Abstract
Marine toxins have potent actions on diverse sodium ion channels regulated by transmembrane voltage (voltage-gated ion channels) or by neurotransmitters (nicotinic acetylcholine receptor channels). Studies of these toxins have focused on varied aspects of venom peptides ranging from evolutionary relationships of predator and prey, biological actions on excitable tissues, potential application as pharmacological intervention in disease therapy, and as part of multiple experimental approaches towards an understanding of the atomistic characterization of ion channel structure. This review examines the historical perspective of the study of conotoxin peptides active on sodium channels gated by transmembrane voltage, which has led to recent advances in ion channel research made possible with the exploitation of the diversity of these marine toxins.
1. An Overview of Bioactive Marine Toxins
A great diversity of marine organisms produce and secrete bioactive toxins, which have apparently evolved as an evolutionary strategy for increasing fitness by enhancing functions ranging from predictable defense against predation, prey capture, and immobilization [1,2]. As marine and other bioactive toxins have been identified and their effects partially or fully characterized, additional research has focused on evolutionary patterns of development of resistance to these toxins [3,4,5,6,7]. The venom of marine cone snails (Conus sp.) has attracted considerable research attention with studies of behavioral strategies, evolution, and toxicity [8,9,10,11,12,13]. A large diversity of species in this molluscan genus produce a plethora of bioactive conopeptides, with over 700 species and more than 10,000 conotoxin peptides identified to date [14]; for review of conotoxin gene families see [15], noting that many additional conotoxin genes have been sequenced [16]. This research has been motivated in part as an element of evolutionary studies as mentioned above, and also by an interest in determining sequence- and disulfide-linkage determinants of toxin binding and isoform-specific bioactivity. Functional characterizations of conopeptides and conotoxins have provided unique opportunities to study ligand-gated receptor channels, voltage-gated channels, transport proteins, and GTP binding proteins in specific tissues (for reviews see [17,18,19,20,21,22]).
In addition, these toxins have been invaluable as part of structural studies (reviewed by [23,24,25]) and provide promise for pharmacological intervention in disease (reviewed by [26,27,28,29,30,31,32,33]). Progress in the development of conopeptides or analogues as analgesics has resulted in FDA approval of Ziconotide, or synthetic ω-conotoxin MVIIA [34]. Investigation of the therapeutic potential of conopeptides for nonopioid analgesia continues (reviewed by [35,36,37]), and a recent study demonstrating agonist activity of Conus venom extracts on CB1 receptors as determined by internalization and resulting in significant analgesic or antinociceptive effects in mice [38]. Acceleration of the development of conopeptide analogues may be facilitated with alternative synthesis approaches as has been shown for the μ-conotoxin KIIIA [39].
Many α-conotoxins have been purified or synthesized, and their actions on nicotinic acetylcholine receptors (nAChRs) have been and continue to be investigated (for reviews see [40,41,42,43,44,45,46]). The diversity of α-conotoxins has been exploited to investigate acetylcholine receptors comprising subunit combinations that are determinants of synaptic communication in the nervous system and periphery (reviewed by [47]). This work has evolved to include robust computational approaches including molecular dynamics and in silico scanning of potential α-conotoxin analogues (reviewed by [48]); recent work includes [49,50,51,52,53,54]. These and functional investigations have explored α-conotoxin specificity in the characterization of nAChR permutations, and the potential therapeutic use of naturally occurring, synthetic, and mutant α-conotoxins to treat neurodegenerative diseases including Parkinson’s Disease [55,56,57,58,59,60], as well as management of peripheral painful neuropathies [51,61,62,63,64,65,66].
2. Toxins Targeting Voltage-Gated Sodium Channels
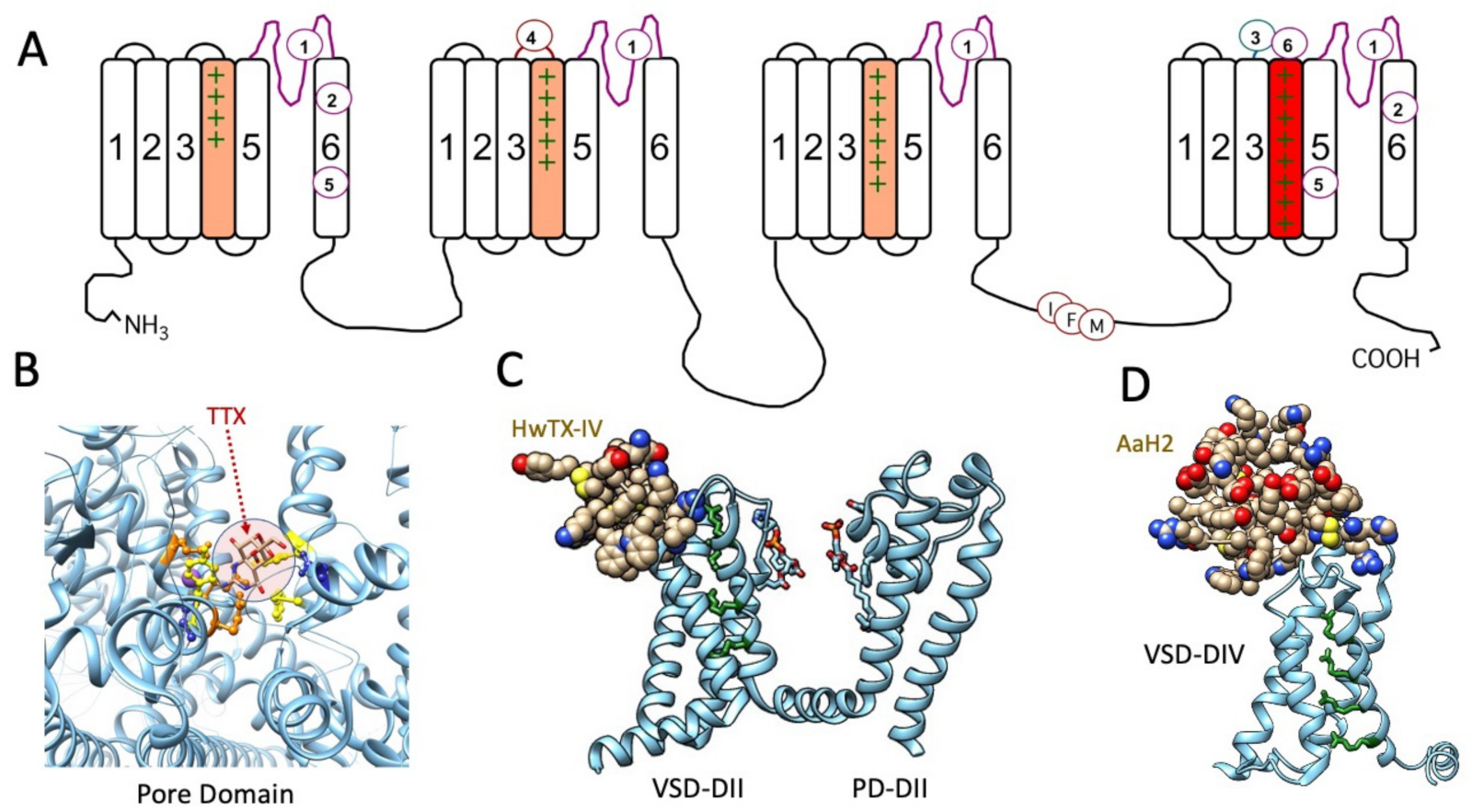
Biological toxins with potent actions on voltage-gated sodium channels act on numerous receptor sites (reviewed by [67]; Figure 1A). Alkaloid guanidinium toxins such as saxitoxin (STX), tetrodotoxin (TTX), and the μ-conotoxins act at site 1 within the outer vestibule of the pore module, to limit sodium permeation during channel opening. Receptor site 2 in S6 of domain I and domain IV, and perhaps extending to domains II and III [68,69], bind plant alkaloids such as veratridine and animal toxins such as batrachotoxin. Other bioactive toxins including dinoflagellate ciguatoxins and brevetoxins act at site 5 in DI S6 and involve DIV S5 (reviewed by [17,70,71,72,73]) to promote activation and slow the entry of sodium channels into the fast inactivated state. Sea anemone, spider, and scorpion toxins bind to extracellular loops comprising receptor sites 3 or 4 in the voltage-sensing module of domains IV and II, respectively. These toxins modify channel gating by trapping the S4 segment voltage sensor in resting or activated states (reviewed by [22,74,75,76,77,78,79]). Conotoxins target sites in each of these S3–S4 extracellular loops. For example, receptor site 6 is, similar to site 3, in the domain IV S3–S4 linker and is targeted by δ-conotoxins that slow the entry of channels into a state of fast inactivation [80]. The μO-conotoxins have actions suggesting they act, in part, at receptor site 4 to limit sodium permeance, through gating modifying actions similar to β scorpion toxins [81]. A revised convention for these binding site models is provided in [67].
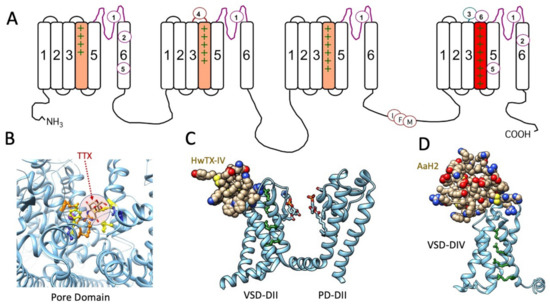
Figure 1.
(A) Diagram of the sequence of the voltage-gated sodium channel, with voltage sensor S4 segments shaded in domains I–III (orange) and in domain IV (red) to denote the role of DIV S4 (domain IV S4 segment) in fast inactivation, with the inactivation particle identified as the IFM motif in the DIII–DIV linker. Receptor sites for marine toxins and extending to that for veratridine/batrachotoxin (site 2) are indicated, excluding insecticide and other receptor binding sites. Cryo-EM structures of toxins bound to sodium channels are shown below. (B) TTX bound within receptor site 1 in the pore domain of hNaV1.4, with the toxin shaded (arrow). Binding site loci include acidic residues glutamate (orange), aspartate (blue), and aromatic residues (yellow): .pdb 6A95 [82]. (C) Spider toxin HwTX-IV bound to receptor site 4 in the extracellular S3–S4 linker of the voltage sensor domain (VSD) of domain II in hNaV1.7, with DII pore domain (PD) included in this view: .pdb 7K48 [83]. (D) α-scorpoin toxin AaH2 bound to receptor site 3 in the extracellular S3–S4 linker of the voltage sensor domain of domain IV of hNaV1.7: .pdb 6NT4 [84]. S4 positively charged residues are shown in green for (A,C,D) and illustrate the activated position of DIIS4 with HwTX-IV bound (C) and resting position of DIVS4 with AaH2 bound (D).
Cryo-EM structures of several voltage-gated sodium channels in complex with guanidinium or animal toxins have supported the functional experiments localizing toxin receptor binding sites, detailing specific interactions of toxins with binding site residues, and confirming toxin actions as pore blockers or gating modifiers. A few of these toxin-bound channel structures are illustrated in Figure 1: tetrodotoxin (TTX) in receptor site 1 (Figure 1B), spider toxin in receptor site 4 (Figure 1C), and α-scorpion toxin in receptor site 3 (Figure 1D).
3. Conotoxins Targeting Voltage-Gated Sodium Channels
The expanding database for conopeptide gene superfamilies, sequence, structure, and biological actions is updated in the ConoServer website (http://www.conoserver.org) described in [85]. Conotoxins are typically classified according to their endoplasmic signal sequence into one of about a dozen superfamilies, and further classified by cysteine framework into a larger set of conotoxin families acting on target channels, receptors, or transporters (reviewed by [86]). The structural diversity of conotoxins acting on voltage-gated sodium channels originates from a limited set of cysteine frameworks, for which folding patterns dictated by sequence are the primary determinant of the structural diversity, as suggested by comparisons of the M and O superfamilies [87] and later extended to the remaining conotoxin gene superfamilies [86]. Structural features for conotoxins targeting voltage-gated sodium channels are shown in Table 1.

