Abstract
The semisynthesis of renieramycin-type derivatives was achieved under mild and facile conditions by attaching a 1,3-dioxole-bridged phenolic moiety onto ring A of the renieramycin structure and adding a 4′-pyridinecarbonyl ester substituent at its C-5 or C-22 position. These were accomplished through a light-induced intramolecular photoredox reaction using blue light (4 W) and Steglich esterification, respectively. Renieramycin M (4), a bis-tetrahydroisoquinolinequinone compound isolated from the Thai blue sponge (Xestospongia sp.), served as the starting material. The cytotoxicity of the 10 natural and semisynthesized renieramycins against non-small-cell lung cancer (NSCLC) cell lines was evaluated. The 5-O-(4′-pyridinecarbonyl) renieramycin T (11) compound exhibited high cytotoxicity with half-maximal inhibitory concentration (IC50) values of 35.27 ± 1.09 and 34.77 ± 2.19 nM against H290 and H460 cells, respectively. Notably, the potency of compound 11 was 2-fold more than that of renieramycin T (7) and equal to those of 4 and doxorubicin. Interestingly, the renieramycin-type derivatives with a hydroxyl group at C-5 and C-22 exhibited weak cytotoxicity. In silico molecular docking and dynamics studies confirmed that the mitogen-activated proteins, kinase 1 and 3 (MAPK1 and MAPK3), are suitable targets for 11. Thus, the structure–cytotoxicity study of renieramycins was extended to facilitate the development of potential anticancer agents for NSCLC cells.
1. Introduction
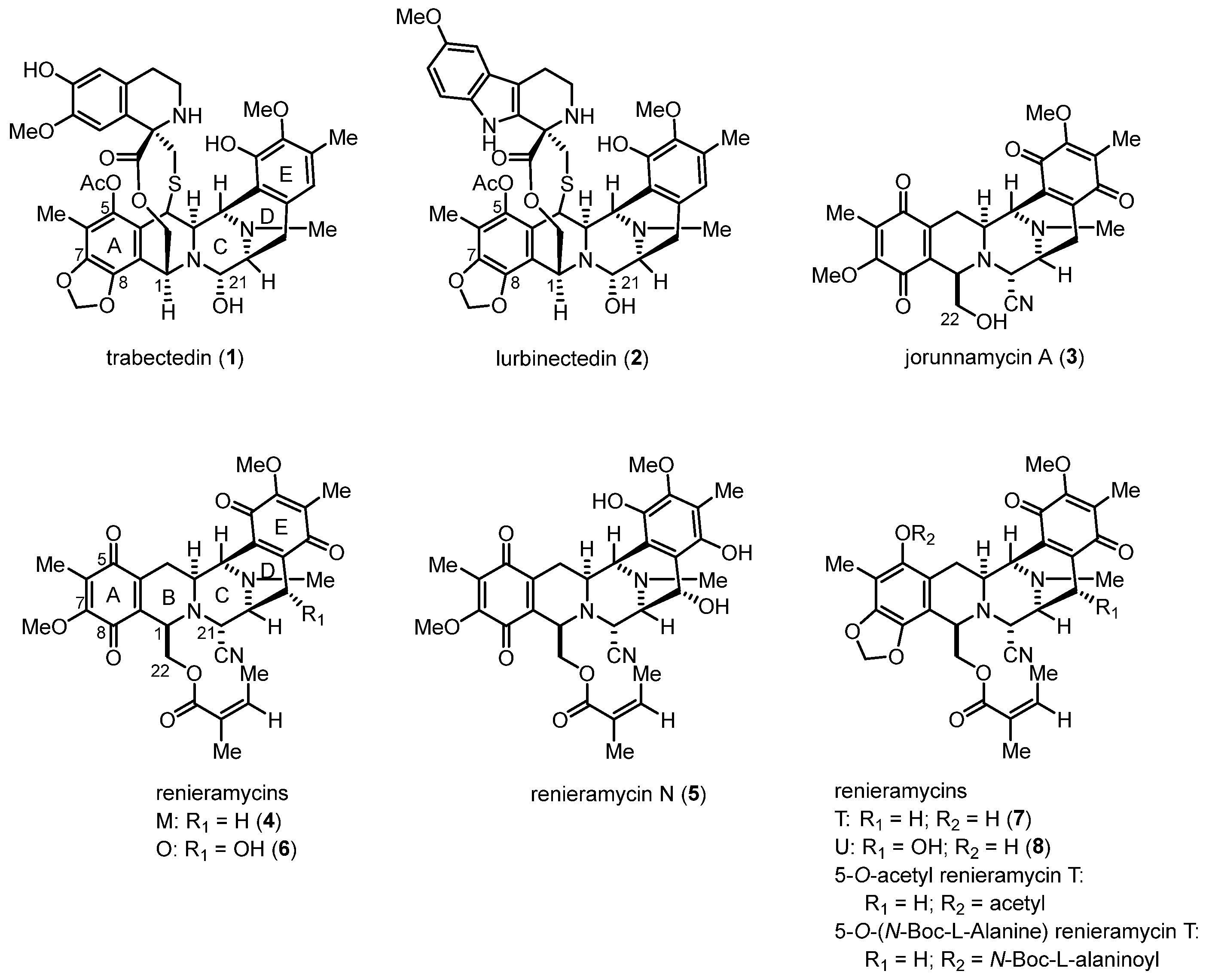
Renieramycins, bis-tetrahydroisoquinolinequinone marine alkaloids, along with ecteinascidins and jorunnamycins, have been reported to exhibit significant anticancer activity (Figure 1) [1,2]. Trabectedin (also known as ecteinascidin 743, 1), which was isolated from the Caribbean tunicate (Ecteinascidia turbinate), was approved by the United States Food and Drug Administration in 2015 for the treatment of advanced soft tissue sarcoma and ovarian carcinoma [3,4,5]. Lurbinectedin (PM01183, 2), a bis-tetrahydroisoquinoline analog of 1, was approved for the second-line treatment of metastatic small-cell lung cancer in 2020 [6,7]. Compounds 1 and 2 are chemotherapeutic drugs that covalently bind specifically to the N2 position of guanine in the DNA minor groove, leading to double-strand breaks and apoptosis in cancer cells [3,6].
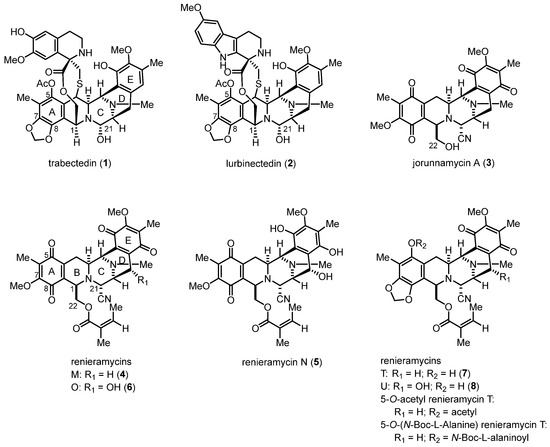
Figure 1.
Structures of the bis-tetrahydroisoquinoline alkaloids as anticancer drugs and promising drug leads.
Renieramycins are isolated from marine sponges, such as genera Reniera [8,9], Cribrochalina [10], and Xestospongia [11,12], and they possess a pentacyclic bis-tetrahydroisoquinolinequinone moiety as the core scaffold (Figure 1). Several renieramycins containing an angelate ester at the C-22 position display potent bioactivities, including antimicrobial [8], antileishmanial [13], and anticancer [11,12,14,15] activities. Interestingly, renieramycins M (4) and N (5) have been successfully isolated in the gram scale of the Thai blue sponge (Xestospongia sp.) [11]. Renieramycins M, N, and O (4–6) have been demonstrated to exhibit potent cytotoxicity against colon (HCT116) and lung carcinoma (QG56) [11,12]. Additionally, compound 4 has been reported to display strong cytotoxicity against various human cancer cell lines, including colon (DLD1), lung (NCI-H460), pancreatic adenocarcinoma (AsPC1), and ductal breast epithelial (T47D) cells [11,12,14]. Renieramycins T (7) and U (8), renieramycin–ecteinascidin hybrid marine natural alkaloids containing a 1,3-dioxole ring that is similar to the left-side carbon framework of trabectedin, exhibit strong in vitro anticancer activity against non-small-cell lung cancer (NSCLC) cells [14]. Furthermore, the cytotoxicity mechanisms of the renieramycin–ecteinascidin hybrid derivatives against NSCLC cells have been revealed. Compound 7 was discovered to enhance apoptosis induction via the degradation of the myeloid cell leukemia 1 (Mcl-1) protein in NSCLC [16,17]. Further, it suppressed mouse melanoma (B16F10) cell metastasis and migration through the downregulation of NF-E2-related factor 2 [18]. Moreover, the 5-O-ester derivatives of 7, which possess acetyl and N-Boc-L-alaninoyl substituents, induced apoptosis and suppressed cancer stem cell markers via protein kinase B (AKT) inhibition in NSCLC cells [19,20].
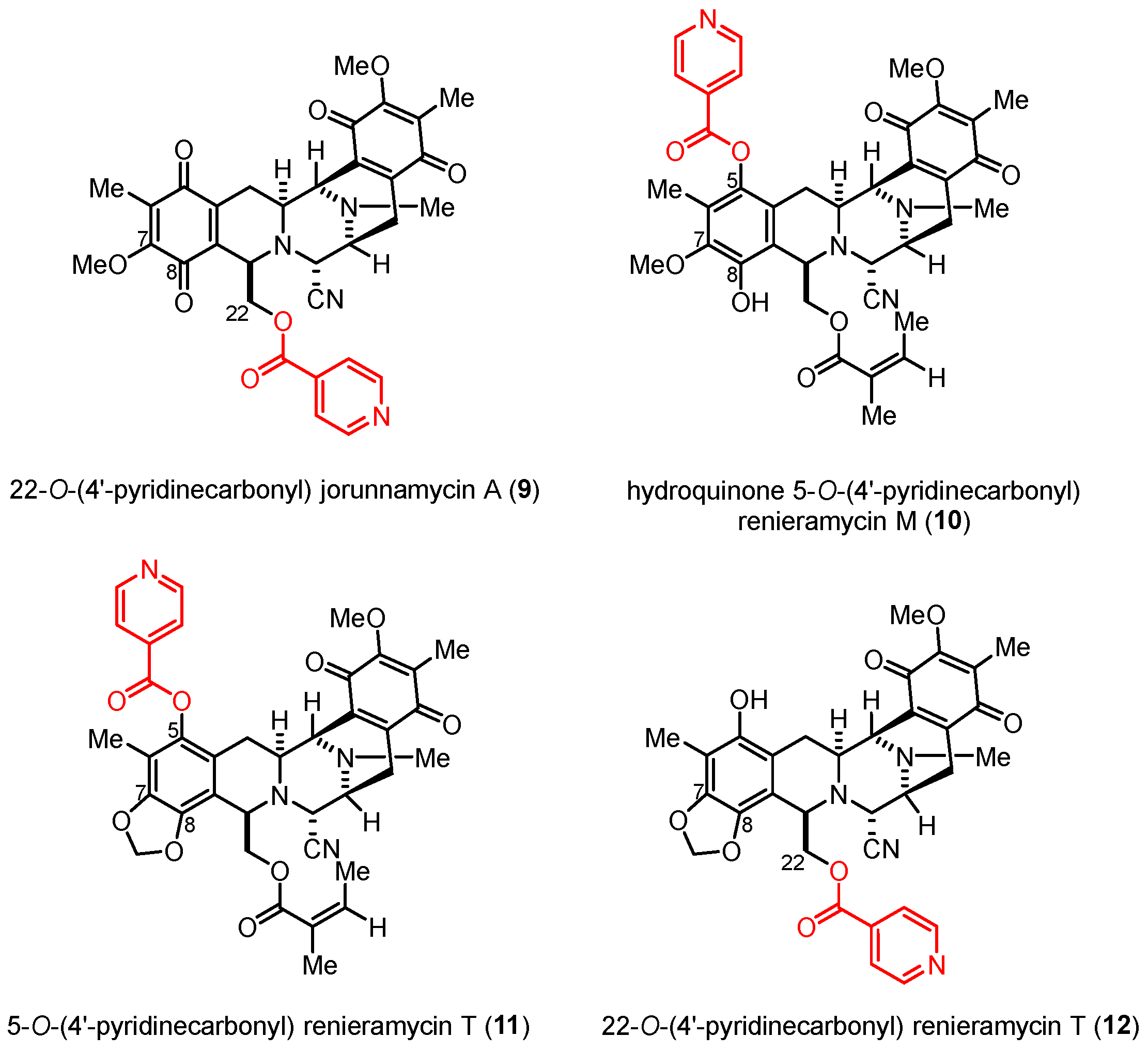
According to the structure–activity relationship (SAR) study, nitrogen-containing heterocyclic ester substituents, particularly 4′-pyridinecarbonyl ester derivatives, as side chains on the renieramycin framework have a remarkable effect on the cytotoxicity of renieramycin against various cancer cells (Figure 2). The 22-O-(4′-pyridinecarbonyl) jorunnamycin A compound (9) exhibited significantly better cytotoxicity than jorunnamycin A (3), its parent compound, against human colon (HCT116), breast (MDA-MB-435), and NSCLC (H292 and H460) cell lines [21,22]. Furthermore, hydroquinone 5-O-(4′-pyridinecarbonyl) renieramycin M (10) was prepared from 4 by a two-step process, including hydrogenation and Steglich esterification. The resulting derivative (10) exhibited cytotoxicity against the highly metastatic H292 and H460 NSCLC cell lines at inhibitory concentration (IC50) values in the nanomolar range [15].
