, H.; Spiers, Z.B.; Place, A.R.; Dorantes-Aranda, J.J.; Ruszczyk, J. A fish kill associated with a bloom of Amphidinium carterae in a coastal lagoon in Sydney, Australia. Harmful Algae 2015, 49, 19–28. [Google Scholar] [CrossRef]
- Rensel, J.E.J.; Haigh, N.; Tynan, T.J. Fraser river sockeye salmon marine survival decline and harmful blooms of Heterosigma akashiwo. Harmful Algae 2010, 10, 98–115. [Google Scholar] [CrossRef]
- Matsuyama, Y. Impacts of the harmful dinoflagellate Heterocapsa circularisquama bloom on shellfish aquaculture in Japan and some experimental studies on invertebrates. Harmful Algae 2012, 14, 144–155. [Google Scholar] [CrossRef]
- Lin, J.N.; Yan, T.; Zhang, Q.C.; Wang, Y.F.; Liu, Q.; Zhou, M.J. In situ detrimental impacts of Prorocentrum donghaiense blooms on zooplankton in the East China Sea. Mar. Pollut. Bull. 2014, 88, 302–310. [Google Scholar] [CrossRef]
- Zeilinger, S.; Gruber, S.; Bansal, R.; Mukherjee, P.K. Secondary metabolites in Trichoderma—Chemistry meets genomics. Fungal Biol. Rev. 2016, 30, 74–90. [Google Scholar] [CrossRef]
- Li, M.-F.; Li, G.-H.; Zhang, K.-Q. Non-volatile metabolites from Trichoderma spp. Metabolites 2019, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Keswani, C.; Mishra, S.; Sarma, B.K.; Singh, S.P.; Singh, H.B. Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. Appl. Microbiol. Biotechnol. 2014, 98, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-L.; Tang, W.-L.; Huang, Q.-R.; Li, Y.-Z.; Wei, M.-L.; Jiang, L.-L.; Liu, C.; Yu, X.; Zhu, H.-W.; Chen, G.-Z.; et al. Trichoderma: A treasure house of structurally diverse secondary metabolites with medicinal importance. Front. Microbiol. 2021, 12, 723828. [Google Scholar] [CrossRef]
- Song, F.; Dai, H.; Tong, Y.; Ren, B.; Chen, C.; Sun, N.; Liu, X.; Bian, J.; Liu, M.; Gao, H.; et al. Trichodermaketones A-D and 7-O-methylkoninginin D from the marine fungus Trichoderma koningii. J. Nat. Prod. 2010, 73, 806–810. [Google Scholar] [CrossRef]
- Miao, F.-P.; Liang, X.-R.; Yin, X.-L.; Wang, G.; Ji, N.-Y. Absolute configurations of unique harziane diterpenes from Trichoderma species. Org. Lett. 2012, 14, 3815–3817. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wu, Z.; Guo, H.; Liu, L.; Chen, S. A review of terpenes from marine-derived fungi: 2015-2019. Mar. Drugs 2020, 18, 321. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.-Z.; Liu, X.-H.; Li, X.-N.; Ji, N.-Y. Antifungal and antimicroalgal trichothecene sesquiterpenes from the marine algicolous fungus Trichoderma brevicompactum A-DL-9-2. J. Agric. Food Chem. 2020, 68, 15440–15448. [Google Scholar] [CrossRef]
- Song, Y.-P.; Shi, Z.-Z.; Miao, F.-P.; Fang, S.-T.; Yin, X.-L.; Ji, N.-Y. Tricholumin A, a highly transformed ergosterol derivative from the alga-endophytic fungus trichoderma asperellum. Org. Lett. 2018, 20, 6306–6309. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Xu, T.-T.; Sun, L.-J.; Cen, R.-H.; Su, S.; Yang, X.-Q.; Yang, Y.-B.; Ding, Z.-T. The chemical diversity, the attractant, anti-acetylcholinesterase, and antifungal activities of metabolites from biocontrol Trichoderma harzianum uncovered by OSMAC strategy. Bioorg. Chem. 2021, 114, 105148. [Google Scholar] [CrossRef]
- Li, B.; Huang, Q.-X.; Gao, D.; Liu, D.; Ji, Y.-B.; Liu, H.-G.; Lin, W.-H. New C13 lipids from the marine-derived fungus Trichoderma harzianum. J. Asian Nat. Prod. Res. 2015, 17, 468–474. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, Q.; Yu, H.; Wang, J.; Wang, H.; Yang, Q.; Zhu, H.; Li, Y. Absolute configuration determination of one new compound trichoderol A from Trichoderma sp. fungus. Chem. J. Chin. Univ.-Chin. 2016, 37, 1972–1976. [Google Scholar]
- Hoye, T.R.; Eklov, B.M.; Jeon, J.; Khoroosi, M. Sequencing of three-component olefin metatheses: Total synthesis of either (+)-gigantecin or (+)-14-deoxy-9-oxygigantecin. Org. Lett. 2006, 8, 3383–3386. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.; Adhikari, A.S.; Majumdar, N. Iridium-catalyzed enantioselective ring opening of alkenyl oxiranes by unactivated carboxylic acids. Org. Lett. 2022, 24, 7388–7393. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, Y.; Fitzsinnons, B.J.; Adams, J.; Perez, F.; Rokach, J. The total synthesis of 12-HETE and 12,20-DiHETE. J. Org. Chem. 1986, 51, 789–793. [Google Scholar] [CrossRef]
- Bae, M.; Kim, H.; Shin, Y.; Kim, B.Y.; Lee, S.K.; Oh, K.-B.; Shin, J.; Oh, D.-C. Separacenes A-D, novel polyene polyols from the marine actinomycete, Streptomyces sp. Mar. Drugs 2013, 11, 2882–2893. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; An, B.; Wu, J.; Li, Y.; Bian, Q.; Wang, M.; Zhong, J. Asymmetric synthesis of sex pheromone of the western hemlock looper, Lambdina fiscellaria lugubrosa (Hulst). Tetrahedron Lett. 2023, 118, 154401. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).