Table 1.
Conotoxins targeting voltage-gated sodium channels. Structural features are listed for cysteine framework, number of inter-cysteine loops, di-sulfide cysteine connectivity, fold pattern, and distinguishing motif; ICK is the inhibitory cysteine knot motif.
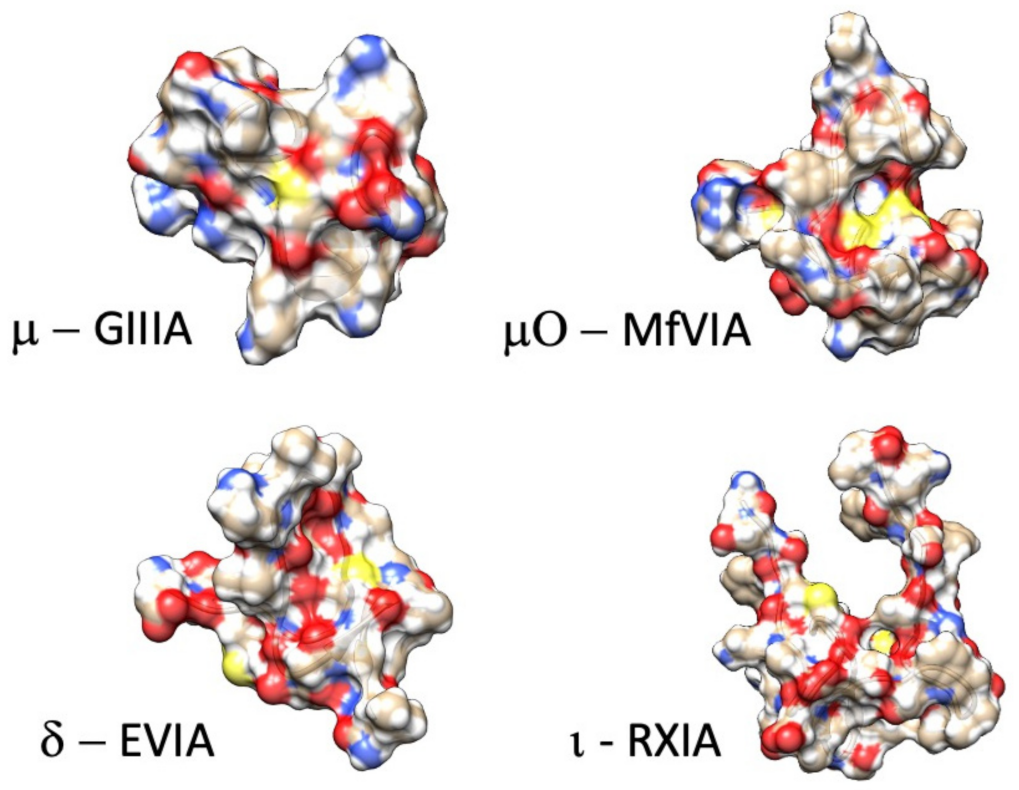
Here, we review from a historical perspective, the isolation, synthesis, and structural determination, with a principal focus on the functional characterization of the actions of conotoxins on voltage-gated sodium channels. In the M superfamily of conotoxins, these peptides act as pore blockers (µ-conotoxins), whereas in the O superfamily, µO-conotoxins prohibit sodium permeance by acting as gating modifiers (for reviews see [81,88,89,90]). Other conotoxins act as gating modifier toxins that influence fast inactivation or activation (δ-conotoxins and ι-conotoxins; for reviews see [19,91]). Example structures for each of these families of conotoxins targeting sodium channels are shown in Figure 2. As shown in Figure 1 and further discussed in this review, their binding sites are consistent with actions to block the sodium channel pore or alter voltage-dependent gating transitions. Recent investigations employing cryo-EM, homology modeling, molecular dynamics, and transcriptome approaches have enhanced our understanding of the structure-to-function relations of these conotoxins and their target sodium channel isoforms.
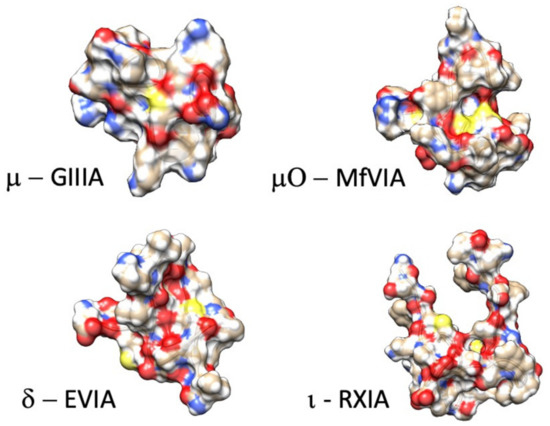
Figure 2.
Example structures of functionally characterized conotoxins (µ-conotoxin GIIIA: .pdb 1TCK [92]; µO-conotoxin MfVIA: .pdb 2N7F [93]; δ-conotoxin EVIA: .pdb 1G1P [94]; ι-conotoxin RXIA: .pdb 2JRY [95]).
4. Isolation and Initial Characterization of Actions of μ-Conotoxins on Nerve and Muscle
Conotoxins acting on voltage-gated sodium channels were discovered several decades ago along with α- and β- scorpion venom toxins, certain alkaloids, and the guanidinium toxins tetrodotoxin and saxitoxin used in biochemical purification of sodium channels from elasmobranch electric organs and mammalian tissue. Venom extracts from Conus geographus were shown to inhibit electrical and contractile activity of skeletal (but not smooth or cardiac) muscle in 1974 [96]. Micro conotoxins from this species were then isolated; purified as GIIIA, GIIIB, and GIIIC; and tested functionally [97,98,99]; initial reports referred to these μ-conotoxins as geographutoxins. Their functional characterization showed their inhibition of muscle action potentials elicited either with stimulation of the motor nerve or by direct muscle stimulation. In those experiments, GIIIA applied during indirect stimulation of the muscle (via motor nerve) blocked the compound muscle action potential muscle leaving the endplate depolarization unaffected, suggesting toxin action on the sarcolemmal voltage-gated sodium channels. This was confirmed with the observation that the muscle fiber action potential was prohibited with application of GIIIA during direct stimulation of the muscle. Additional experiments in these and other studies showed that neuronal sodium channel activity (synaptosome preparations) was not blocked by GIII toxins, suggesting that μ-conotoxins specifically target skeletal muscle sodium channels [100,101,102].
The isolation of μ-conotoxins from Conus geographus was accomplished in the same period as that for the cloning of sodium channels from electric fish [103]. Sodium channel genes were subsequently identified in mammalian brain [104,105], and in cardiac [106,107,108] and developing [109] or adult [110,111] skeletal muscle. Functional expression of these genes in heterologous expression with messenger RNA [110,112,113,114,115,116] was typically associated with tests for the actions of STX and TTX, and often with GIIIA. These and additional results supported the hypothesis that GIIIA targeted TTX-sensitive skeletal muscle sodium channels (NaV1.4) with little effect on TTX-resistant sodium channels of developing skeletal muscle or the myocardium (NaV1.5), or sodium channels in brain (i.e., NaV1.2). As additional sodium channel genes from brain and peripheral nervous tissues were cloned and functionally characterized, this specificity was modified slightly as GIIIA, GIIIB, and GIIIC were shown to compete, albeit weakly, with STX in radiolabeling binding assays in brain tissue [117] and produce partial inhibition of certain neuronal sodium channels, i.e., TTX-sensitive NaV1.6 [118].
Importantly, cloning of voltage-gated sodium channels facilitated investigations into the mechanism of μ-conotoxin binding to the sodium channel, for which specific regions had now been associated with channel functions of activation and fast inactivation [113]. Characterization of the effects of saxitoxin and tetrodotoxin showed that these bind to receptor site 1 in the pore to block sodium entry through open channels, whereas sea anemone toxins such as the anthopleurins bind to receptor site 3 (extracellular S3–S4 loop in the domain IV voltage sensor module, impeding the translocation of the domain S4 segment, to slow the entry of sodium channels into a state of fast inactivation [119,120] and reviewed by [78] (Figure 3). Use of GIII μ-conotoxins in the aforementioned studies to characterize sodium channel genes with their functional expression showed that these μ-conotoxins acted, similar to the guanidinium toxins STX or TTX, as pore blockers. Descriptions of the initial characterization and tests on expressed sodium channels for these and additional conotoxins follows.
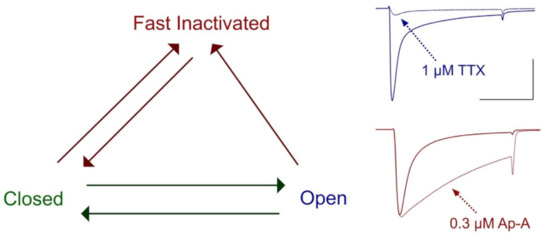
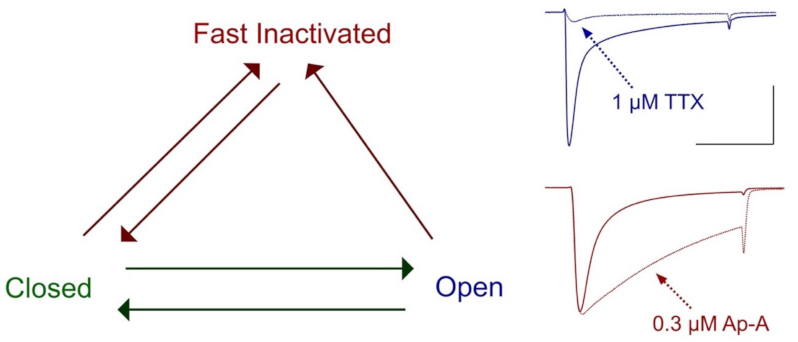
Figure 3.
Voltage-dependent state transitions in sodium channels. Channels respond to weak depolarization by inactivating, or to strong depolarization by opening prior to inactivation. At right is shown the action of the site-1 toxin tetrodotoxin (TTX) to block sodium permeance in hNaV1.4 channels (dotted line, blue) and is compared to the action of the site-3 toxin Anthopleurin-A (Ap-A) to slow the entry of these channels into fast inactivation (dotted line, red). Calibration; vertical 1 μA, horizontal 10 ms. Groome, unpublished.
Since TTX binds effectively to the pore of neuronal as well as skeletal muscle sodium channels, the finding that GIIIA block was specific to skeletal muscle channels revealed differences in the site 1 receptor in these channel isoforms [121], and that the developmental stage of embryonic muscle is critical for GIIIA potency [100,101]. Experiments with radiolabeled TTX, STX, or GIIIA in competition binding assays suggested that the guanidinium toxins and μ-conotoxins share an overlapping region of binding in receptor site 1 [121,122,123,124]. This hypothesis was supported by the results of single channel recordings illustrating differential wash out under conditions of single or co-occupancy of guanidinium and GIIIA toxin and gating current recordings showing that TTX and GIIIA have direct and distinguishable actions on S4 voltage sensor translocation [125,126,127].