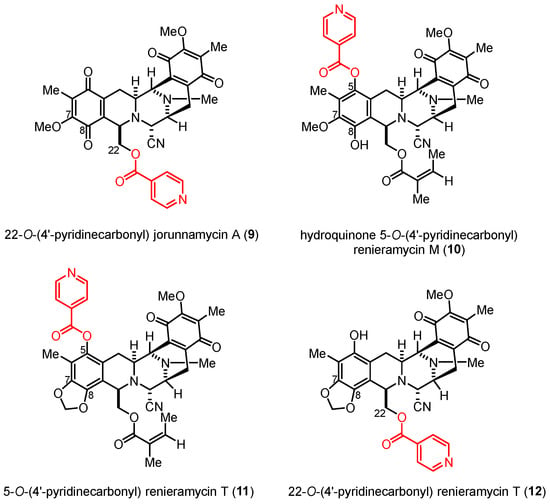
Figure 2.
Semisynthetic derivatives of the renieramycin-type derivatives containing a 4′-pyridinecarbonyl ester substituent.
Regarding the continuous structure–cytotoxicity relationship study of the renieramycin-type derivatives, the semisynthesis of the renieramycin–ecteinascidin hybrid derivatives of natural bis-tetrahydroisoquinolinequinones (4–6) was conducted via a facile, light-induced intramolecular photoredox reaction to transform quinone ring A into the 1,3-dioxole-bridged phenolic moiety. The mild photoredox reaction enables the selective one-step modification of the methoxy-substituted quinone unit located on ring A of renieramycins, resulting in the formation of 5-hydroxy-tetrahydroisoquinol-1,3-dioxoles with excellent yields [23]. This transformation yields a series of renieramycin–ecteinascidin derivatives that closely resemble the structure of ring A found in Trabectedin and Lurbinectedin, well-known tetrahydroisoquinoline-based chemotherapeutic drugs. Next, a series of 4′-pyridinecarbonyl esters for the renieramycin-type derivatives were prepared by mild and selective Steglich esterification. The new 4′-pyridinecarbonyl esters, 11 and 12, were obtained (Figure 2). The in vitro cytotoxicity of the compounds against human H292 and H460 NSCLC cell lines was evaluated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Additionally, the in silico prediction of the target genes and molecular pathways associated with the activity of the compounds was achieved by virtual network pharmacology study, including molecular docking and molecular dynamics studies.
2. Results and Discussion
2.1. Light-Induced Intramolecular Photoredox Reaction of Renieramycins
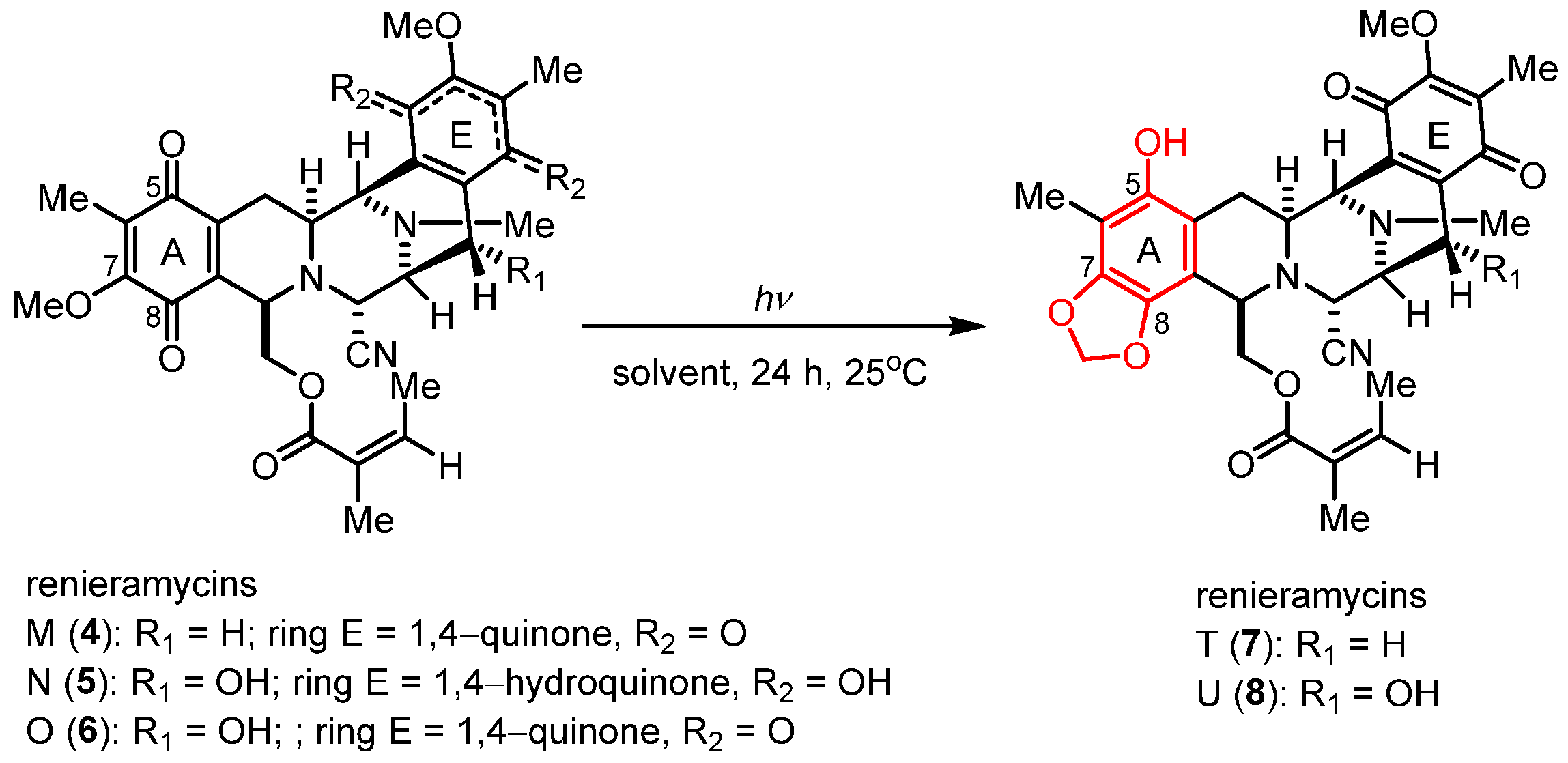
Renieramycins 4–6 were isolated from the Thai blue sponge (Xestospongia sp.) by pretreatment with 10% potassium cyanide following a previously reported protocol [11,12]. The photoredox reactions occurring through the light-induced radical formation and intramolecular cyclization of the natural bis-tetrahydroisoquinolinequinone alkaloids (4–6) were investigated (Scheme 1 and Table 1) [23]. To determine the optimal reaction conditions, we experimented with compound 4, along with various solvents and light sources. Through irradiation by an 18-W fluorescent lamp (white light) in dichloromethane (CH2Cl2) for 24 h, compound 4 was smoothly transformed into compound 7 in 64% yield (Table 1, entry 1). The application of chloroform (CHCl3) and tetrahydrofuran (THF) as solvents for the photoredox reaction of compound 4 furnished compound 7 in 46% and 47% yields, respectively (Table 1, entries 2 and 3). Further improvement in the yield was achieved when the reaction solution was exposed to irradiation by a 4-W light-emitting diode (LED) lamp (blue light) in CH2Cl2, affording 7 in excellent yield (81% yield, entry 4). However, using CHCl3 and THF with blue light irradiation decreased the yields of 7 to 51% and 54%, respectively (Table 1, entries 5 and 6). Thus, optimized photoredox reactions using blue light and CH2Cl2 were further performed with natural alkaloids 5 and 6 (Table 1, entries 7 and 8).
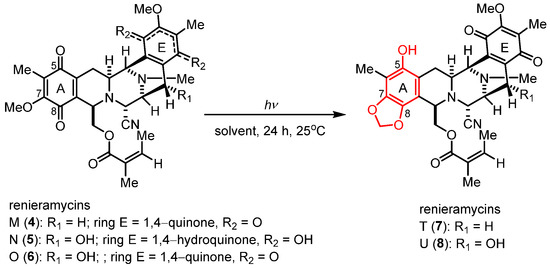
Scheme 1.
Photoredox reaction of the natural renieramycins.

Table 1.
Optimization of the conditions for the photoredox of natural renieramycins.
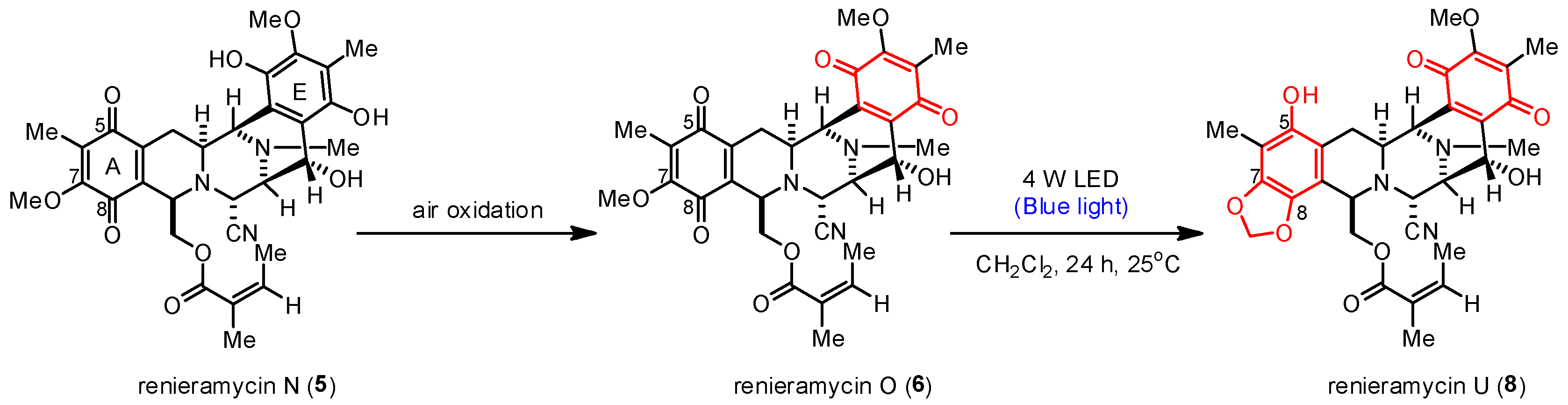
Unexpectedly, the photoredox reaction of compound 5 yielded a mixture of bis-tetrahydroisoquinolinequinones 6 and 8 (Table 1, entry 7). Compound 8 was obtained as the major product in 37% yield, two times higher than the yield of compound 6 (18% yield). Meanwhile, the photoredox reaction of compound 6 under the optimized condition yielded compound 8 in 48% yield (Table 1, entry 8). Therefore, the light-induced radical formation and intramolecular cyclization of compound 5 to yield compounds 6 and 8 were proposed to occur via stepwise transformations. The mechanism involved air oxidation to convert the unstable 1,4-hydroquinone into the 1,4-quinone moiety, followed by the photoredox reaction [24] (Scheme 2). The results of the light-mediated transformation highlight the chemical diversity of the natural renieramycins found around the blue Xestospongia sponge habitat.
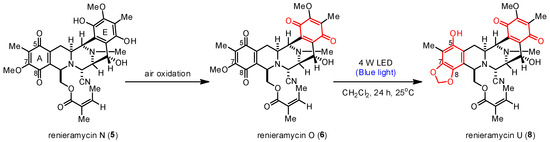
Scheme 2.
Proposed transformation mechanism of 5 into 6 and 8.
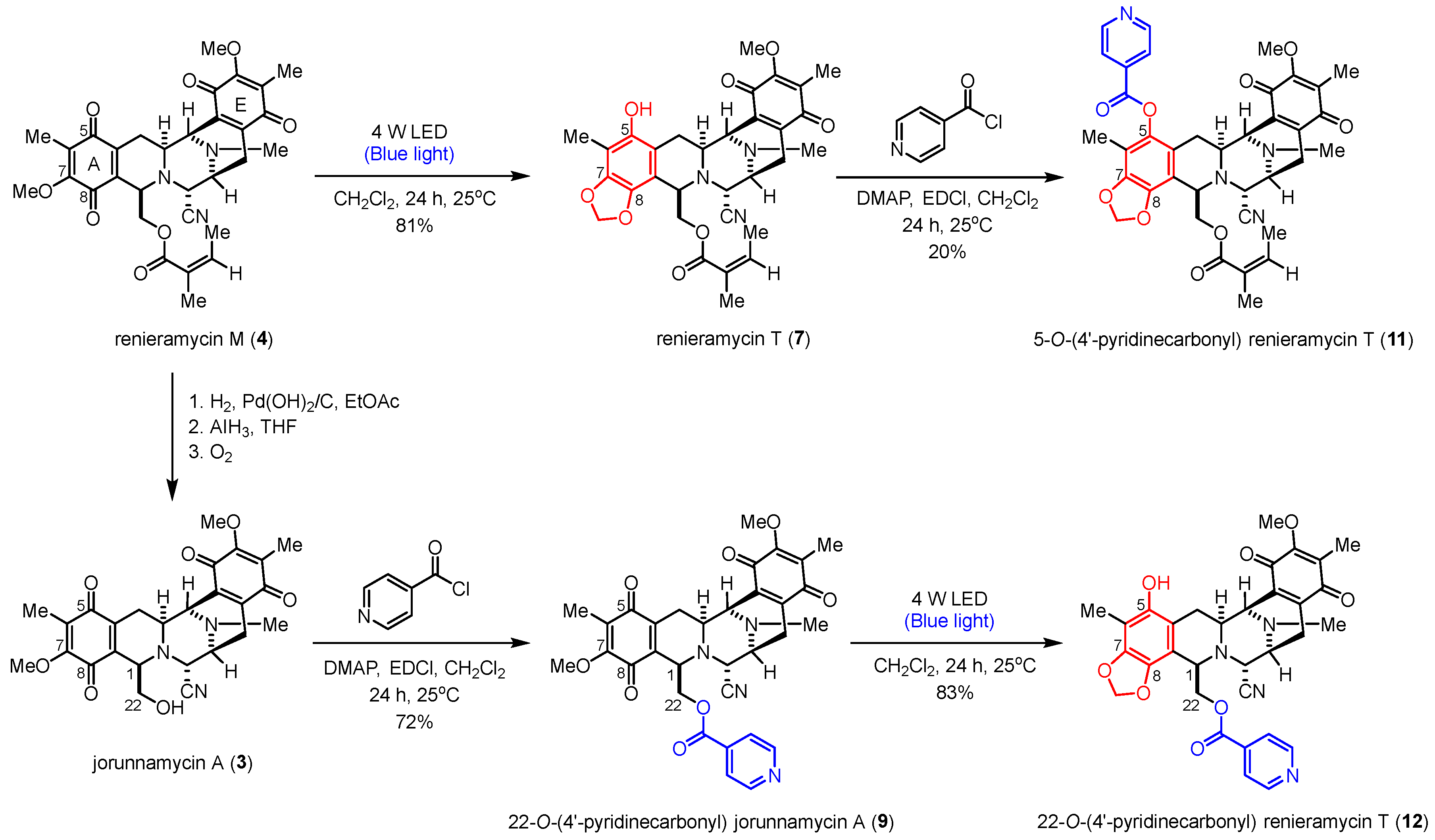
2.2. Semisynthesis of 4′-Pyridinecarbonyl-Substituted Renieramycin-Type Derivatives
The semisynthesis of the 4′-pyridinecarbonyl-substituted renieramycin–ecteinascidin hybrid derivative (11) started with the photoredox transformation and esterification (Scheme 3). The irradiation of 4 under the 4-W LED light in CH2Cl2 furnished 7. Subsequently, a 4′-pyridinecarbonyl motif of 7 was installed by Steglich esterification using isonicotinoyl chloride as an acylating agent, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDCI) as a coupling reagent, and 4-dimethylaminopyridine (DMAP) as a nucleophilic base catalyst to obtain the desired product (11) in acceptable yield. The semisynthesis protocol of the renieramycin-type derivative (12) was envisioned. Compound 3 was prepared from compound 4 by hydrogenation, hydride reduction, and air oxidation [21,24]. Ester compound 12 was obtained in excellent yield by attaching the 4′-pyridinecarbonyl substituent to the C-22 position of renieramycin by Steglich esterification, followed by the formation of the 1,3-dioxole-bridged phenolic moiety via light-induced radical formation and intramolecular cyclization.
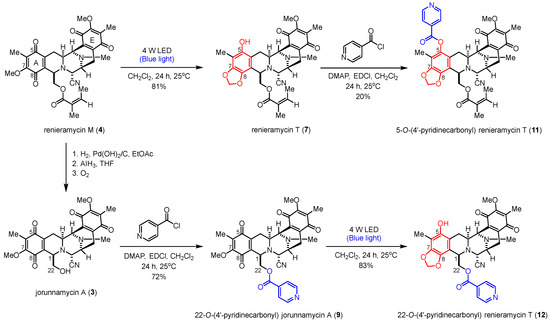
Scheme 3.
Semisynthesis of the 4′-pyridinecarbonyl-substituted renieramycin-type derivatives.
The chemical structures of all semisynthetic renieramycin-type derivatives were characterized by spectroscopic techniques (see Supporting Information for the spectra, Figures S1–S15). The spectral data of compounds 7–9 were consistent with previous reports [14,22]. Regarding the 4′-pyridinecarbonyl-substituted renieramycin–ecteinascidin hybrid derivatives (11 and 12), the characteristic chemical shift of the methylene moiety (–CH2–) at the newly constructed 1,3-dioxole ring showed a proton signal as a pair of doublets at 5.95 ± 0.07 ppm and a carbon signal at 101.6 ± 0.3 ppm. Moreover, the C-7 and C-8 quaternary carbon signals of both compounds 11 and 12 shifted upfield, compared with the signals of the parent compound 4. The carbonyl carbon signals of the resulting 4′-pyridinecarbonyl ester were confirmed at 167.0 and 164.2 ppm for 11 and 12, respectively. Moreover, the chemical shifts of the carbonyl carbon at C-15 and C-18 for both 11 and 12 are located at 186.0 ± 0.2 ppm and 182.5 ± 0.1 ppm, respectively, indicating the presence of the quinone group on ring E.
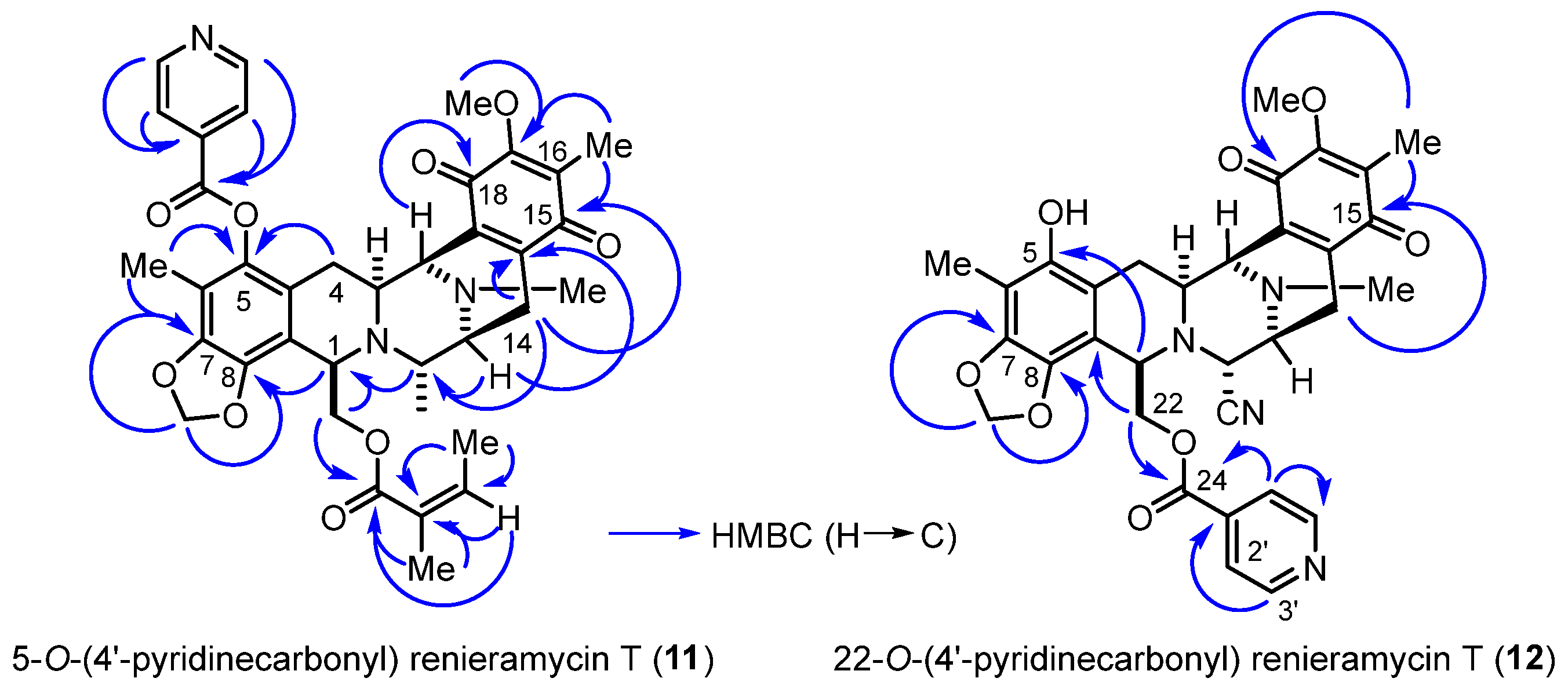
The heteronuclear multiple bond correlations (HMBCs) between the methylene proton at the 1,3-dioxole ring and the quaternary aromatic carbons at C-7 and C-8 clearly confirmed the fused ring structure at ring A in compounds 11 and 12 (Figure 3). The key HMBCs of compound 11 included the correlations between the C-5 quaternary carbon and both the C-4 methylene protons and the C-6 methyl protons, along with those of the C-1′ pyridine carbon, all the protons at the C-2′ and C-3′ position of pyridine, and C-6 methyl motifs. Compound 12 exhibited significant HMBCs between the C-24 carbonyl carbon and both the C-22 methylene protons and the C-2′ pyridine proton.