NMR determination of the three-dimensional structures of GIIIA [128] and GIIIB [129] revealed similar, compact structures, stabilized by three disulfide bridges, and helices orienting positively charged arginine or lysine residues into the aqueous solvent, suggesting their potential interaction of the outer vestibule region in the pore of the skeletal muscle sodium channel. Studies by [130] using rat diaphragm muscle bioassay and [131] using radiolabeling binding assay and single channel recordings showed that the guanidinium side chain of Arg-13 was essential for μ-conotoxin binding and channel occlusion, with other arginine, lysine, or hydroxyproline residues providing lesser but significant determinants of toxin binding. An NMR study of GIIIA mutations supported the notion that Arg-13 substitution did not (indirectly) inhibit binding as the result of conformational alteration of the toxin, and it also further supported roles for other arginine, lysine, and hydroxyproline residues in channel interaction and overlap with the TTX and STX binding site [92]. However, conformational effects as determined with NMR were correlated with significant decrease in toxin potency for glutamate substitution at Asp-12 [132]. Conformational perturbation with alanine substitutions at individual cysteine residues significantly decreased binding efficacy of GIIIA, for each of the three disulfide linkages [133]. The NMR determination of the structure of GIIIC [118] revealed that naturally occurring differences at position 16 between GIIIC (Leu-16) and the other μ-conotoxins affected both the conformation of this toxin and its differential block of NaV1.4 (skeletal muscle) versus NaV1.6 (brain).
The role of Arg-13 and other residues in GIIIA in toxin binding were assessed in several investigations collectively utilizing a variety of approaches to test for interaction with toxin and pore residues. Sodium permeation is dependent on outer vestibule acidic residues and the constricted inner pore selectivity filter ([134,135,136,137,138,139]; Figure 4). Specific skeletal muscle sodium channel pore acidic residues were suggested as the target for GIIIA Arg-13. Point mutations in the outer vestibule of the pore and extending to the selectivity filter negatively impacted GIIIA affinity [140,141,142,143]. These and additional studies suggested that binding of GIIIA involves the concerted action of multiple toxin residues interacting with outer vestibule pore residues and favoring the binding pose of Arg-13 inserted more deeply into the pore [144,145,146,147,148] and included at least one neutral toxin residue, A22 [149]. Interestingly, differences between GIIIA and GIIIB binding were correlated with naturally occurring differences in toxin residues that mediate their differential interactions with the sodium channel pore [150,151].
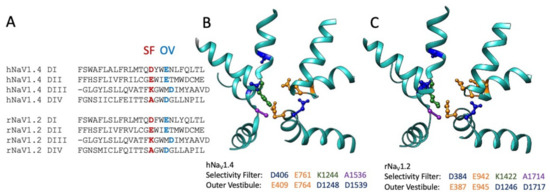
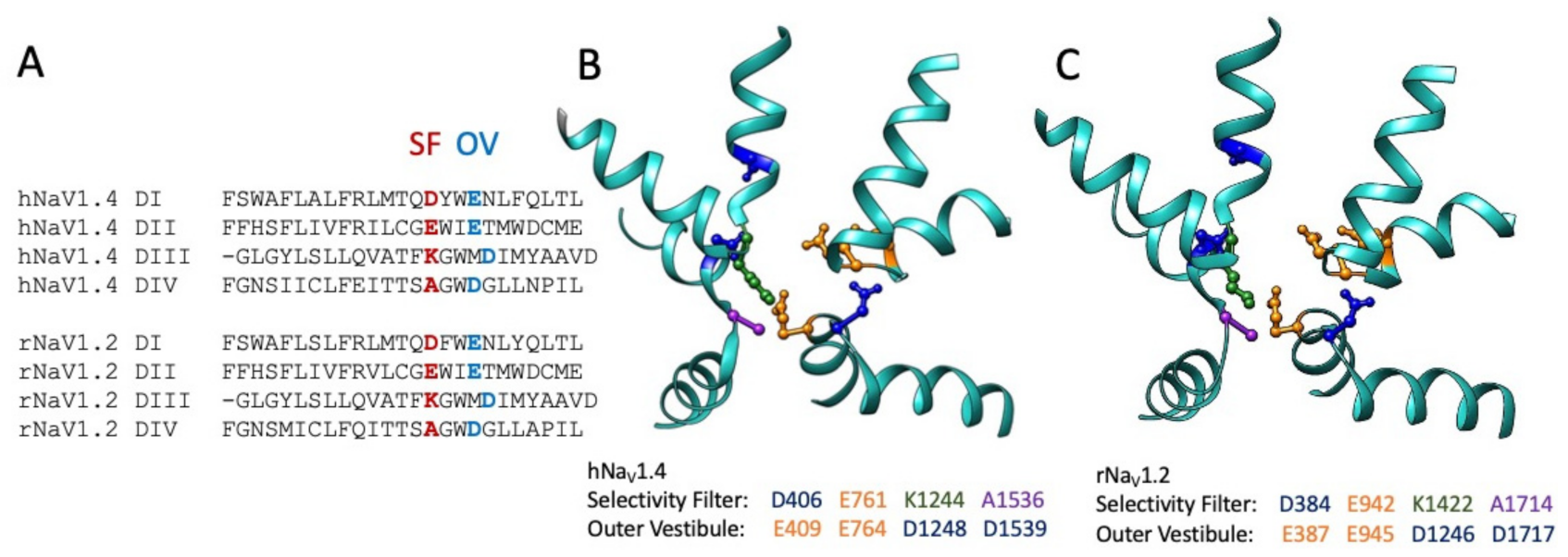
Figure 4.
Sequence alignment (A) of the pore helices for sodium channels hNaV1.4 (B): .pdb 6AGF [82]) and rNaV1.2 (C): .pdb 6J8E [152]). The selectivity filter (SF) is shown in red for the alignment, with acidic residues of the outer pore vestibule (OV) shown in sky blue. Residues within the pore helical structures in (B,C) are listed as acidic aspartate and glutamate, positive lysine, and neutral alanine. Sequence alignments were performed in Seaview 5 [153].
Thiosulfonate reagents were used with cysteine-substituting mutations to show that residue charge and structure (bulkiness of side chains as for tryptophan) contribute to GIIIA binding in the pore region [142,154] and furthered our understanding of the anionic “DEKA” selectivity filter [148,155,156] and of slow inactivation [157]. These mutant cycle analyses of several GIIIA or GIIIB Arg-13 mutants, sodium channel pore mutants including but not restricted to the selectivity filter, and their respective double toxin/channel mutational perturbation were used to calculate free energy (differences) for toxin interaction with the pore. These studies also identified electrostatic and other interactions of the toxin with outer pore vestibule residues outside of the selectivity filter, and they elucidated the tilted orientation of GIIIA as it bound to the pore. Functional characterizations of the effects of mutations of negatively charged residues in the outer pore ring on GIIIA binding were used to direct molecular dynamics simulations that largely confirmed the emerging consensus for μ-conotoxin binding in the outer vestibule of the pore module, and with its relative bulkiness, binding less deeply in the pore than STX or TTX [146].
An additional, interesting result from investigation of the binding of GIIIA to the skeletal muscle sodium channel pore was the determination of the organization of domains in this channel [158]. The clockwise orientation of hNaV1.4 domains is of considerable relevance in investigations of sodium channel gating for which perturbations in voltage sensor or pore domains may directly or allosterically influence function in their counterpart domain, as many voltage-gated sodium channels exhibit pairwise, swapped domains (i.e., the pore module in DI is closest to the voltage sensor module in DIV in hNaV1.4). Most importantly, these studies gave an early view of the structure of the sodium channel pore, in addition to an understanding of μ-conotoxin action.
5. Isolation and Characterization of μ-Conotoxins Targeting Neuronal Sodium Channels
Following the isolation of GIII μ-conotoxins, the characterization of their action on NaV1.4 and their use in structure to function studies, nearly a decade and a half passed prior to the isolation of additional μ-conotoxin peptides. The timeline for the subsequent discovery of novel μ-conotoxins (also) displaying three inter-cysteine loops (CC---C---C---CC) and with C1-C4, C2-C5, and C3-C6 disulfide linkages, but with significant sequence diversity, is elegantly depicted in [90], and it resulted in the classification of the μ-conotoxin family as the M4 and M5 branches of the M-superfamily of Conus peptides (Table 2). Conotoxins without sequence similarity to the μ-conotoxins, and with four inter-cysteine loops (C---C---CC---C---C), were also isolated during that time [159] and classified in the O-superfamily as μO-conotoxins; their characterization on skeletal muscle and neuronal sodium channels is described later, along with a description of the characterization of ι-conotoxins with five inter-cysteine loops (C---C---CC---CC---C---C).

Table 2.
Sequence alignments of μ-conotoxins blocking permeance in voltage-gated sodium channels. M4 and M5 branches distinguished in part by number of residues in inter-cysteine loop 3. O = hydroxyproline, Z = pyroglutamate.
The isolation of PIIIA from Conus purpurascens [160] and its NMR solution structure [117] extended known μ-conotoxin targets to include neuronal voltage-gated sodium channels. PIIIA displayed a disulfide linkage similar to GIIIA, its competitive inhibition of STX binding for skeletal muscle was similar to GIIIA/B/C [117,173], and PIIIA Arg-14, homologous to the critical Arg-13 of Conus geographus μ-conotoxins, was an important determinant for inhibition of the skeletal muscle sodium channel. However, PIIIA was found to be a more competitive inhibitor than was GIIIA/B/C in rat or human brain. This peptide also blocked rat brain type II (NaV1.2) sodium channels with high affinity, unlike GIIIA, and displayed sequence divergence from GIIIA-C. These studies and investigations by [174,175] revealed several significant differences between GIIIA and PIIIA in their block of sodium channels. First, native PIIIA exhibited 10-fold less affinity for the NaV1.4 muscle channel compared to GIIIA. Second, alanine substitution produced more decrement of channel block for GIIIA R13-A compared to PIIIA R14-A, with approximately two-fold more residual current observed in the PIIIA mutant. Charge-altering substitutions of several residues in PIIIA also decreased the voltage dependence of channel block. In combination with molecular dynamics simulations, these results suggest that PIIIA R12, R20, R14, and K17 residues insert deep into the outer vestibule, with R14 and K17 entering into the transmembrane electric field and thus in proximity to the selectivity filter [174], consistent with the results from functional characterization and simulations of GIIIA/pore residue interactions [146,147]. Interestingly, a nonconserved histidine (H19) in PIIIA may also favor binding towards neuronal channels [175], as glutamine substitution at that residue increased PIIIA affinity towards NaV1.4. In GIIIA, that locus is a native glutamine, and GIIIA shows increased selectivity towards NaV1.4 with respect to PIIIA.
As neuronal sodium channel genes were cloned and associated with pathologies including epilepsy, familial hemiplegic migraine, and peripheral neuropathies, an important focus of the research on PIIIA and subsequently isolated μ-conotoxins was the determination of the affinities of native or mutant μ-conotoxin peptides to specific brain or peripheral nerve sodium channel isoforms. For example, soon after its isolation, PIIIA was shown to inhibit brain sodium channel NaV1.2 more effectively than PN1 of peripheral nerve [176], and this difference was utilized to track the developmental profile of brain versus peripheral nerve sodium channels in PC12 cells.
The GIIIA/B/C and PIIIA μ-conotoxins described above share a common pro-peptide sequence and cysteine framework and are classified in the M4 branch of the M superfamily of conotoxins [87,89,90]. The isolation of SmIIIA [164] extended the μ-conotoxins to a second, M5 branch of this superfamily. Functional characterization of SmIIIA actions on sensory (dorsal root ganglion) and autonomic (sympathetic chain ganglia) neurons revealed that this μ-conotoxin preferentially inhibited TTX-resistant sodium current, and that this action was not readily reversible. A chimeric approach was used [177] to investigate the locus for toxin affinity to TTX-resistant sodium channels in these neuronal populations, by swapping toxin regions between PIIIA, which does not affect TTX-resistant current, and SmIIIA, which does. Their results localized that site to residues in inter-cysteine loop 3 SmIIIA C-terminal half residues swapped into PIIIA conferred the toxin with ability to block TTX-resistant current, whereas the complementary swap of C-terminal half residues of PIIIA into SmIIIA abolished the capacity of that toxin to block TTX-resistant current.