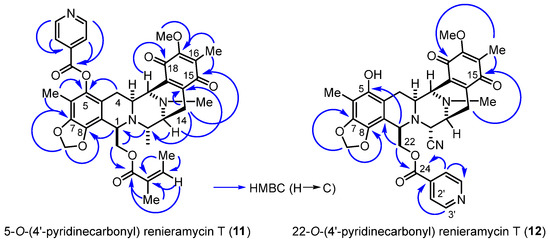
Figure 3.
HMBCs (blue arrows) for the 4′-pyridinecarbonyl-substituted derivatives (11 and 12).
2.3. Cytotoxicity of Compounds 3–12 against NSCLC Cell Lines
The in vitro cytotoxic activities of the series of semisynthesized renieramycin-type derivatives and the natural renieramycins analogs were analyzed by MTT assay against the human H292 and H460 NSCLC cell lines (Table 2). The positive controls were cisplatin and doxorubicin, which were the first-line chemotherapeutic drugs. The results showed that compound 3, which has a primary alcohol at the C-22 position, and renieramycins 5–8, which possess unique structures (1,4-quinone or 1,3-dioxole-bridged phenol at ring A, methylene or secondary alcohol at C-14 of ring D, and 1,4-quinone or 1,4-hydroquinone at ring E), exhibited weak cytotoxicity against both H292 and H460 NSCLC cell lines at the IC50 values of 72.64–183.37 nM (Table 2, entries 1 and 3–6). However, compound 4 exhibited the strongest cytotoxicity among the series of natural renieramycins at the IC50 values of 35.36 and 33.86 nM against H292 and H460 cells, respectively (Table 2, entry 2). The semisynthesized renieramycin-type derivatives (9–11) exhibited substantially high cytotoxic activity at nanomolar concentrations against both cell lines (Table 2, entries 7–9). The known semisynthesized alkaloid (9), which possesses bis-tetrahydroisoquinolinequinone and 4′-pyridinecarbonyl moieties, exhibited the strongest cytotoxicity among all the renieramycin derivatives in this study. The compound 9 was approximately 10-fold and 9-fold more potent than the parent compound (4) against H292 and H460 NSCLC cell lines, respectively [22]. Both compound 10, the known 8-hydroquinone, and compound 11, the renieramycin–ecteinascidin hybrid derivative, contained the 5-O-(4′-pyridinecarbonyl) ester. They exhibited significant cytotoxicity, similar to those of 4 and docxorubicin, at the same IC50 level. Moreover, compound 11 was 3-fold and 2-fold more potent than 7, the parent compound, against the H292 and H460 cell lines. Interestingly, compound 12, a renieramycin-type derivative with a hydroxyl group at C-5, lost its cytotoxic activity and inhibited both the H292 and H460 cell lines at micromolar IC50 values (Table 2, entry 10). However, compound 12 exhibited 3-fold and 2-fold stronger potency than cisplatin against the H292 and H460 NSCLC cell lines, respectively. Therefore, the presence of the 4′-pyridinecarbonyl ester and the chemical structure of ring A constitute the essential pharmacophore controlling the cytotoxic potency [15,22,25]. The cytotoxicity of the renieramycin-type derivatives having a hydroxyl group at either the C-5 or C-22 position was weak.

Table 2.
Cytotoxicity of the natural renieramycins and semisynthesized renieramycin derivatives against non-small-cell lung cancer cell lines.
2.4. In Silico Prediction of the Target Genes and Molecular Pathways
The series of the 4′-pyridinecarbonyl ester of the renieramycin-type derivatives (10 and 11) exhibited significant cytotoxicity against NSCLC cells. The anticancer mechanisms of compounds 10 and 11 were not investigated because of their limited quantity. Regarding data mining from several databases, pharmaco-mapping, and computational strategies, network pharmacology has recently emerged as a comprehensive tool for elucidating the putative biomolecular targets, signaling pathway complexities, and protein–protein interactions (PPIs) of potential leads [26,27]. Therefore, the inhibition mechanism and molecular pathways of both 10 and 11 against NSCLC were investigated by molecular docking and molecular dynamics studies, enabling the identification of the primary target genes.
2.4.1. Investigation of the Potential Targets against NSCLC
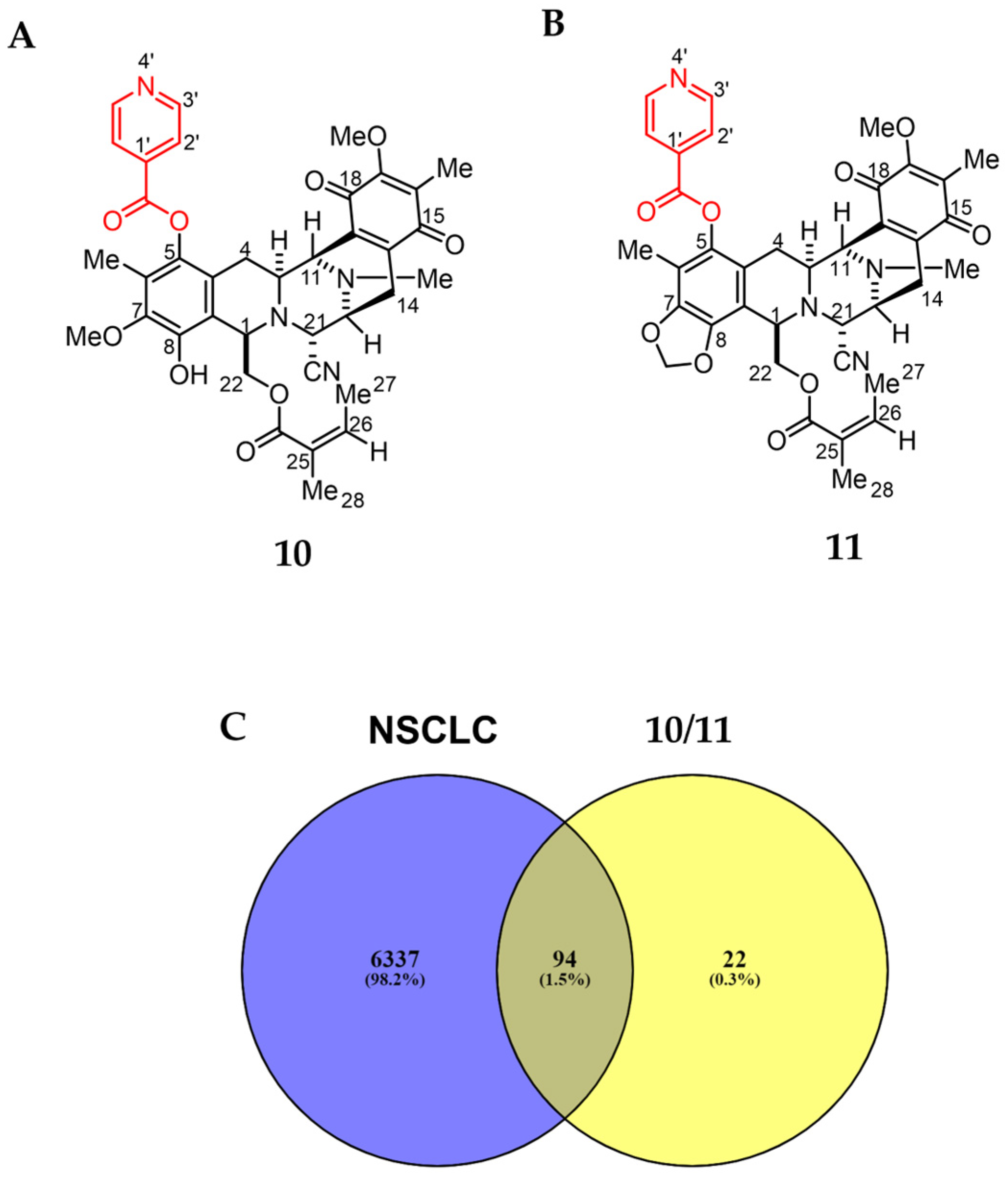
The Swiss Target Prediction online tool was employed to compare the renieramycin-type derivatives (10 and 11) to a massive internal pharmacophore model database to identify prospective drug targets using reverse pharmacophore alignment (Figure 4) [28]. One hundred and sixteen potential targets related to 10 and 11 were found. From the following four databases: GeneCards, Therapeutic Targets Database (TTD), Online Mendelian Inheritance in Man (OMIM), and DisGeNET, 6431 genes were found in association with NSCLC. Overlapping the corresponding genes using a Venn diagram, 94 prospective targets of 10 and 11 against NSCLC were generated (Figure 4C).
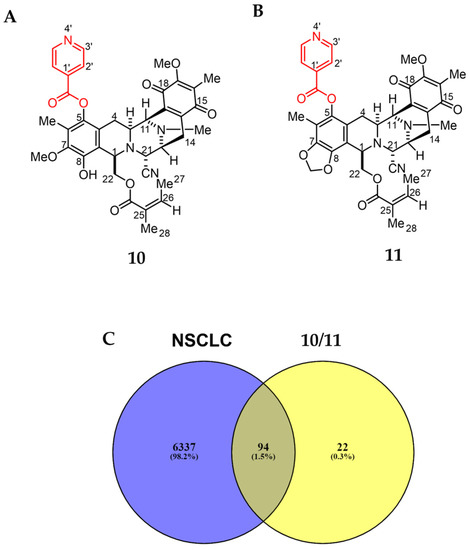
Figure 4.
Structure and putative analysis of 10 and 11 against NSCLC. (A) Structure of 10. (B) Structure of 11. (C) Venn diagram analysis showing both 10 and 11 associated with 116 targets, whereas the NSCLC cells are associated with 6431 targets. The interception analysis showing 94 overlapping targets between 10/11 and NSCLC.
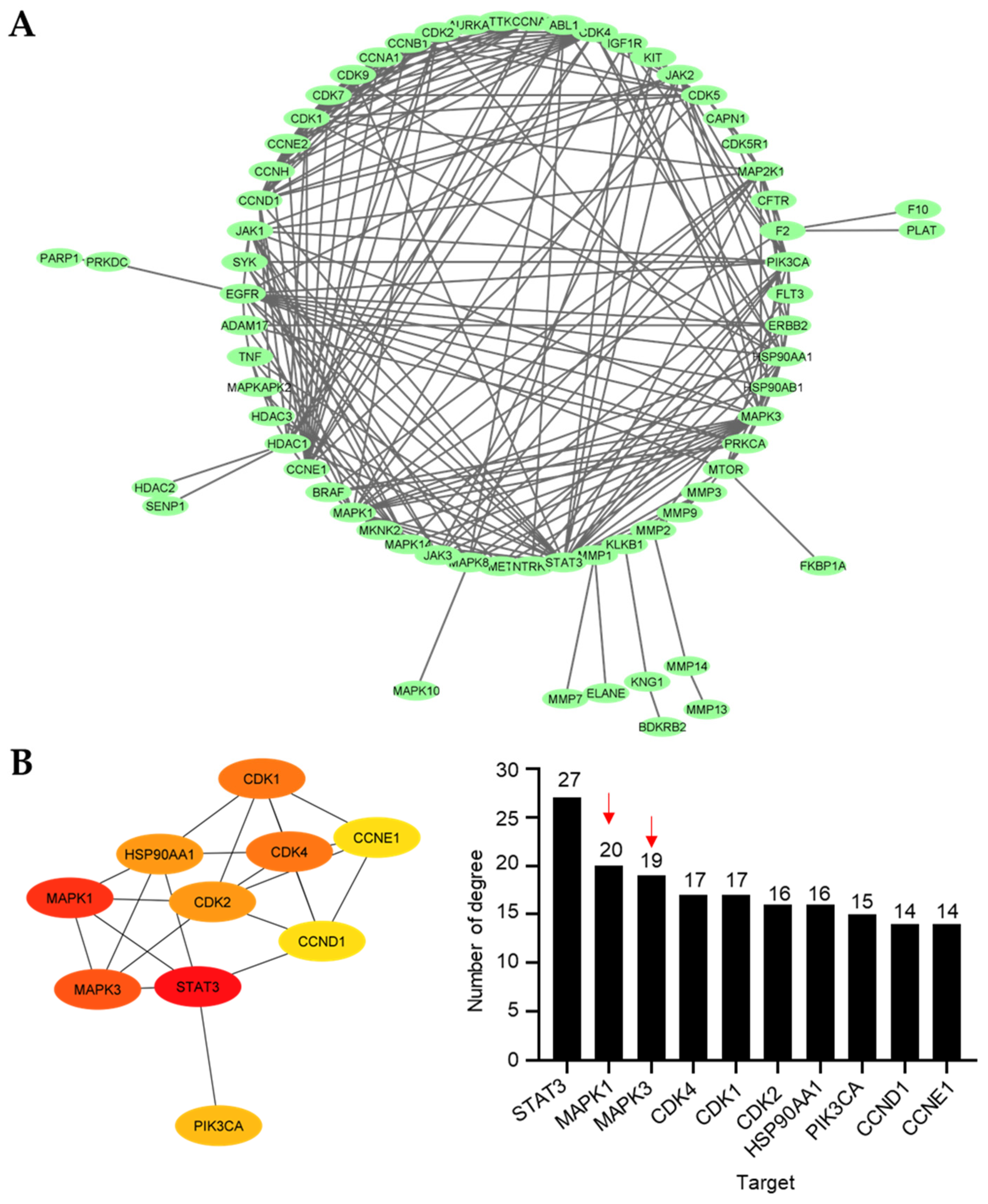
2.4.2. Protein–Protein Interaction Network Construction and Core Target Identification
To envision the relationship between the 94 prospective targets matched with 10 and 11 against NSCLC, a PPI network was retrieved and visualized using Cytoscape v3.9.1 after the resulting putative targets were loaded into the STRING platform (Figure 5A). The PPI network illustrates the interactions between the targets, with highly linked proteins in the network having a comparatively high number of interactions. CytoHubba, a plugin for Cytoscape, was employed to perform a topological analysis, which revealed the top 10 proteins linked to the number of degrees (Figure 5B). The colors from yellow to red represent small to large degrees. The results revealed several proteins, including signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 1 (MAPK1), mitogen-activated protein kinase 3 (MAPK3), cyclin-dependent kinase 4 (CDK4), cyclin-dependent kinase 1 (CDK1), cyclin-dependent kinase 2 (CDK2), 90 kDa heat shock protein (HSP90AA1), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), cyclin D1 (CCND1), and cyclin E1 (CCNE1), as the 10 core hub proteins serving as potential therapeutic targets against NSCLC. Regarding the previous virtual study, gene ontology and KEGG pathway enrichment analyses depicted that MAPKs were potential targets of 9 toward the induction of NSCLC apoptosis [29]. Thus, we focused on MAPK1 and MAPK3 for further analysis due to the structural similarities between 9 and 11, which are the 4′-pyridinecarbonyl esters of the tetrahydroisoquinolinequinone alkaloids.
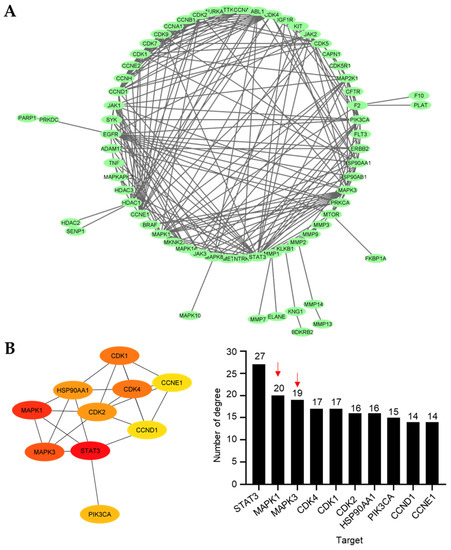
Figure 5.
Protein–protein interaction (PPI). (A) The main cluster of the PPI obtained from the 94 overlapping targets and visualized using Cytoscape 3.9.1. (B) Top 10 targets based on the number of degrees visualized by the CytoHubba plugin. More than 10 degrees are displayed for all the main targets. The intensity of the colors represents the degree values, where red, orange, and yellow correspond to large, medium, and small values, respectively. Red arrow indicates the MAPK family.
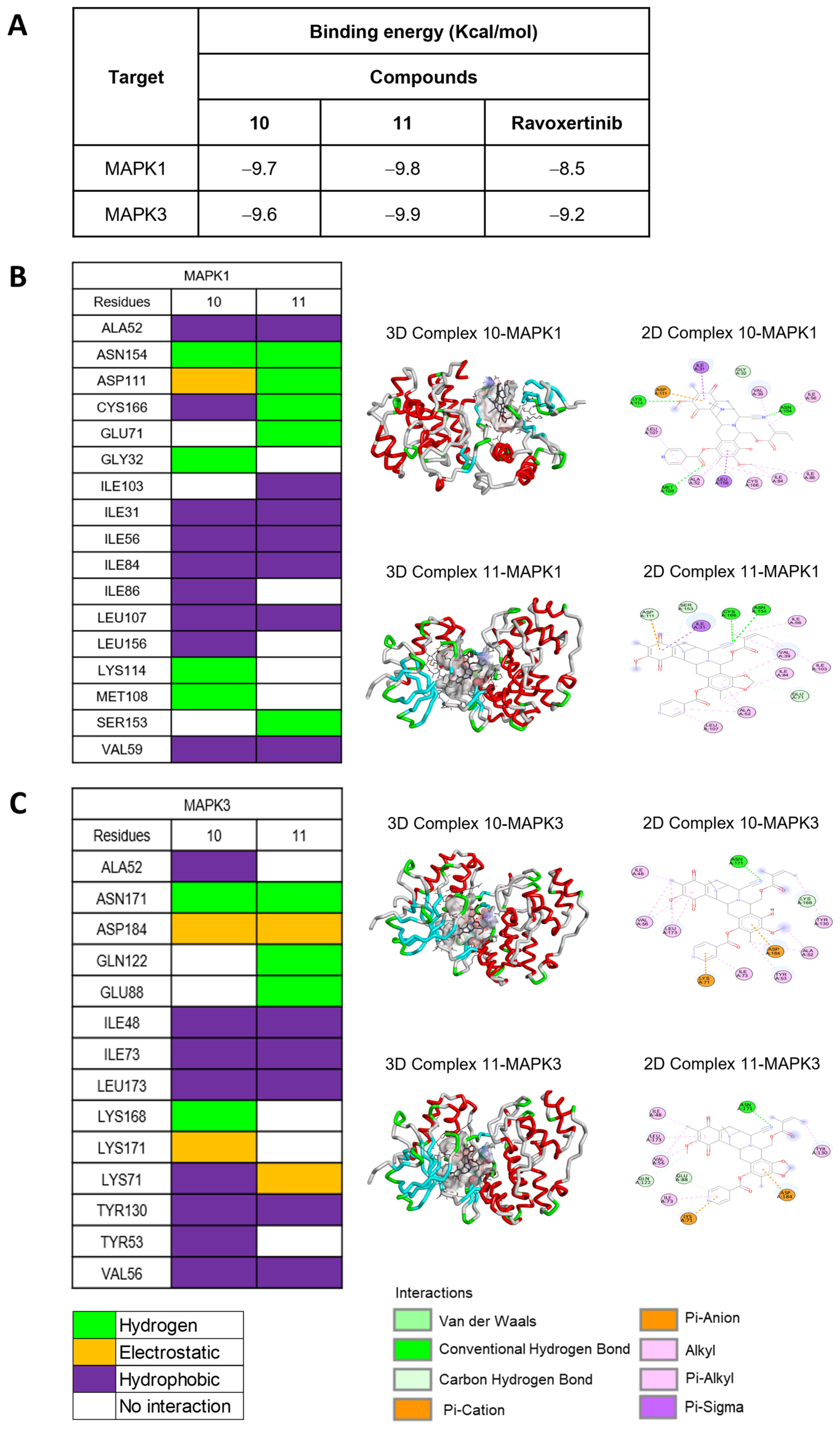
2.4.3. Molecular Docking of MAPK1 and MAPK3
To explore the binding relationships between the MAPK1 and MAPK3 proteins, compounds 10 and 11 were docked in comparison with Ravoxertinib, a known inhibitor of both MAPK1 and MAPK3 (see Supporting Information, Figure S16). From the binding energy calculations, 11 exhibited the lowest binding energies of −9.8 kcal/mol and −9.9 kcal/mol, respectively, for MAPK1 and MAPK3, which was better than those of 10 and Ravoxertinib (Figure 6A). Furthermore, docking studies were conducted for the protonated forms of compounds 10 and 11, specifically protonated at the nitrogen at position 12, as shown in the Supporting Information (Figure S17). Both the neutral and protonated forms of compounds 10 and 11 exhibited similar binding energy values. Docking with MAPK-1 revealed that both the neutral and protonated forms of compounds 10 and 11 predominantly displayed comparable binding interactions with amino acid residues. Interestingly, the protonated forms of compounds 10 and 11 exhibited hydrogen bonding interactions rather than hydrophobic interactions. Docking with MAPK-3 demonstrated that the protonated forms of compounds 10 and 11 could engage in additional interactions at the enzyme site. However, these additional interactions primarily resulted from hydrophobic interactions, which are generally weaker compared to hydrogen bonding interactions. Upon considering the binding interactions of amino acid residues at the enzyme active site, the neutral forms of compounds 10 and 11 exhibited a closer match with the known ligand, Ravoxertinib, compared to the protonated forms. Specifically, for MAPK-1, the neutral forms of compounds 10 and 11 demonstrated interactions with amino acid residues, including ASP111, ILE31, ILE103, and LEU156. For MAPK-3, the neutral forms of compounds 10 and 11 displayed interactions with LEU173 and VAL56. The number of interacting bonds involving hydrogen, hydrophobic interactions, and electrostatic interactions demonstrated the stability of both 10 and 11 in the binding pocket of both MAPK1 and MAPK3. However, compound 11 interacted with both MAPK1 and MAPK3 with stronger hydrogen binding than that of 10 (Figure 6B,C). Regarding the amino acid residues of MAPK1 that interacted with 11, the key binding site of MAPK1 comprised ALA52, ASN154, ASP111, CYS166, GLU71, and ILE103. The interactions between 11 and MAPK3 at the specific amino acid residues involved ASN171, GLN122, ILE48, LEU173, LYS71, and TYR130. Interestingly, compounds 10 and 11, along with compound 9, had similar binding sites, which confirmed that MAPK1 and MAPK3 are the biomolecular targets. The docking conformations of 11 with MAPK1 and MAPK3 demonstrated stable ligand–receptor binding and increased interaction, thereby affirming the necessity of validating the interaction stability through molecular dynamics simulation.