Soon thereafter, two additional M5 conotoxins, KIIIA and SIIIA, were isolated and also found to inhibit the TTX-resistant sodium current in dorsal root and sympathetic ganglia [165]. Interestingly, while SmIIIA potently inhibited the compound action potential in frog skeletal muscle, this effect was not observed for KIIIA or SIIIA. Several differences in the sequences of the M5 conotoxins were noted as targets of investigation for the observed specificity of KIIIA and SIIIA towards neuronal sodium channels. KIIIA and SIIIA each exhibit a reduced number of residues in loop 1, the absence of heretofore conserved positively charged Arg or Lys residues in this loop, and the “substitution” at the critical Arg-13 position in loop 2 by lysine (R13 in GIIIA, PIIIA, and SmIIIA, but K7 for KIIIA, and K11 for SIIIA).
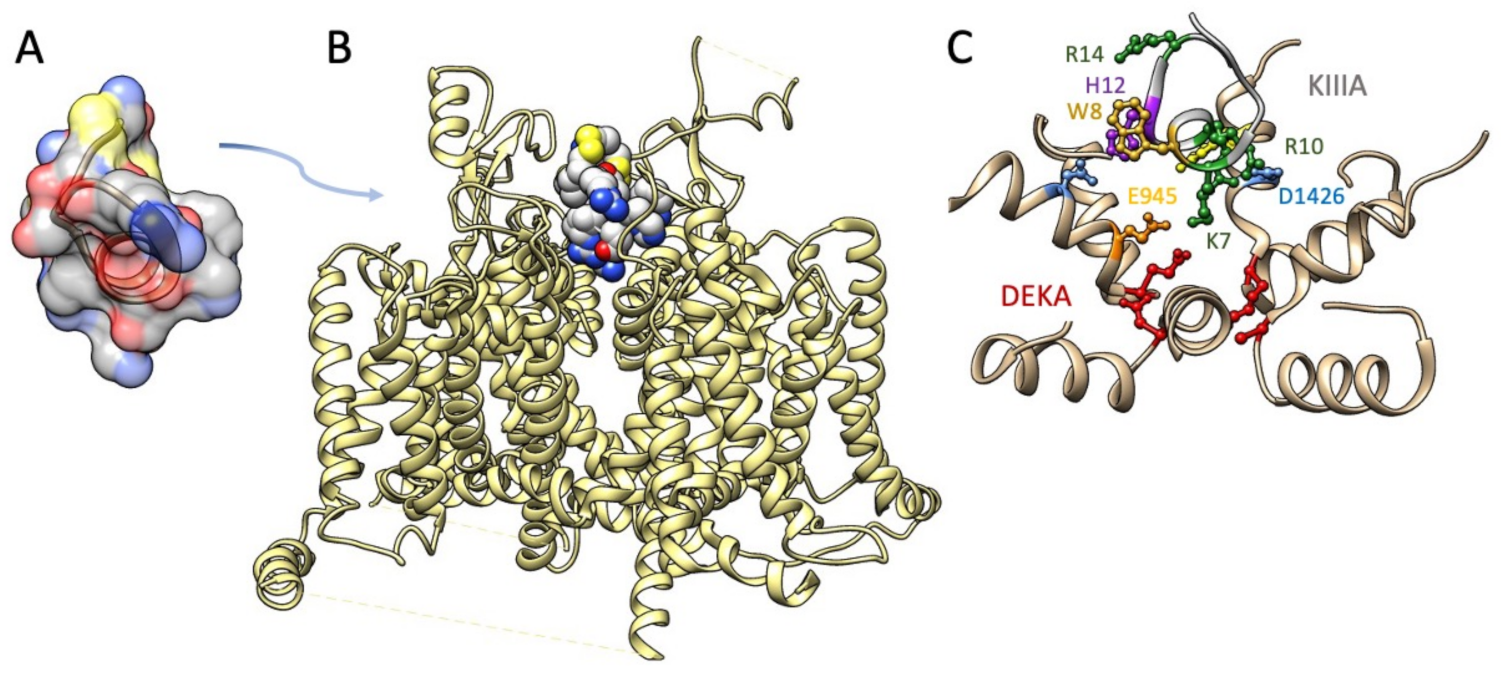
The M5 conotoxin KIIIA interaction with NaV1.2 has been determined in cryo-EM [152] (Figure 5). Several features of toxin interaction with its sodium channel target are captured in that structural determination. The critical Lys-7 (K7) in the helical portion of KIIIA extends deep into the pore towards the DEKA selectivity filter, similar to the poses proposed for Arg-13 from functional and computational investigations with GIIIA. K7 and other positively charged toxin residues including R10 interact with outer pore residues above the selectivity filter. Additional toxin residues including aromatic residues W8 and H12 interact with other pore and extracellular residues. This structural investigation supported a mode of μ-conotoxin binding in general as comprising electrostatic, polar, and H-bond interactions of positive residues on one face of the toxin with pore and other residues, and occlusion of the pore by KIIIA distinguished from that by TTX or STX with the KIIIA binding pose extending higher from the selectivity filter to occupy the outer pore vestibule. These differential binding poses are also supported by functional experiments showing that KIIIA could trap TTX or STX in its position lower in the pore and resulting in a slower rate of dissociation [178].
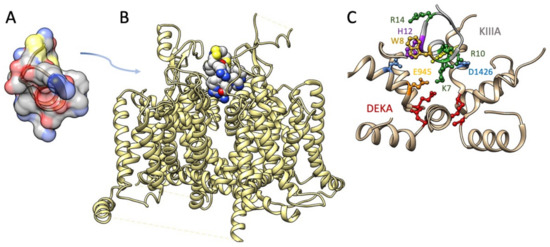
Figure 5.
(A) Structure of KIIIA (.pdb 7SAV; [179]) (B) Cryo-EM structure of NaV1.2 bound by KIIIA (.pdb 6J8E; [152]). (C) Enlargement of the P-loops of NaV1.2 (tan helices) with KIIIA toxin residue K7 interacting with outer pore residues E945 and D1426, and R10 (shown in two possible orientations) interacting with D1426. Toxin residues W8, H12, and R14 are shown for which additional interactions within the P-loops, or with extracellular residues, were noted. DEKA selectivity filter residues are shown in red.
Functional characterization of μ-conotoxins with bioassays continued with the discovery of additional peptides in the M superfamily. Four M5 μ-conotoxins (CIIIA from Conus catus, CnIIIA, CnIIIB from Conus consors, and MIIIA from Conus magus were isolated and characterized [167]. CIIIA and CnIIIA blocked the TTX-resistant sodium current in dorsal root ganglion neurons with potency similar to KIIIA whereas SIIIA, MIIIA, and CnIIIB showed reduced potency. Interestingly, CIIIA was nonselective, as it blocked skeletal muscle action potentials, reminiscent of the profile exhibited by SmIIIA. CnIIIC was isolated [171] and exhibited similar potency on skeletal muscle contraction compared to TIIIA, and it inhibited the compound action potential elicited in myelinated and unmyelinated nerve preparations; inhibition of nerves by CnIIIC was not readily reversible. As additional M4 and M5 μ-conotoxins were isolated, their functional characterization typically utilized heterologous expression systems to test for isoform specificity of toxin action, as described below.
The isolation of μ-conotoxins other than GIIIA/B/C and PIIIA in the M4 branch, and the discovery and isolation of M5 branch μ-conotoxins, provided a wealth of native μ-conotoxins as potential specific antagonists of sodium permeance of sodium channel isoforms in the central and peripheral nervous system. The development of heterologous expression systems (Xenopus oocytes and mammalian embryonic cells) and incorporation of sodium channel genes into bacterial plasmids for expression of those sodium channels provided an important approach to determine the specificity of μ-conotoxins. Sodium channel nomenclature for isoforms was established [180], with NaV1.1, NaV1.2, and NaV1.3 found in brain; NaV1.4 in skeletal muscle; NaV1.5 in cardiac muscle; NaV1.6 in brain including the cerebellum, and peripheral nerve; and NaV1.7, NaV1.8, and NaV1.9 in peripheral nerve and including their relative sensitivity to TTX (NaV 1.1–1.4, 1.6, 1.7) or resistance to TTX (NaV1.5, 1.8, 1.9). Together with the cloning and functional expression of each of the sodium channel isoforms in muscle, brain, and peripheral nerve, an important focus of investigation was to establish the pharmacological profile of novel conotoxins across neuronal, skeletal, and cardiac muscle sodium channels. These studies supported previous radiolabeling competition studies of the effects of μ-conotoxins on brain and muscle tissues, as well as bioassays of muscle and nerve preparations. In addition, they provided a reductionist determination of the relative affinities of M4 and M5 μ-conotoxins for specific sodium channel isoforms in these tissues.
As the complement of isolated μ-conotoxins grew, their functional characterization revealed a diversity of target sodium channels. Development of the pharmacological profile of μ-conotoxin action on channels expressed in oocytes or in mammalian cells was needed to provide a path forward in the development of μ-conotoxin analogues for treatment of channelopathies of brain, muscle, and peripheral nerve (reviewed by [26]). For the M4 branch of μ-conotoxins, GIIIA exhibited a 100-fold increase in potency on NaV1.4 compared to its actions NaV1.2 (reviewed by [19,20]). Interestingly, GIIIB produced significant block of NaV1.3 [161]. Similar to GIIIA, PIIIA also blocked NaV1.4 with greater potency than that observed for NaV1.2, but to a lesser extent [176]. The M4 μ-conotoxin TIIIA isolated from Conus tulipa produced effects similar to PIIIA, with significant but lesser block of NaV1.2 compared to that of NaV1.4 [161], and it was without action on NaV1.3, NaV1.5, NaV1.7, or NaV1.8 expressed in Xenopus oocytes. However, TIIIA did inhibit NaV1.7 endogenously expressed in neuroblastoma cells [181] at the relatively high dose of 1 μM. SxIIIA action was first assessed on NaV1.4, where it produced a rapidly reversible block [162] such as observed for GIIIA. The SxIIIA toxin was later shown to block NaV1.1, Nav1.6, and NaV1.2 with less potency compared to that for NaV1.4 [182]. More recently, the M4 μ-conotoxin TsIIIA was isolated from Conus tessulatus and found to inhibit TTX-resistant sodium current in dorsal root ganglion neurons [163]. When tested on sodium channels expressed in mammalian cells, TsIIIA had no effect on TTX-sensitive channels NaV1.1, NaV1.2, NaV1.3, NaV1.4, NaV1.6, or NaV1.7; it did not block TTX-resistant NaV1.5 but blocked TTX-resistant NaV1.8 with an IC50 at 2.1 μM [183].
The M5 conotoxin KIIIA blocked NaV1.2 to a slightly greater extent compared to NaV1.4 [184], and its block of NaV1.2 was not readily reversible, as is typically observed for the M4 branch. Alanine substitution of KIIIA Trp-8 (W8) resulted in a significant decrease in toxin block, and structural determination of KIIIA interaction with NaV1.2 revealed a strong interaction of W8 with DII extracellular Tyr-362 [152]; that aromatic residue is conserved in TTX-sensitive channels but not conserved in TTX-resistant channels NaV1.5 or NaV1.8. KIIIA partially blocked NaV1.1, NaV1.3, NaV1.6, and NaV1.7, extending the determination of neuronal isoforms targeted by μ-conotoxins [184,185]. The N-terminally extended isomer of KIIIA, KIIIB, was also shown to be a potent inhibitor of NaV1.2, with significant effects on NaV1.7 [185]. Certain isomers influencing disulfide connectivity of KIIIA had significant impact on its binding to NaV1.2, NaV1.4, and NaV1.7 [179]. The structural determination of SIIIA and the novel μ-conotoxin SIIIB revealed their sequence diversity, with an alpha helical motif conferring binding affinity for this toxin of equivalent potency on NaV1.2 and NaV1.4 [168]. That study, and [186], showed that both KIIIA and SIIIA blocked NaV1.2 irreversibly. Structural data on SIIIA in both studies suggested the importance of the C-terminal half of the peptide to promote neuronal sodium channel affinity; a later study concluded that N-terminal residues also contribute [187]. SIIIA was shown to act on other neuronal sodium channels [186,188] including NaV1.6 and to a lesser extent NaV1.7. The chimeric (NaV1.4 and NaV1.5 domain-swapping) and mutagenesis approaches utilized in [188] localized pore loop residues in domains I and II as structural determinants of SIIIA discrimination between sodium channel isoforms. While heterologously expressed TTX-resistant channels were not affected by KIIIA or SIIIA in these studies, each of these μ-conotoxins produced a persistent decrement of TTX-resistant sodium current in dorsal root ganglion neurons [165,166].