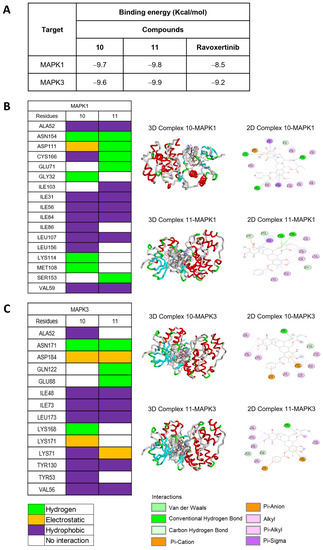
Figure 6.
Molecular docking of 10 and 11 with both MAPK1 (ERK2) and MAPK3 (ERK1). (A) Binding energies indicate that the interactions of 10 and 11 with both MAPK1 and MAPK3, compared to Ravoxertinib, a known inhibitor. (B) Interactions between the amino acid residues of MAPK1 and both 10 and 11. (C) Interactions between the amino acid residues of MAPK3 and both 10 and 11.
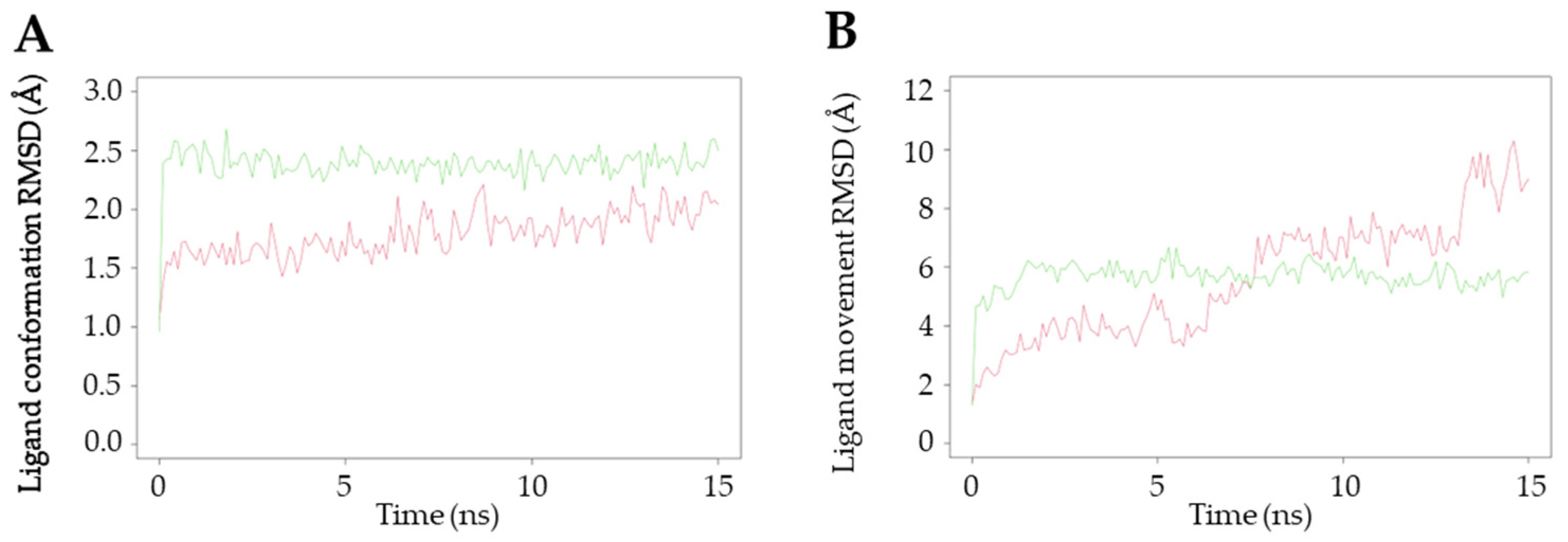
2.4.4. Molecular Dynamics Analysis of MAPK1 and MAPK3 with 11
To gain an in-depth understanding of the protein–ligand interaction of 11, a molecular dynamics study was conducted on the core proteins of MAPK1 and MAPK3, utilizing the protein–ligand complexes obtained from the results of the molecular docking simulations. When the trajectory exhibits steep changes, it indicates that the system has undergone a significant transition, whereas a smooth trajectory signifies that the system has reached equilibrium. One crucial parameter in molecular dynamics simulations is the root mean square deviation (RMSD), which serves as a measure of the stability of the trajectory. In this simulation, the conformation and movement of the ligand were analyzed using the RMSD value. The RMSD (Å) values of the ligand conformations for MAPK1 and MAPK3 were found to be 2.38 ± 0.15 and 1.79 ± 0.19, respectively, as depicted in Figure 7A. The ligand conformation analysis revealed that both MAPK1 and MAPK3 maintained consistent proximity throughout the entire 15 ns duration of the experiment. However, when comparing the RMSD values, the interaction between 11 and MAPK3 exhibited greater stability in the ligand conformation, and both protein targets did not show significant differences in terms of their interactions. Additionally, the RMSD (Å) of the ligand movement between 11 and these targets was assessed. The results indicated that the average RMSD values for MAPK1 and MAPK3 were 5.68 ± 0.52 and 5.63 ± 2.04, respectively (Figure 7B). A relatively small RMSD of the ligand movement indicates good proximity between the ligand and the targets. The stability of the interaction between compound 11 and MAPK3 showed a greater tendency to fluctuate at 5, 7.5, and 13.2 ns compared to the interaction with MAPK1, particularly at 5 ns intervals.
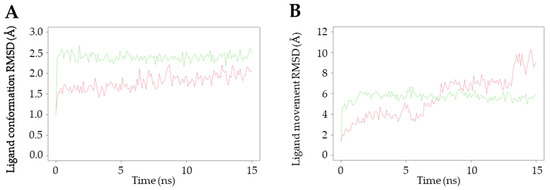
Figure 7.
Molecular dynamics study of the interactions between 11 and MAPK1 and MAPK3. Green line shows the interaction with MAPK1, whereas the red line shows the interaction with MAPK3. (A) The ligand conformation and (B) ligand movement interaction of 11 with MAPK1 and MAPK3.
Considering all these findings, we hypothesized that 11 interacts with multiple targets in the MAPK family. Among these targets, MAPK1 demonstrated the highest potential and consistently engaged in strong interactions with 11, aligning with our previous investigation. According to previous studies investigating the structure-cytotoxicity relationship, which employed both in vitro and in silico approaches, the mechanistic pathways underlying the anti-lung cancer effects of marine alkaloids from the tetrahydroisoquinolinequinone family have been elucidated. Specifically, compounds such as 5-O-(N-Boc-L-alanine)-renieramycin T, a renieramycin-ecteinascidin derivative [20], (1R,4R,5S)-10-(benzyloxy)-9-methoxy-8,11-dimethyl-3-(thiazol-5-ylmethyl)-1,2,3,4,5,6-hexahydro-1,5-epimi- nobenzo [d]azocine-4-carbonitrile (DH_25), a right-half C–E ring analog of renieramycins [30], and 22-O-(4′-pyridinecarbonyl) jorunnamycin A, a 4′-pyridinecarbonyl substituted renieramycin-type derivative [29], have demonstrated efficacy against non-small cell lung cancer (NSCLC) cells through modulation of the protein kinase B (Akt), myeloid cell leukemia-1 (MCL-1), and mitogen-activated protein kinase (MAPK) signaling pathways, leading to the induction of apoptosis. The proteins Akt, MCL-1, and MAPK are interconnected targets of cellular signaling networks involved in the regulation of apoptosis. While the MAPK pathway exerts pro-apoptotic effects, Akt signaling generally promotes cell survival. Furthermore, MCL-1, functioning as an anti-apoptotic protein, is influenced by both the MAPK and Akt pathways, thereby contributing to the delicate balance between cell survival and apoptosis in cancer cells [31,32]. Therefore, targeting these signaling pathways and their interactions holds promise for therapeutic interventions aimed at inducing apoptosis in cancer therapy.
3. Materials and Methods
3.1. General Experimental Procedures
All commercial reagents were purchased from Tokyo Chemical Industry, Tokyo, Japan. The glassware was dried in an oven (105 °C) before the experiment. Photochemical reactions were carried out using a commercially supplied LED (4W LED Filament GLS Blue, 220–240 V, EVE) and 18-W fluorescent light (100 V, HATAYA, Tokyo, Japan). All reactions were monitored by thin-layer chromatography (TLC) using aluminum silica gel 60 F254 (Merck, Darmstadt, Germany), and the TLC chromatogram was recorded under UV light at 254 and 365 nm. Flash column chromatographic purification was performed with silica gel (230–400 mesh) using a mixture of ethyl acetate and hexane as the mobile phase. Moreover, 1H and 13C NMR spectra were recorded on a Bruker ADVANCE NEO NMR spectrometer (Bruker, Massachusetts, MA, USA) at 400 and 100 MHz, respectively. Deuterated chloroform (CDCl3) was used as the internal standard for the 13C (77.0 ppm) and 1H (7.27 ppm) spectroscopic analyses. Infrared spectra were measured using a Perkin Frontier Fourier-Transform Infrared Spectrometer (PerkinElmer, Massachusetts, MA, USA). High-resolution mass spectrometry was conducted using a microTOF Bruker Daltonics mass spectrometer. Optical rotations were measured with a JASCO P-2000 polarimeter using a 1-mL cell with a cell path length of 1 dm. The spectra of electronic circular dichroism were obtained using a JASCO J-815 CD spectrometer (JASCO, Maryland, MD, USA).
3.2. Extraction and Isolation
With assistance from the Aquatic Resources Research Institute, Chulalongkorn University and approval from the Department of Fisheries, Ministry of Agriculture and Cooperatives, Thailand (0510.2/8234, Date: 28 October 2019), a Thai blue sponge (Xestospongia sp.) was collected during a SCUBA diving mission at a depth of 3–5 m at Sichang Island, the Gulf of Thailand. The fresh blue sponge was mashed and subsequently pre-treated with 10% potassium cyanide solution in a phosphate-buffered solution at pH 7. Next, the suspension was macerated with methanol. The extraction process was repeated three times. The methanolic extracts were filtered, combined, and concentrated at reduced pressure. The condensed aqueous-methanolic extract was partitioned with hexane and ethyl acetate. The ethyl acetate extract was further concentrated and purified by silica gel column chromatography using hexane and ethyl acetate as the eluent. Natural renieramycins, including renieramycin M (4), renieramycin N (5) [11], and renieramycin O (6) [12], were obtained. Thereafter, compound 4 was transformed into Jorunnamycin A (3) under a three-step semisynthesis including hydrogenation, hydride reduction, and air oxidation [21,24].
3.3. Screening of the Photoredox Reaction Conditions with the Renieramycins
Natural renieramycins 4–6 were employed as the starting material for the light-induced intramolecular photoredox reaction. A solution of the renieramycin alkaloid (15 mg) with 20 mL of the selected solvent (CH2Cl2, CHCl3, and THF) was stirred vigorously under a light source comprising white light (18 W fluorescent) and blue light (4 W LED) for 24 h at room temperature under a nitrogen atmosphere. The reaction was monitored by TLC using a solution of ethyl acetate and hexane (1:1 v/v) as the mobile phase. After the completion of the reaction, the solvent was evaporated under reduced pressure. The resultant crude product was purified by flash column chromatography using silica gel as the stationary phase and the mixed solvents of ethyl acetate and hexane as the mobile phase to afford the corresponding products, 7 and 8.
3.4. Procedure for the Semisynthesis of Compound 11
Compound 4 (15 mg, 0.03 mmol) in dry CH2Cl2 (20 mL) was irradiated by a 4 W LED lamp (blue light) at room temperature under a nitrogen atmosphere for 24 h. The reaction was monitored by TLC using a hexane and ethyl acetate solution (1:1 v/v) as the mobile phase. After the completion of the reaction, the solvent was removed under reduced pressure. The crude product was precipitated using hexane, affording 7 as a yellow amorphous solid. Compound 7 was employed in the next step without further purification. Next, compound 7 (21 mg, 0.04 mmol), 4-dimethylaminopyridine (DMAP) (5 mg, 0.04 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDCI) (8 mg, 0.04 mmol), and isonicotinoyl chloride hydrochloride (32 mg, 0.18 mmol) were dissolved in CH2Cl2 (20 mL). The mixture was stirred for 24 h at room temperature under a nitrogen atmosphere. After the completion of the reaction, as determined by TLC, the reaction mixture was quenched by adding distilled H2O (20 mL) and extracted with CH2Cl2 (20 mL, three times). The organic layers were combined and dried over anhydrous magnesium sulfate (MgSO4), filtered, and concentrated under reduced pressure. After purification by silica gel flash chromatography using the hexane and ethyl acetate solution as the eluent, 5-O-(4′-pyridinecarbonyl) renieramycin T (11) was obtained in 20% yield as a yellow amorphous powder.
3.5. Procedure for the Semisynthesis of Compound 12
Jorunnamycin A (3) (25 mg, 0.05 mmol) was placed into an oven-dried, round-bottomed flask and dissolved in dry CH2Cl2 (10 mL). Thereafter, DMAP (31 mg, 0.25 mmol), EDCI (49 mg, 0.25 mmol), and isonicotinoyl chloride hydrochloride (45 mg, 0.25 mmol) were added. The mixture was stirred for 24 h at room temperature under a nitrogen atmosphere and monitored by TLC using a mixture solvent of ethyl acetate and hexane (1:1 v/v) as the mobile phase. After completion, the solution was concentrated under reduced pressure. Next, the resultant crude product was purified by flash column chromatography using silica gel as the stationary phase and the mixed solvent of ethyl acetate and hexane as the mobile phase to yield a yellow product (9) in 72% yield. Subsequently, compound 9 (6 mg, 0.01 mmol) was dissolved in CH2Cl2 (15 mL) and irradiated by a 4 W LED lamp (blue light) at room temperature under a nitrogen atmosphere for 24 h. The reaction mixture was monitored by TLC and further purified following the procedure described above to obtain 22-O-(4′-pyridinecarbonyl) renieramycin T (12) as an amorphous yellow powder in 83% yield.
3.6. Cytotoxic Evaluations against NSCLC Cell Lines
The in vitro cytotoxicity of compounds 3–12 against H292 and H460 NSCLC cell lines was determined by MTT colorimetric assay. The H292 and H460 cultured cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The Roswell Park Memorial Institute (RPMI) medium 1640, fetal bovine serum (FBS), L-glutamine, a penicillin/streptomycin solution, and Albumax I were purchased from Gibco (Gaithersburg, MA, USA). Dimethyl sulfoxide (DMSO) was acquired from Merck Millipore (Billerica, MA, USA) or Sigma-Aldrich. All cell lines were cultured in the RPMI 1640 medium, supplemented with 10% FBS, 2 mM L-glutamine, and 100 units/mL of penicillin/streptomycin, and the temperature was maintained at 37 °C under 5% CO2. Cells were trypsinized and seeded at a density of 5000 cells/well with the RPMI media for 24 h in a 96-well plate. A 10 mm stock solution of each test compound in DMSO was prepared and diluted into the serial concentrations at 1–250 nM, with a DMSO concentration of less than 0.2% v/v. The cells were treated with various concentrations of each derivative for 72 h. Next, the cells were incubated with 0.5 mg/mL of MTT for 2 h. After solubilizing the resulting formazan salt with 100 µL of DMSO, the absorbance of the formed formazan was measured by a spectrophotometric microtiter plate reader (Perkin Elmer Victor 3 1420 Multilabel Plate Counter, Massachusetts, MA, USA) at 570 nm. The experiment was performed in three replicate wells with at least five concentrations of the tested compounds. The cell viability was determined using the GraphPad Prism software (version 5), calculated as a percentage of non-treated control cells and the mean of the half-maximal inhibitory concentration (IC50) values. Cisplatin and doxorubicin were used as the positive controls, and 0.2% DMSO was used as the negative control.
3.7. Network Pharmacology Study
3.7.1. Database Mining and Identification of 10, 11, and NSCLC-Related Targets
The Swiss Target Prediction database (http://www.swisstargetprediction.ch), which was accessed on 14 January 2023, was utilized to gather information on the probable targets of 10 and 11. To identify potential targets associated with NSCLC cells, various human genetic databases, such as GeneCards, TTD, OMIM, and DisGeNET, were screened. A Venn diagram illustrating the overlapping targets between 10 and 11 with respect to NSCLC can be viewed at this link (https://bioinfogp.cnb.csic.es/tools/venny/, accessed on 15 March 2023). The Venn diagram was generated using Venny 2.1.0 (Centro Nacional de Biotecnología, Madrid, Spain), https://bioinfogp.cnb.csic.es/tools/venny/, accessed on 15 March 2020.
3.7.2. Construction of Protein–Protein Interaction Network and Identification of Potential Core Targets
The overlapping targets derived from the Venn diagram were inputted into the STRING database. For STRING analysis, the organism parameter was set to Homo sapiens with the highest confidence level of 0.900. The obtained results were gathered and imported into Cytoscape v3.9.1 (The Cytoscape Consortium, San Diego, CA, USA) for visualization and further analysis. To examine the top 10 core targets, the CytoHubba plugin was employed, using the number of degrees as the primary parameter.
3.8. Molecular Docking Study
The ligand structures (10, 11, and Ravoxertinib) were prepared using ChemDraw Ultra 15.0 (Perkin Elmer, MA, USA). The protonation state of 10 and 11 were prepared using Open Babel GUI and Command Prompt with pH 7.4. To predict the potential binding interactions between the compounds and the core targets, an in silico assay was conducted using PyRx Virtual Screening Tools v0.8. The X-ray crystal structures of the proteins were obtained from the Protein Data Bank (https://www.rcsb.org/, accessed on 16 March 2023), e.g., 6GES for MAPK3 (ERK1) and 1WZY for MAPK1 (ERK2). Prior to the analysis, the proteins were prepared by removing water molecules and any other ligands attached to the main protein using Pymol v2.5 (https://pymol.org/2/, accessed on 16 March 2023). The free binding energies (ΔG), interaction types, and amino acid residues were calculated and analyzed using BIOVIA Discovery Studio Visualizer 2022 (Biovia, CA, USA).
3.9. Molecular Dynamics Study
The stability of the interactions between ligand 11 and the proteins was assessed using Yasara Dynamic, following the previously described methodology [29]. In summary, the optimal binding between ligand 11 and the proteins was selected for further analysis, incorporating the AMBER14 force field with standard parameters. The simulation was conducted at a temperature of 298 K and a pH of 7.4 for 15 ns. Two RMSD parameters were employed: RMSD ligand conformation and RMSD ligand movement. The evaluation utilized the default scripts: md_run.mcr and md_analyse.mcr. The RMSD plots were generated using the Rstudio program (https://posit.co/products/open-source/rstudio/, accessed on 17 March 2023).
4. Conclusions
Here, 4′-pyridinecarbonyl-substituted renieramycin-type derivatives were prepared from compound 4 using light-induced intramolecular photoredox reactions and Steglich esterification. The optimal conditions for the photoredox reaction were achieved by employing 4 W blue light and dichloromethane. The natural renieramycins (4–6) were successfully transformed into their corresponding derivatives (7 and 8, which contained a 1,3-dioxole-bridged phenolic moiety on ring A) in excellent–moderate yields (yields: 81%–37%). The attachment of the 4′-pyridinecarbonyl ester at the C-5 and C-22 positions was accomplished smoothly. Cytotoxicity study revealed that compounds 4 and 9 exhibited the most potent cytotoxicity against the human H292 and H460 NSCLC cell lines among the natural renieramycins and semisynthesized renieramycin-type derivatives, respectively. Interestingly, compound 11, which contained both the 4′-pyridinecarbonyl ester at C-5 and the 1,3-dioxole-bridged phenolic motifs on ring A, showed better cytotoxicity than its parent compound (7). In addition, the renieramycin-type derivatives with a hydroxyl group at C-5 and C-22 lost their cytotoxicity against the NSCLC cells. Thus, the conformation of ring A featuring the 4′-pyridinecarbonyl ester at C-5 is crucial for the cytotoxic activity of renieramycins. Due to the limited quantity of the compound, a virtual network pharmacology study was conducted to simulate the target genes and molecular pathways. Molecular docking and dynamics studies predicted that the target proteins of compound 11 were MAPK1 and MAPK3. In future studies, the renieramycin-type derivatives will be further explored for their potential as anti-lung cancer agents.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21070400/s1, Figures S1–S15: 1H NMR, 13C NMR and 2D NMR spectra of compounds 7–9 and 11–12, Figures S16 and S17: Molecular docking of Ravoxertinib and the protonated 10 and 11 with both MAPK1 (ERK2) and MAPK3 (ERK1), Tables S1 and S2: Theoretical level of minimization of the 3D structures of 10 and 11.
Author Contributions
Conceptualization, S.C.; methodology, S.S., K.B., K.P., B.I., K.S., M.Y., N.S., P.C. and S.C.; software, V.P., I.I. and S.C.; validation, K.S., M.Y., N.S. and S.C.; formal analysis, S.S., K.B., I.I., K.P., B.I., C.C. and S.C.; investigation, S.S., K.B., I.I., K.P., B.I. and S.C.; resources, P.C. and S.C.; data curation, S.C.; writing—original draft preparation, S.S., I.I. and K.B.; writing—review and editing, S.C.; visualization, S.C.; supervision, S.C.; project administration, S.C.; funding acquisition, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Thai Research Fund, Thailand (RSA 6280009) to S.C.
Data Availability Statement
The data described and analyzed in this study are available from the corresponding author upon reasonable request.