CnIIIC effectively and irreversibly blocked NaV1.4 expressed in mammalian cells [171,189]. Similar to SIIIA, its potency on NaV1.2 was similar to that of NaV1.4, with partial block of NaV1.6 and NaV1.7, with no effect on TTX-resistant sodium channels NaV1.5 and NaV1.8, and surprisingly with nanomolar block of the well-characterized α3β2 nicotinic acetylcholine receptor. BuIII A, BuIIIB, and BuIIIC isolated from Conus bullatus each blocked NaV1.4 to the same extent as M5 μ-conotoxin KIIIA [169]. Similar to the action of KIIIA, a BuIIIB block of NaV1.4 was not readily reversible. A subsequent study showed that BuIIIB also produced significant block of NaV1.1, NaV1.2, and NaV1.3 [182], with partial and lesser block for NaV1.5 and NaV1.6, respectively.
6. Structural Divergence in M4 and M5 Conotoxins Affecting Channel Selectivity
The above studies on M5 μ-conotoxins extended the neuronal actions of conotoxins from NaV1.2 to several additional TTX-sensitive sodium channel isoforms. Shorter peptide analogues of BuIIIC and KIIIA suggested promise as candidates for increased selectivity towards block of neuronal sodium channels [190]. All of the conotoxins of the M5 branch possess a unique tryptophan residue in the third inter-cysteine loop, for which potential π/π and cation/π interactions with neighboring histidine and arginine residues were suggested as a possible structural determinant of the potency of KIIIA or SmIIIA interaction with neuronal channels [152,177,184,191]. These studies provided strong evidence that this aromatic residue is a determinant in isoform specific actions of M5 μ-conotoxins. Comparison of the actions of the M4 μ-conotoxin TIIIA isolated from Conus tulipa [161] to those of M5 μ-conotoxins SIIIA and SIIIB isolated from Conus striatus [168] provided additional insight into stable μ-conotoxin conformations and the important role of these homologous loci. SxIIIC was isolated from Conus striolatus [172], and its characterization revealed its block of several neuronal sodium channel isoforms including NaV1.3, NaV1.1, NaV1.6, NaV1.7, and NaV1.2 in order of potency. A subsequent comparison of relative potencies of μ-conotoxins on NaV1.7 showed that three μ-conotoxins with reported block of NaV1.7, SxIIIC, KIIIA, and SmIIIA blocked that sodium channel isoform with the greatest variability compared to actions on other neuronal channels [192]. That study explored structural differences in KIIIA, SmIIIA, and SxIIIC, focused on N-terminal residue extension in SmIIIA and SxIIIC compared to KIIIA, number of residues in loop 1, and charged residues in loop 3. Their findings revealed that number of loop 1 residues are a determinant in the relative potencies of SmIIIA, SxIIIC, and KIIIA, and that charged residues in loop 3, as contributing to overall net charge of toxin surface of M4 versus M5 μ-conotoxins, are an important factor in selectivity for NaV1.7.
The aforementioned studies showed that μ-conotoxins of the M superfamily have affinity to the outer vestibule of the pore module in voltage-gated sodium channels, with select residues extending to the selectivity filter, with pore-blocking actions such as those of saxitoxin and tetrodotoxin, through their interactions with binding sites overlapping with those of marine toxins in receptor site 1. The initial characterization of Conus geographus μ-conotoxins determined critical interaction between toxin residues and those of the skeletal muscle sodium channel pore. Subsequent, isolation, structural determination and functional characterization of additional M4 and M5 μ-conotoxins shown in Table 2 revealed a diversity of toxin sequences and neuronal sodium channel isoforms targeted, enhancing our understanding of toxin action and expanding the possibilities for development of isoform specific μ-conotoxin analogues to investigate neuropathic sodium channelopathies, and provide alternative intervention in these diseases.
7. Isolation and Characterization of μO-Conotoxins
As μ-conotoxins were being isolated and studied, μO- and δ-conotoxins of the O superfamily were also discovered and functionally characterized for their actions on voltage-gated sodium channels (Table 3). μO-conotoxins were isolated from Conus marmoreus as MrVIA and MrVIB [159].
These peptides exhibit sequence similarity to δ-conotoxins (targeting sodium channels), ω-conotoxins (targeting calcium channels), and κ-conotoxins (targeting potassium channels). Their cysteine framework is distinguished from that of the μ-conotoxins by a disulfide cysteine pattern within the peptide, named the inhibitory cysteine knot motif (ICK), with the terminal disulfide bridge passing through the polypeptide backbone and imparting a compact globular structure [193]. This pattern has been identified as a common structural determinant of peptide toxins acting on a diversity of ion channels (reviewed by [194,195]) and comprises four inter-cysteine loops (C---C---CC---C---C), compared to three such loops as determined for μ-conotoxins (Table 1). MrVIA and MrVIB were thus classified as μO-conotoxins with their general tertiary structure as defined by their polypeptide precursor sequence, the resulting cysteine knot pattern and with their ability to inhibit sodium current.

Table 3.
Sequences of μO- and δ-conotoxin gating modifiers of voltage-gated sodium channels. Sequences characterized with cysteine framework ---C---C---CC---C---C---.
Table 3.
Sequences of μO- and δ-conotoxin gating modifiers of voltage-gated sodium channels. Sequences characterized with cysteine framework ---C---C---CC---C---C---.
| Toxin | Conus Species | Sequence | References |
|---|---|---|---|
| μ-O | ---C------C-------------CC----C----C--- | ||
| MrVIA | C. marmoreus | --ACRKKWEYC----IVPIIGFIYCCPGLICGPFVCV---- | [159] |
| MrVIB | C. marmoreus | --ACSKKWEYC----IVPILGFVYCCPGLICGPFVCV---- | [159] |
| MfVIA | C. magnificus | -RDCQEKWEYC----IVPILGFVYCCPGLICGPFVCV---- | [196] |
| δ | |||
| Tx(V)IA | C. textile | --WCKQSGEMCN---LLD--QN--CCDGY-CIVLVCT---- | [197,198,199] |
| TxIB | C. textile | --WCKQSGEMCN---VLD--QN--CCDGY-CIVFVCT---- | [198] |
| GmVIA | C. gloriamus | VKPCRKEGQLCD-----PIFQN--CCRGWNCV-LFCV---- | [200] |
| NgVIA | C. nigropunctatus | -SKCFSOGTFCG---IKO--GL--CCSVR-CFSLFCISFE- | [201] |
| PVIA | C. purpurascens | -EACYAOGTFCG---IKO--GL--CCSEF-CLPGVCFG--- | [202] |
| SVIE | C. striatus | -DGCSSGGTFCG---IHO--GL--CCSEF-CF-LWCITFID | [203] |
| CnVIA | C. consors | -YECYSTGTFCG---ING--GL--CCSNL-CLFFVCLTFS | [203] |
| Am2766 | C. amadis | ---CKQAGESCD---IFS--QN--CCVGT-CA-FICIE--- | [204] |
| EVIA | C. ermineus | -DDCIKOYGFCSLPILKN--GL--CCSGA-CV-GVCADL-- | [205] |
| CnVIB | C. consors | -DECFSOGTFCG---TKO--GL--CCSAR-CFSFFCISLEF | [206] |
| CnVIC | C. consors | -DECFSOGTFCG---IKO--GL--CCSAR-CLSFFCISLEF | [206] |
| CnVID | C. consors | -DECFSOGTFCG---FKO--GL--CCSAR-CFSLFCISLEF | [206] |
| TsVIA | C. tessulatus | ---CAAFGSFCG---L-P--GLVDCCSGR-CF-IVCLL--- | [207] |
| SuVIA | C. suturatus | ---CAGIGSFCG---L-P--GLVDCCSDR-CF-IVCLP--- | [2] |
MrVIA blocked sodium current in molluscan (Aplysia) neurons and produced ataxia and/or coma when injected at nanomole doses (or less) into the central nervous system (intra-cranial injection) of mice [159]. Interestingly, MrVIA was without effect when injected into the periphery and thus accessible to muscle (inter-peritoneal injection). In contrast, the effects of μ-conotoxin GIIIB are observed for peripheral, but not central, injections [99]. The μO-conotoxins were of considerable interest for their apparent neuronal specificity, as noted several years before the discovery of M5 μ-conotoxins acting on voltage-gated sodium channels in brain and peripheral nerve. MrVIA inhibited TTX-sensitive sodium currents in cultured hippocampal neurons [208], and NaV1.2 expressed in Xenopus oocytes with nanomolar potency. This effect on NaV1.2 was the result of a hyperpolarized shift of the steady-state fast inactivation curve, suggesting its action was not to block permeation as for GIIIA. Indeed, MrVIA did not displace STX from brain or electric eel tissue in radiolabel binding assay, suggesting the toxin does not inhibit sodium channels through interaction with receptor site 1.
Receptor site 4 is targeted by β-scorpion toxins [209,210] and the inhibitory cysteine knot tarantula toxin ProTx-II [211]. This site includes residues of domain II S1–S2 and S3–S4 extracellular loops [212,213,214] and extends to the N-terminal pore loop in domain III, proximal to the DII VSD. β-scorpion and spider toxins act as gating modifiers by “trapping” the voltage sensor in domain II in the activated position (for reviews of voltage sensor trapping see [76,77]; Figure 1C,D). MrVIA inhibited NaV1.4 DII voltage sensor translocation [215], and a chimeric approach revealed its interaction with the N-terminal pore loop in domain III [216]. These findings suggested overlapping binding sites for MrVIA, β-scorpion, and spider toxins. MrVIA trapping of the domain II voltage sensor in NaV1.4 was effectively competed against by β-scorpion toxin, which did not compete against pore block by GIIIA [215]. MrVIA, but not GIIIA inhibition of NaV1.4 was relieved by repetitive depolarization, an action also observed for the β-scorpion toxins acting on receptor site 4. Thus, the μO-conotoxins prohibit sodium permeance as a consequence of their effect on DII voltage sensor translocation rather than through pore occlusion, as observed for the μ-conotoxins.
Attention was focused on the specificity of MrVIA and MrVIB μO-conotoxins for neuronal sodium channel isoforms and its potential use as a pharmacological intervention in peripheral neuropathies associated with TTX-resistant sodium channels NaV1.8 and NaV1.9. Both MrVIA [217] and MrVIB [218] inhibited action potentials in nociceptive C fibers and produced analgesic effects in pain assays in mice. MrVIA was shown to be more effective at inhibiting TTX-resistant versus TTX-sensitive sodium current in dorsal root ganglion neurons [219], similar to the actions of μ-conotoxins SMIIIA [164] and KIIIA [165]. MrVIB also blocked TTX-resistant sodium current in these neurons, and it specifically inhibited NaV1.8 in heterologous expression [218]. Recovery of NaV1.8 current amplitude was prolonged, unless strong depolarization was used to condition the channels during that recovery, presumably due to displacement of the toxin from receptor site 4 with voltage sensor translocation. Interestingly, rate of MrVIB block was accelerated by the presence of any of the β 1–4 subunits co-expressed with the α subunit of the sodium channel [220]. A similar effect was subsequently noted for β subunit modulation of the rate of pore block of NaV1.1, NaV1.2, NaV1.6, or NaV1.7 by μ-conotoxins TIIIA, PIIIA, SmIIIA, and KIIIA [221]. Thus, while STX block of the pore is insensitive to β subunit presence [220], β subunits do modulate the kinetics of sodium channel inhibition mediated either by μ-conotoxin pore blockers or μO-conotoxin gating modifiers.