Acknowledgments
This research is supported by the 90th anniversary of Chulalongkorn university fund No. GCUGR1125661018M (Ratchadaphiseksomphot Endowment Fund) for S.S. and the Second Century Fund (C2F), Chulalongkorn University for postdoctoral research fellowship to K.B. We gratefully acknowledge the Natural Products and Nanoparticles Research Unit (NP2) and the Department of Pharmacognosy and Pharmaceutical Botany and the Department of Pharmacology and Physiology, Faculty of Pharmaceutical Sciences, Chulalongkorn University for their scientific support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fang, Y.; Li, H.; Ji, B.; Cheng, K.; Wu, B.; Li, Z.; Zheng, C.; Hua, H.; Li, D. Renieramycin-type alkaloids from marine-derived organisms: Synthetic chemistry, biological activity and structural modification. Eur. J. Med. Chem. 2021, 210, 113092. [Google Scholar] [CrossRef] [PubMed]
- Saito, N. Chemical Research on Antitumor Isoquinoline Marine Natural Products and Related Compounds. Chem. Pharm. Bull. 2021, 69, 155–177. [Google Scholar] [CrossRef] [PubMed]
- D’Incalci, M.; Galmarini, C.M. A Review of Trabectedin (ET-743): A Unique Mechanism of Action. Mol. Cancer Ther. 2010, 9, 2157–2163. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.M.; Sankhala, K.K.; Chawla, N.; Chawla, S.P. Trabectedin for Soft Tissue Sarcoma: Current Status and Future Perspectives. Adv. Ther. 2016, 33, 1055–1071. [Google Scholar] [CrossRef]
- Cesne, A.L.; Martín-Broto, J.; Grignani, G. A review of the efficacy of trabectedin as second-line treatment of advanced soft tissue sarcoma. Future Oncol. 2022, 18, 5–11. [Google Scholar] [CrossRef]
- Patel, S.; Petty, W.J.; Sands, J.M. An overview of lurbinectedin as a new second-line treatment option for small cell lung cancer. Ther. Adv. Med. Oncol. 2021, 13, 17588359211020529. [Google Scholar] [CrossRef]
- Manzo, A.; Sforza, V.; Carillio, G.; Palumbo, G.; Montanino, A.; Sandomenico, C.; Costanzo, R.; Esposito, G.; Laudato, F.; Mercadante, E.; et al. Lurbinectedin in small cell lung cancer. Front. Oncol. 2022, 12, 932105. [Google Scholar] [CrossRef]
- Frincke, J.M.; Faulkner, D.J. Antimicrobial metabolites of the sponge Reniera sp. J. Am. Chem. Soc. 1982, 104, 265–269. [Google Scholar] [CrossRef]
- He, H.Y.; Faulkner, D.J. Renieramycins E and F from the sponge Reniera sp.: Reassignment of the stereochemistry of the renieramycins. J. Org. Chem. 1989, 54, 5822–5824. [Google Scholar] [CrossRef]
- Pettit, G.R.; Knight, J.C.; Collins, J.C.; Herald, D.L.; Pettit, R.K.; Boyd, M.R.; Young, V.G. Antineoplastic Agents 430. Isolation and Structure of Cribrostatins 3, 4, and 5 from the Republic of Maldives Cribrochalina Species. J. Nat. Prod. 2000, 63, 793–798. [Google Scholar] [CrossRef]
- Suwanborirux, K.; Amnuoypol, S.; Plubrukarn, A.; Pummangura, S.; Kubo, A.; Tanaka, C.; Saito, N. Chemistry of Renieramycins. Part 3. Isolation and Structure of Stabilized Renieramycin Type Derivatives Possessing Antitumor Activity from Thai Sponge Xestospongia Species, Pretreated with Potassium Cyanide. J. Nat. Prod. 2003, 66, 1441–1446. [Google Scholar] [CrossRef]
- Amnuoypol, S.; Suwanborirux, K.; Pummangura, S.; Kubo, A.; Tanaka, C.; Saito, N. Chemistry of Renieramycins. Part 5. Structure Elucidation of Renieramycin-Type Derivatives O, Q, R, and S from Thai Marine Sponge Xestospongia Species Pretreated with Potassium Cyanide. J. Nat. Prod. 2004, 67, 1023–1028. [Google Scholar] [CrossRef]
- Nakao, Y.; Shiroiwa, T.; Murayama, S.; Matsunaga, S.; Goto, Y.; Matsumoto, Y.; Fusetani, N. Identification of Renieramycin A as an Antileishmanial Substance in a Marine Sponge Neopetrosia sp. Mar. Drugs 2004, 2, 55–62. [Google Scholar] [CrossRef]
- Daikuhara, N.; Tada, Y.; Yamaki, S.; Charupant, K.; Amnuoypol, S.; Suwanborirux, K.; Saito, N. Chemistry of renieramycins. Part 7: Renieramycins T and U, novel renieramycin–ecteinascidin hybrid marine natural products from Thai sponge Xestospongia sp. Tetrahedron Lett. 2009, 50, 4276–4278. [Google Scholar] [CrossRef]
- Chamni, S.; Sirimangkalakitti, N.; Chanvorachote, P.; Saito, N.; Suwanborirux, K. Chemistry of Renieramycins. 17. A New Generation of Renieramycins: Hydroquinone 5-O-Monoester Analogues of Renieramycin M as Potential Cytotoxic Agents against Non-Small-Cell Lung Cancer Cells. J. Nat. Prod. 2017, 80, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Petsri, K.; Chamni, S.; Suwanborirux, K.; Saito, N.; Chanvorachote, P. Renieramycin T Induces Lung Cancer Cell Apoptosis by Targeting Mcl-1 Degradation: A New Insight in the Mechanism of Action. Mar. Drugs 2019, 17, 301. [Google Scholar] [CrossRef] [PubMed]
- Petsri, K.; Yokoya, M.; Tungsukruthai, S.; Rungrotmongkol, T.; Nutho, B.; Vinayanuwattikun, C.; Saito, N.; Matsubara, T.; Sato, R.; Chanvorachote, P. Structure–Activity Relationships and Molecular Docking Analysis of Mcl-1 Targeting Renieramycin T Analogues in Patient-derived Lung Cancer Cells. Cancers 2020, 12, 875. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liang, J.; Li, X.; Liu, L.; Yao, J.; Chen, X.; Chen, R. Renieramycin T Inhibits Melanoma B16F10 Cell Metastasis and Invasion via Regulating Nrf2 and STAT3 Signaling Pathways. Molecules 2022, 27, 5337. [Google Scholar] [CrossRef]
- Chantarawong, W.; Chamni, S.; Suwanborirux, K.; Saito, N.; Chanvorachote, P. 5-O-Acetyl-Renieramycin T from Blue Sponge Xestospongia sp. Induces Lung Cancer Stem Cell Apoptosis. Mar. Drugs 2019, 17, 109. [Google Scholar] [CrossRef]
- Suksamai, D.; Racha, S.; Sriratanasak, N.; Chaotham, C.; Aphicho, K.; Lin, A.C.K.; Chansriniyom, C.; Suwanborirux, K.; Chamni, S.; Chanvorachote, P. 5-O-(N-Boc-l-Alanine)-Renieramycin T Induces Cancer Stem Cell Apoptosis via Targeting Akt Signaling. Mar. Drugs 2022, 20, 235. [Google Scholar] [CrossRef]
- Charupant, K.; Daikuhara, N.; Saito, E.; Amnuoypol, S.; Suwanborirux, K.; Owa, T.; Saito, N. Chemistry of renieramycins. Part 8: Synthesis and cytotoxicity evaluation of renieramycin M–jorunnamycin A analogues. Bioorg. Med. Chem. 2009, 17, 4548–4558. [Google Scholar] [CrossRef] [PubMed]
- Sirimangkalakitti, N.; Chamni, S.; Charupant, K.; Chanvorachote, P.; Mori, N.; Saito, N.; Suwanborirux, K. Chemistry of Renieramycins. 15. Synthesis of 22-O-Ester Derivatives of Jorunnamycin A and Their Cytotoxicity against Non-Small-Cell Lung Cancer Cells. J. Nat. Prod. 2016, 79, 2089–2093. [Google Scholar] [CrossRef]
- Yokoya, M.; Yamazaki-Nakai, M.; Nakai, K.; Sirimangkalakitti, N.; Chamni, S.; Suwanborirux, K.; Saito, N. Transformation of Renieramycin M into Renieramycins T and S by Intramolecular Photoredox Reaction of 7-Methoxy-6-methyl-1,2,3,4-tetrahydroisoquinoline-5,8-dione Derivatives. J. Nat. Prod. 2023, 86, 222–231. [Google Scholar] [CrossRef]
- Saito, N.; Tanaka, C.; Koizumi, Y.-I.; Suwanborirux, K.; Amnuoypol, S.; Pummangura, S.; Kubo, A. Chemistry of renieramycins. Part 6: Transformation of renieramycin M into jorumycin and renieramycin J including oxidative degradation products, mimosamycin, renierone, and renierol acetate. Tetrahedron 2004, 60, 3873–3881. [Google Scholar] [CrossRef]
- Chamni, S.; Sirimangkalakitti, N.; Chanvorachote, P.; Suwanborirux, K.; Saito, N. Chemistry of Renieramycins. Part 19: Semi-Syntheses of 22-O-Amino Ester and Hydroquinone 5-O-Amino Ester Derivatives of Renieramycin M and Their Cytotoxicity against Non-Small-Cell Lung Cancer Cell Lines. Mar. Drugs 2020, 18, 418. [Google Scholar] [CrossRef] [PubMed]
- Xi, P.; Niu, Y.; Zhang, Y.; Li, W.; Gao, F.; Gu, W.; Kui, F.; Liu, Z.; Lu, L.; Du, G. The mechanism of dioscin preventing lung cancer based on network pharmacology and experimental validation. J. Ethnopharmacol. 2022, 292, 115138. [Google Scholar] [CrossRef]
- Iksen, I.; Witayateeraporn, W.; Wirojwongchai, T.; Suraphan, C.; Pornputtapong, N.; Singharajkomron, N.; Nguyen, H.M.; Pongrakhananon, V. Identifying molecular targets of Aspiletrein-derived steroidal saponins in lung cancer using network pharmacology and molecular docking-based assessments. Sci. Rep. 2023, 13, 1545. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Iksen, I.; Sinsook, S.; Wattanathamsan, O.; Buaban, K.; Chamni, S.; Pongrakhananon, V. Target Identification of 22-(4-Pyridinecarbonyl) Jorunnamycin A, a Tetrahydroisoquinoline Derivative from the Sponge Xestospongia sp., in Mediating Non-Small-Cell Lung Cancer Cell Apoptosis. Molecules 2022, 27, 8948. [Google Scholar] [CrossRef]
- Petsri, K.; Yokoya, M.; Racha, S.; Thongsom, S.; Thepthanee, C.; Innets, B.; Ei, Z.Z.; Hotta, D.; Zou, H.; Chanvorachote, P. Novel Synthetic Derivative of Renieramycin T Right-Half Analog Induces Apoptosis and Inhibits Cancer Stem Cells via Targeting the Akt Signal in Lung Cancer Cells. Int. J. Mol. Sci. 2023, 24, 5345. [Google Scholar] [CrossRef]
- Kitazumi, I.; Tsukahara, M. Regulation of DNA fragmentation: The role of caspases and phosphorylation. FEBS J. 2011, 278, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Pithi, C.; Supakarn, C.; Chuanpit, N.; Preeyaporn Plaimee, P. Potential Anti-metastasis Natural Compounds for Lung Cancer. Anticancer Res. 2016, 36, 5707. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).