The related μO-conotoxin MfVIA was isolated from Conus magnificus [196]. This conotoxin, similar to MrVIA and MrVIB, blocked TTX-resistant sodium current in dorsal root ganglion neurons, as well as TTX-sensitive NaV1.4 and TTX-resistant NaV1.8 sodium channels in heterologous expression. The study by [196] also noted the increased potency of these μO-conotoxins on TTX-resistant current in neurons compared to that expressed in oocytes or mammalian cells, hypothesizing that this difference could be due to the demonstrated action of β subunits on the rate of block, as shown for MrVIB [220]. MfVIA μO-conotoxin was used in a study combining mutagenesis and automated patch clamp technology [93]. Here, the double mutant MrVIA E5K/E8K produced a significant increase in potency and provided analgesia in a formalin-based behavioral assay for pain. Two additional μO-conotoxins were isolated from Conus litteratus [222,223], each of which inhibit sodium current in dorsal root ganglion neurons but have not yet been tested in heterologous expression for isoform specificity.
8. Delta (δ-)Conotoxins
Delta (δ)-conotoxins share sequence similarity to the ω-conotoxins, κ-conotoxins, and μO-conotoxins but not the μ-conotoxins (reviewed by [81,224]; Table 3), with the C---C---CC---C---C--- cysteine framework and four inter-cysteine loops and considerable hydrophobicity of amino acid residues of those loops, in contrast to the μ-conotoxins ([94]. For mammalian sodium channel isoforms targeted, the impact of δ-conotoxins was observed as a pronounced slowing of the entry of channels into a state of fast inactivation, similar to the action of α-scorpion (reviewed by [225]) or sea anemone toxins (reviewed by [58,59]; Figure 3). The S4 segment in domain IV acts as the voltage sensor for fast inactivation [226], and toxins acting at receptor site-3 trap the domain IV S4 segment in a resting or intermediate configuration ([119,227,228]; Figure 1D).
Several investigations of the effect of δ-conotoxins on sodium current or action potentials in molluscan neurons [198,200,229,230] preceded electrophysiological characterization using mammalian sodium channels in heterologous expression. Am2766 from Conus amadis [204,231] slowed the entry of NaV1.2 into the fast-inactivated state; the related peptide Am2755 differs from Am2766 in the C-terminus and has no effect on NaV1.2 fast inactivation. EVIA (Conus ermineus) prolonged fast inactivation of NaV1.2, NaV1.3, and NaV1.6 sodium channels without affecting NaV1.4 or NaV1.5 [205]. SVIE (Conus striatus), first characterized by its effect to slow sodium current inactivation in frog sympathetic ganglion neurons [203,232], was later employed in studies to define the binding site. Alpha scorpion toxin Lqh-2 and alpha-like toxin Lqh-3 have overlapping but distinct site-3 binding sites in the domain IV S3–S4 linker [233]. A comparison of sodium channel NaV1.4 response to Lqh-2, Lqh-3, and SVIE, and incorporating cysteine scanning mutagenesis of the S3–S4 linker, revealed an important hydrophobic interaction of the δ-conotoxin with a conserved YFV motif as part of the receptor 6 binding site for δ-conotoxins [80]. A comparison of the kinetic block and gating modifying actions of μO- and δ-conotoxins is provided in the review by [81].
CnVIB, CnVIC and CnVID (Conus consors) slowed fast inactivation in several TTX-sensitive channels, with effects pronounced for CnVIC/D on NaV1.2, CnVID on NaV1.3, CnVIB/C on NaV1.4, and each prohibiting fast inactivation of NaV1.6 [206]. While TTX-resistant channels NaV1.5 and NaV1.8 were largely unaffected, CnVIC did elicit a slight slowing of fast inactivation in NaV1.5, illustrating significant sodium channel isoform specificity for these δ-conotoxins. TsVIA (Conus tessulatus) enhanced the depolarizing influence of the site-2 toxin veratridine and slowed fast inactivation of NaV1.6 [207]. SuVIA (Conus suturatus) was shown to enhance veratridine depolarization for NaV1.3, NaV1.4, NaV1.6, and NaV1.7, using a fluorescence-based membrane potential assay [2]. In whole cell recordings, 5 nM SuVIA produced an (excitatory) left shift of the conductance curve for on NaV1.7, without significant effect on fast inactivation kinetics. Thus, δ-conotoxins typically act as gating modifiers with toxin specificity towards TTX-sensitive channels, promoting their excitatory effect by slowing the entry of sodium channels into the fast-inactivated state or enhancing activation.
9. Isolation and Characterization of Iota (ι)-Conotoxins
Conotoxins of the I superfamily exhibit a structure characterized by eight cysteine residues as C---C---CC---CC---C---C with five inter-cysteine loops [234,235]. Of five similar “iota-conotoxin” peptides in this superfamily purified from Conus radiatus, r11a, r11b, r11c, and r11e elicited repetitive action potentials in nerve or muscle when applied to the frog cutaneous pectoris neuromuscular preparation. These and other I-1 conotoxins isolated from Conus figulinus, Conus magus, and Conus striatus were isolated, and posttranslational modification of r11a was shown to increase its biological activity [236].
The first of these I-1 conotoxins characterized on mammalian sodium channels was ι-RXIA (r11a), which increased current amplitude and produced an excitatory left shift of the activation probability curve for NaV1.6 expressed in Xenopus oocytes [95] without affecting steady-state fast inactivation. ι-RXIA was shown to produce similar effects on NaV1.2 and NaV1.7, at higher doses [237], and it promoted increased action potential frequency in A (proprioceptive) fibers known to express NaV1.6, and C (nociceptive) fibers known to express NaV1.7. Thus, both the δ- and ι-conotoxins are excitatory in their effect, albeit typically acting on different gating transitions of voltage-gated sodium channels.
10. Summary
In conclusion, conotoxins have provided unique opportunities to study the structure-to-function relationships of voltage-gated sodium channels, within the context of an intriguing evolutionary background of considerable interest to many in the field of toxin biology. In addition to the conotoxins of the M, O, and I superfamilies characterized by functional experiments described in this review, other conotoxins appear to target voltage-gated sodium channels. For example, conotoxin GS was isolated, with a sequence exhibiting six cysteine residues and with ability to displace TTX from skeletal muscle homogenate [238]. The conotoxin TvIIA isolated from Conus tulipa displays structural similarities to μO-conotoxins, δ-conotoxins, and to GS conotoxin, but has not been tested for actions on sodium channels [239]. A four cysteine conotoxin from the T superfamily, sr5a, was isolated from Conus spurius [240]. This conotoxin was subsequently tested on expressed sodium channels and found to produce partial block of TTX-resistant NaV1.5 at 0.2 μM, with lesser block of TTX-sensitive NaV1.6 or NaV1.7 [241] and named μ-SrVa. These studies illustrate the importance of continued discovery and characterization of novel conotoxins targeting voltage-gated sodium channels.
A few of the other approaches and questions that have shared this historical path towards discovery should be noted, albeit in brief. Computational approaches such as docking studies using homology models, or molecular dynamics, have been instrumental in understanding sodium channel function and their interactions with conotoxins [165,242,243]; for reviews see [24,244,245], including prokaryotic sodium channels as part of this effort [246,247,248,249] and extending to examine the roles of toxin residues [250] or disulfide linkage connectivity on μ-conotoxin binding and block of sodium permeance [179,251,252].
Proteome and transcriptome analyses have played an increasingly important role in conotoxin research. The venomics approach has revealed numerous and interesting details with respect to the diversity, plasticity, post-translational modification, and evolution of conopeptides (for reviews see [253,254,255,256,257]). Along with computational approaches, these larger scale analyses provide opportunities for success in future investigations employing structural and functional characterization of these peptides. Finally, targeting post-translational modification with existing and new technologies may promote more rapid development of conotoxin analogues suitable for specific pharmacological intervention (reviewed by [258,259,260]) including new strategies for antimicrobials [261,262,263].
Funding
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant P20GM103408.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflict of interest.
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Israel, M.R.; Inserra, M.C.; Smith, J.J.; Lewis, R.J.; Alewood, P.F.; Vetter, I.; Dutertre, S. δ-Conotoxin SuVIA Suggests an Evolutionary Link between Ancestral Predator Defence and the Origin of Fish-Hunting Behaviour in Carnivorous Cone Snails. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150817. [Google Scholar] [CrossRef] [PubMed]
- Jost, M.C.; Hillis, D.M.; Lu, Y.; Kyle, J.W.; Fozzard, H.A.; Zakon, H.H. Toxin-Resistant Sodium Channels: Parallel Adaptive Evolution across a Complete Gene Family. Mol. Biol. Evol. 2008, 25, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Brodie, E.D., III; Brodie, E.D., Jr. Predictably Convergent Evolution of Sodium Channels in the Arms Race between Predators and Prey. Brain. Behav. Evol. 2015, 86, 48–57. [Google Scholar] [CrossRef]
- Feldman, C.R.; Durso, A.M.; Hanifin, C.T.; Pfrender, M.E.; Ducey, P.K.; Stokes, A.N.; Barnett, K.E.; Brodie, E.D.; Brodie, E.D. Is There More than One Way to Skin a Newt? Convergent Toxin Resistance in Snakes Is Not Due to a Common Genetic Mechanism. Heredity 2017, 119, 468. [Google Scholar] [CrossRef]
- Geffeney, S.L.; Williams, B.L.; Rosenthal, J.J.C.; Birk, M.A.; Felkins, J.; Wisell, C.M.; Curry, E.R.; Hanifin, C.T. Convergent and Parallel Evolution in a Voltage-Gated Sodium Channel Underlies TTX-Resistance in the Greater Blue-Ringed Octopus: Hapalochlaena Lunulata. Toxicon 2019, 170, 77–84. [Google Scholar] [CrossRef]
- Van Thiel, J.; Khan, M.A.; Wouters, R.M.; Harris, R.J.; Casewell, N.R.; Fry, B.G.; Kini, R.M.; Mackessy, S.P.; Vonk, F.J.; Wüster, W.; et al. Convergent Evolution of Toxin Resistance in Animals. Biol. Rev. 2022, 97, 1823–1843. [Google Scholar] [CrossRef]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-Capture Strategies of Fish-Hunting Cone Snails: Behavior, Neurobiology and Evolution. Brain. Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef]
- Olivera, B.M.; Showers Corneli, P.; Watkins, M.; Fedosov, A. Biodiversity of Cone Snails and Other Venomous Marine Gastropods: Evolutionary Success Through Neuropharmacology. Annu. Rev. Anim. Biosci. 2014, 2, 487–513. [Google Scholar] [CrossRef]
- Olivera, B.M.; Raghuraman, S.; Schmidt, E.W.; Safavi-Hemami, H. Linking Neuroethology to the Chemical Biology of Natural Products: Interactions between Cone Snails and Their Fish Prey, a Case Study. J. Comp. Physiol. A 2017, 203, 717–735. [Google Scholar] [CrossRef]
- Olivera, B.M.; Walker, C.; Cartier, G.E.; Hooper, D.; Santos, A.D.; Schoenfeld, R.; Shetty, R.; Watkins, M.; Bandyopadhyay, P.; Hillyard, D.R. Speciation of Cone Snails and Interspecific Hyperdivergence of Their Venom Peptides: Potential Evolutionary Significance of Intronsa. Ann. N. Y. Acad. Sci. 1999, 870, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Kapil, S.; Hendriksen, S.; Cooper, J.S. Cone Snail Toxicity. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Yu, C.; Yu, H.; Li, P. Highlights of Animal Venom Research on the Geographical Variations of Toxin Components, Toxicities and Envenomation Therapy. Int. J. Biol. Macromol. 2020, 165, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.; Norton, R. Conotoxin Gene Superfamilies. Mar. Drugs 2014, 12, 6058–6101. [Google Scholar] [CrossRef]
- Phuong, M.A.; Mahardika, G.N. Targeted Sequencing of Venom Genes from Cone Snail Genomes Improves Understanding of Conotoxin Molecular Evolution. Mol. Biol. Evol. 2018, 35, 1210–1224. [Google Scholar] [CrossRef]
- Benoit, E. Mechanism of action of neurotoxins acting on the inactivation of voltage-gated sodium channels. Comptes Rendus Seances Soc. Biol. Fil. 1998, 192, 409–436. [Google Scholar]
- Terlau, H.; Olivera, B.M. Conus Venoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus Venom Peptide Pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- Green, B.R.; Olivera, B.M. Venom Peptides From Cone Snails. In Current Topics in Membranes; Elsevier: Amsterdam, The Netherlands, 2016; Volume 78, pp. 65–86. ISBN 978-0-12-805386-7. [Google Scholar]
- Duque, H.M.; Campos Dias, S.; Franco, O. Structural and Functional Analyses of Cone Snail Toxins. Mar. Drugs 2019, 17, 370. [Google Scholar] [CrossRef]
- Mackieh, R.; Abou-Nader, R.; Wehbe, R.; Mattei, C.; Legros, C.; Fajloun, Z.; Sabatier, J.M. Voltage-Gated Sodium Channels: A Prominent Target of Marine Toxins. Mar. Drugs 2021, 19, 562. [Google Scholar] [CrossRef]
- Daly, N.L.; Craik, D.J. Structural Studies of Conotoxins. IUBMB Life 2009, 61, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Zhorov, B.S.; Tikhonov, D.B. Computational Structural Pharmacology and Toxicology of Voltage-Gated Sodium Channels. In Current Topics in Membranes; Elsevier: Amsterdam, The Netherlands, 2016; Volume 78, pp. 117–144. ISBN 978-0-12-805386-7. [Google Scholar]
- Tikhonov, D.B.; Zhorov, B.S. Predicting Structural Details of the Sodium Channel Pore Basing on Animal Toxin Studies. Front. Pharmacol. 2018, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Knapp, O.; McArthur, J.R.; Adams, D.J. Conotoxins Targeting Neuronal Voltage-Gated Sodium Channel Subtypes: Potential Analgesics? Toxins 2012, 4, 1236–1260. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.; Lewis, R.J. Therapeutic Potential of Cone Snail Venom Peptides (Conopeptides). Curr. Top. Med. Chem. 2012, 12, 1546–1552. [Google Scholar] [CrossRef]
- Munasinghe, N.; Christie, M. Conotoxins That Could Provide Analgesia through Voltage Gated Sodium Channel Inhibition. Toxins 2015, 7, 5386–5407. [Google Scholar] [CrossRef]
- Tosti, E.; Boni, R.; Gallo, A. Μ-Conotoxins Modulating Sodium Currents in Pain Perception and Transmission: A Therapeutic Potential. Mar. Drugs 2017, 15, 295. [Google Scholar] [CrossRef]
- Twede, V.D.; Miljanich, G.; Olivera, B.M.; Bulaj, G. Neuroprotective and Cardioprotective Conopeptides: An Emerging Class of Drug Leads. Curr. Opin. Drug Discov. Devel. 2009, 12, 231–239. [Google Scholar]
- Coulter-Parkhill, A.; McClean, S.; Gault, V.A.; Irwin, N. Therapeutic Potential of Peptides Derived from Animal Venoms: Current Views and Emerging Drugs for Diabetes. Clin. Med. Insights Endocrinol. Diabetes 2021, 14, 117955142110060. [Google Scholar] [CrossRef]
- De Castro Figueiredo Bordon, K.; Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Júnior, E.L.; Cerni, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cardoso, I.A.; et al. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef]
- Livett, B.G.; Gayler, K.R.; Khalil, Z. Drugs from the Sea: Conopeptides as Potential Therapeutics. Curr. Med. Chem. 2004, 11, 1715–1723. [Google Scholar] [CrossRef]
- Miljanich, G.P. Ziconotide: Neuronal Calcium Channel Blocker for Treating Severe Chronic Pain. Curr. Med. Chem. 2004, 11, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.; Clark, R. G-Protein Coupled Receptors Targeted by Analgesic Venom Peptides. Toxins 2017, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Pérez de Vega, M.J.; Ferrer-Montiel, A.; González-Muñiz, R. Recent Progress in Non-Opioid Analgesic Peptides. Arch. Biochem. Biophys. 2018, 660, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Hemami, H.; Brogan, S.E.; Olivera, B.M. Pain Therapeutics from Cone Snail Venoms: From Ziconotide to Novel Non-Opioid Pathways. J. Proteom. 2019, 190, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Jergova, S.; Perez, C.; Imperial, J.S.; Gajavelli, S.; Jain, A.; Abin, A.; Olivera, B.M.; Sagen, J. Cannabinoid Receptor Agonists from Conus Venoms Alleviate Pain-Related Behavior in Rats. Pharmacol. Biochem. Behav. 2021, 205, 173182. [Google Scholar] [CrossRef]
- Han, T.S.; Zhang, M.-M.; Gowd, K.H.; Walewska, A.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Disulfide-Depleted Selenoconopeptides: Simplified Oxidative Folding of Cysteine-Rich Peptides. ACS Med. Chem. Lett. 2010, 1, 140–144. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Santos, A.D.; Olivera, B.M. Conus Peptides Targeted to Specific Nicotinic Acetylcholine Receptor Subtypes. Annu. Rev. Biochem. 1999, 68, 59–88. [Google Scholar] [CrossRef]
- Nicke, A.; Wonnacott, S.; Lewis, R.J. Alpha-Conotoxins as Tools for the Elucidation of Structure and Function of Neuronal Nicotinic Acetylcholine Receptor Subtypes. Eur. J. Biochem. 2004, 271, 2305–2319. [Google Scholar] [CrossRef]
- Janes, R.W. α-Conotoxins as Selective Probes for Nicotinic Acetylcholine Receptor Subclasses. Curr. Opin. Pharmacol. 2005, 5, 280–292. [Google Scholar] [CrossRef]
- Dutertre, S.; Nicke, A.; Tsetlin, V.I. Nicotinic Acetylcholine Receptor Inhibitors Derived from Snake and Snail Venoms. Neuropharmacology 2017, 127, 196–223. [Google Scholar] [CrossRef]
- Abraham, N.; Lewis, R. Neuronal Nicotinic Acetylcholine Receptor Modulators from Cone Snails. Mar. Drugs 2018, 16, 208. [Google Scholar] [CrossRef]
- Bekbossynova, A.; Zharylgap, A.; Filchakova, O. Venom-Derived Neurotoxins Targeting Nicotinic Acetylcholine Receptors. Molecules 2021, 26, 3373. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.; Kudryavtsev, D.; Shelukhina, I.; Nikolaev, G.; Utkin, Y.; Tsetlin, V. Marine Origin Ligands of Nicotinic Receptors: Low Molecular Compounds, Peptides and Proteins for Fundamental Research and Practical Applications. Biomolecules 2022, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.; Peigneur, S.; Wijesekara, I.; Tytgat, J. Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview. Mar. Drugs 2014, 12, 2970–3004. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.W.; Marquart, L.A.; Phillips, P.D.; McDougal, O.M. Mutagenesis of α-Conotoxins for Enhancing Activity and Selectivity for Nicotinic Acetylcholine Receptors. Toxins 2019, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Adams, D.J.; Hung, A. Interactions of the A3β2 Nicotinic Acetylcholine Receptor Interfaces with α-Conotoxin LsIA and Its Carboxylated C-Terminus Analogue: Molecular Dynamics Simulations. Mar. Drugs 2020, 18, 349. [Google Scholar] [CrossRef]
- Xu, Q.; Tae, H.-S.; Wang, Z.; Jiang, T.; Adams, D.J.; Yu, R. Rational Design of α-Conotoxin RegIIA Analogues Selectively Inhibiting the Human A3β2 Nicotinic Acetylcholine Receptor through Computational Scanning. ACS Chem. Neurosci. 2020, 11, 2804–2811. [Google Scholar] [CrossRef] [PubMed]
- Hone, A.J.; Servent, D.; McIntosh, J.M. A9-containing Nicotinic Acetylcholine Receptors and the Modulation of Pain. Br. J. Pharmacol. 2018, 175, 1915–1927. [Google Scholar] [CrossRef]
- Hone, A.J.; Kaas, Q.; Kearns, I.; Hararah, F.; Gajewiak, J.; Christensen, S.; Craik, D.J.; McIntosh, J.M. Computational and Functional Mapping of Human and Rat A6β4 Nicotinic Acetylcholine Receptors Reveals Species-Specific Ligand-Binding Motifs. J. Med. Chem. 2021, 64, 1685–1700. [Google Scholar] [CrossRef]
- Wu, X.; Craik, D.J.; Kaas, Q. Interactions of Globular and Ribbon [Γ4E]GID with A4β2 Neuronal Nicotinic Acetylcholine Receptor. Mar. Drugs 2021, 19, 482. [Google Scholar] [CrossRef]
- Liang, J.; Tae, H.-S.; Zhao, Z.; Li, X.; Zhang, J.; Chen, S.; Jiang, T.; Adams, D.J.; Yu, R. Mechanism of Action and Structure–Activity Relationship of α-Conotoxin Mr1.1 at the Human A9α10 Nicotinic Acetylcholine Receptor. J. Med. Chem. 2022, 65, 16204–16217. [Google Scholar] [CrossRef] [PubMed]
- Quik, M.; Polonskaya, Y.; Kulak, J.M.; McIntosh, J.M. Vulnerability of 125I-α-Conotoxin MII Binding Sites to Nigrostriatal Damage in Monkey. J. Neurosci. 2001, 21, 5494–5500. [Google Scholar] [CrossRef]
- Quik, M.; Bordia, T.; Forno, L.; McIntosh, J.M. Loss of Alpha-ConotoxinMII- and A85380-Sensitive Nicotinic Receptors in Parkinson’s Disease Striatum. J. Neurochem. 2004, 88, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Quik, M.; McIntosh, J.M. Striatal A6 * Nicotinic Acetylcholine Receptors: Potential Targets for Parkinson’s Disease Therapy. J. Pharmacol. Exp. Ther. 2006, 316, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Bordia, T.; Grady, S.R.; McIntosh, J.M.; Quik, M. Nigrostriatal Damage Preferentially Decreases a Subpopulation of A6β2 * NAChRs in Mouse, Monkey, and Parkinson’s Disease Striatum. Mol. Pharmacol. 2007, 72, 52–61. [Google Scholar] [CrossRef]
- Azam, L.; McIntosh, J.M. Alpha-Conotoxins as Pharmacological Probes of Nicotinic Acetylcholine Receptors. Acta Pharmacol. Sin. 2009, 30, 771–783. [Google Scholar] [CrossRef]
- Turner, M.; Eidemiller, S.; Martin, B.; Narver, A.; Marshall, J.; Zemp, L.; Cornell, K.A.; McIntosh, J.M.; McDougal, O.M. Structural Basis for α-Conotoxin Potency and Selectivity. Bioorg. Med. Chem. 2009, 17, 5894–5899. [Google Scholar] [CrossRef]
- Satkunanathan, N.; Livett, B.; Gayler, K.; Sandall, D.; Down, J.; Khalil, Z. Alpha-Conotoxin Vc1.1 Alleviates Neuropathic Pain and Accelerates Functional Recovery of Injured Neurones. Brain Res. 2005, 1059, 149–158. [Google Scholar] [CrossRef]
- Livett, B.G.; Sandall, D.W.; Keays, D.; Down, J.; Gayler, K.R.; Satkunanathan, N.; Khalil, Z. Therapeutic Applications of Conotoxins That Target the Neuronal Nicotinic Acetylcholine Receptor. Toxicon 2006, 48, 810–829. [Google Scholar] [CrossRef]
- Pacini, A.; Micheli, L.; Maresca, M.; Branca, J.J.V.; McIntosh, J.M.; Ghelardini, C.; Di Cesare Mannelli, L. The A9α10 Nicotinic Receptor Antagonist α-Conotoxin RgIA Prevents Neuropathic Pain Induced by Oxaliplatin Treatment. Exp. Neurol. 2016, 282, 37–48. [Google Scholar] [CrossRef]
- Liu, C.; Wu, P.; Zhu, H.; Grieco, P.; Yu, R.; Gao, X.; Wu, G.; Wang, D.; Xu, H.; Qi, W. Rationally Designed α-Conotoxin Analogues Maintained Analgesia Activity and Weakened Side Effects. Molecules 2019, 24, 337. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Zhangsun, D.; Yu, G.; Su, R.; Luo, S. The A9α10 Nicotinic Acetylcholine Receptor Antagonist AO-Conotoxin GeXIVA [1,2] Alleviates and Reverses Chemotherapy-Induced Neuropathic Pain. Mar. Drugs 2019, 17, 265. [Google Scholar] [CrossRef] [PubMed]
- Dyachenko, I.A.; Palikova, Y.A.; Palikov, V.A.; Korolkova, Y.V.; Kazakov, V.A.; Egorova, N.S.; Garifulina, A.I.; Utkin, Y.N.; Tsetlin, V.I.; Kryukova, E.V. α-Conotoxin RgIA and Oligoarginine R8 in the Mice Model Alleviate Long-Term Oxaliplatin Induced Neuropathy. Biochimie 2022, 194, 127–136. [Google Scholar] [CrossRef]
- Stevens, M.; Peigneur, S.; Tytgat, J. Neurotoxins and Their Binding Areas on Voltage-Gated Sodium Channels. Front. Pharmacol. 2011, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Nau, C.; Wang, G.K. Residues in Na+ Channel D3-S6 Segment Modulate Both Batrachotoxin and Local Anesthetic Affinities. Biophys. J. 2000, 79, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Barile, M.; Wang, G.K. Disparate Role of Na+ Channel D2-S6 Residues in Batrachotoxin and Local Anesthetic Action. Mol. Pharmacol. 2001, 59, 1100–1107. [Google Scholar] [CrossRef]
- Catterall, W.A.; Trainer, V.; Baden, D.G. Molecular Properties of the Sodium Channel: A Receptor for Multiple Neurotoxins. Bull. Soc. Pathol. Exot. 1990 1992, 85, 481–485. [Google Scholar]
- Trainer, V.L.; Baden, D.G.; Catterall, W.A. Identification of Peptide Components of the Brevetoxin Receptor Site of Rat Brain Sodium Channels. J. Biol. Chem. 1994, 269, 19904–19909. [Google Scholar] [CrossRef]
- Lewis, R.J. Ion Channel Toxins and Therapeutics: From Cone Snail Venoms to Ciguatera. Ther. Drug Monit. 2000, 22, 61–64. [Google Scholar] [CrossRef]
- Wang, S. Voltage-Gated Sodium Channels as Primary Targets of Diverse Lipid-Soluble Neurotoxins. Cell. Signal. 2003, 15, 151–159. [Google Scholar] [CrossRef]
- Norton, R.S. Structure and Structure-Function Relationships of Sea Anemone Proteins That Interact with the Sodium Channel. Toxicon 1991, 29, 1051–1084. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.; Martin-Eauclaire, M.-F.; Cestèle, S.; Kopeyan, C.; Carlier, E.; Khalifa, R.B.; Pelhate, M.; Rochat, H. Scorpion Toxins Affecting Sodium Current Inactivation Bind to Distinct Homologous Receptor Sites on Rat Brain and Insect Sodium Channels. J. Biol. Chem. 1996, 271, 8034–8045. [Google Scholar] [CrossRef] [PubMed]
- Cestèle, S. Molecular Mechanisms of Neurotoxin Action on Voltage-Gated Sodium Channels. Biochimie 2000, 82, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A.; Cestèle, S.; Yarov-Yarovoy, V.; Yu, F.H.; Konoki, K.; Scheuer, T. Voltage-Gated Ion Channels and Gating Modifier Toxins. Toxicon 2007, 49, 124–141. [Google Scholar] [CrossRef]
- Hanck, D.A.; Sheets, M.F. Site-3 Toxins and Cardiac Sodium Channels. Toxicon 2007, 49, 181–193. [Google Scholar] [CrossRef]
- Moran, Y.; Gordon, D.; Gurevitz, M. Sea Anemone Toxins Affecting Voltage-Gated Sodium Channels—Molecular and Evolutionary Features. Toxicon 2009, 54, 1089–1101. [Google Scholar] [CrossRef]
- Leipold, E.; Hansel, A.; Olivera, B.M.; Terlau, H.; Heinemann, S.H. Molecular Interaction of δ-Conotoxins with Voltage-Gated Sodium Channels. FEBS Lett. 2005, 579, 3881–3884. [Google Scholar] [CrossRef]
- Heinemann, S.H.; Leipold, E. Conotoxins of the O-Superfamily Affecting Voltage-Gated Sodium Channels. Cell. Mol. Life Sci. 2007, 64, 1329–1340. [Google Scholar] [CrossRef]
- Shen, H.; Li, Z.; Jiang, Y.; Pan, X.; Wu, J.; Cristofori-Armstrong, B.; Smith, J.J.; Chin, Y.K.Y.; Lei, J.; Zhou, Q.; et al. Structural Basis for the Modulation of Voltage-Gated Sodium Channels by Animal Toxins. Science 2018, 362, eaau2596. [Google Scholar] [CrossRef]
- Wisedchaisri, G.; Tonggu, L.; Gamal El-Din, T.M.; McCord, E.; Zheng, N.; Catterall, W.A. Structural Basis for High-Affinity Trapping of the NaV1.7 Channel in Its Resting State by Tarantula Toxin. Mol. Cell 2021, 81, 38–48.e4. [Google Scholar] [CrossRef]
- Clairfeuille, T.; Cloake, A.; Infield, D.T.; Llongueras, J.P.; Arthur, C.P.; Li, Z.R.; Jian, Y.; Martin-Eauclaire, M.-F.; Bougis, P.E.; Ciferri, C.; et al. Structural Basis of α-Scorpion Toxin Action on Nav Channels. Science 2019, 363, eaav8573. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated Content, Knowledge, and Discovery Tools in the Conopeptide Database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, Synthesis, and Structure–Activity Relationships of Conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Fuller, E.; Green, B.R.; Catlin, P.; Buczek, O.; Nielsen, J.S.; Olivera, B.M.; Bulaj, G. Oxidative Folding of Conotoxins Sharing an Identical Disulfide Bridging Framework: Oxidative Folding of Conotoxins. FEBS J. 2005, 272, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Corpuz, G.P.; Jacobsen, R.B.; Jimenez, E.C.; Watkins, M.; Walker, C.; Colledge, C.; Garrett, J.E.; McDougal, O.; Li, W.; Gray, W.R.; et al. Definition of the M-Conotoxin Superfamily: Characterization of Novel Peptides from Molluscivorous Conus Venoms. Biochemistry 2005, 44, 8176–8186. [Google Scholar] [CrossRef]
- Jacob, R.B.; McDougal, O.M. The M-Superfamily of Conotoxins: A Review. Cell. Mol. Life Sci. 2010, 67, 17–27. [Google Scholar] [CrossRef]
- Green, B.R.; Bulaj, G.; Norton, R.S. Structure and Function of μ-Conotoxins, Peptide-Based Sodium Channel Blockers with Analgesic Activity. Future Med. Chem. 2014, 6, 1677–1698. [Google Scholar] [CrossRef]
- Ekberg, J.; Craik, D.J.; Adams, D.J. Conotoxin Modulation of Voltage-Gated Sodium Channels. Int. J. Biochem. Cell Biol. 2008, 40, 2363–2368. [Google Scholar] [CrossRef]
- Wakamatsu, K.; Kohda, D.; Hatanaka, H.; Lancelin, J.M.; Ishida, Y.; Oya, M.; Nakamura, H.; Inagaki, F.; Sato, K. Structure-Activity Relationships of.Mu.-Conotoxin GIIIA: Structure Determination of Active and Inactive Sodium Channel Blocker Peptides by NMR and Simulated Annealing Calculations. Biochemistry 1992, 31, 12577–12584. [Google Scholar] [CrossRef]
- Deuis, J.R.; Dekan, Z.; Inserra, M.C.; Lee, T.-H.; Aguilar, M.-I.; Craik, D.J.; Lewis, R.J.; Alewood, P.F.; Mobli, M.; Schroeder, C.I.; et al. Development of a ΜO-Conotoxin Analogue with Improved Lipid Membrane Interactions and Potency for the Analgesic Sodium Channel NaV1.8. J. Biol. Chem. 2016, 291, 11829–11842. [Google Scholar] [CrossRef]
- Volpon, L.; Lamthanh, H.; Barbier, J.; Gilles, N.; Molgó, J.; Ménez, A.; Lancelin, J.-M. NMR Solution Structures of δ-Conotoxin EVIA from Conus Ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels. J. Biol. Chem. 2004, 279, 21356–21366. [Google Scholar] [CrossRef] [PubMed]
- Buczek, O.; Wei, D.; Babon, J.J.; Yang, X.; Fiedler, B.; Chen, P.; Yoshikami, D.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structure and Sodium Channel Activity of an Excitatory I1 -Superfamily Conotoxin. Biochemistry 2007, 46, 9929–9940. [Google Scholar] [CrossRef]
- Endean, R.; Parish, G.; Gyr, P. Pharmacology of the Venom of Conus Geographus. Toxicon 1974, 12, 131–138. [Google Scholar] [CrossRef]
- Nakamura, H.; Kobayashi, J.; Ohizumi, Y.; Hirata, Y. Isolation and Amino Acid Compositions of Geographutoxin I and II from the Marine SnailConus Geographus Linné. Experientia 1983, 39, 590–591. [Google Scholar] [CrossRef]
- Sato, S.; Nakamura, H.; Ohizumi, Y.; Kobayashi, J.; Hirata, Y. The Amino Acid Sequences of Homologous Hydroxyproline-Containing Myotoxins from the Marine Snal Conus Geographus Venom. FEBS Lett. 1983, 155, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.J.; Gray, W.R.; Olivera, B.M.; Zeikus, R.D.; Kerr, L.; Yoshikami, D.; Moczydlowski, E. Conus Geographus Toxins That Discriminate between Neuronal and Muscle Sodium Channels. J. Biol. Chem. 1985, 260, 9280–9288. [Google Scholar] [CrossRef] [PubMed]
- Gonoi, T.; Ohizumi, Y.; Nakamura, H.; Kobayashi, J.; Catterall, W. The Conus Toxin Geographutoxin IL Distinguishes Two Functional Sodium Channel Subtypes in Rat Muscle Cells Developing in Vitro. J. Neurosci. 1987, 7, 1728–1731. [Google Scholar] [CrossRef]
- Gonoi, T.; Hagihara, Y.; Kobayashi, J.; Nakamura, H.; Ohizumi, Y. Geographutoxin-Sensitive and Insensitive Sodium Currents in Mouse Skeletal Muscle Developing in Situ. J. Physiol. 1989, 414, 159–177. [Google Scholar] [CrossRef]
- Kobayashi, M.; Wu, C.H.; Yoshii, M.; Narahashi, T.; Nakamura, H.; Kobayashi, J.; Ohizumi, Y. Preferential Block of Skeletal Muscle Sodium Channels by Geographutoxin II, a New Peptide Toxin FromConus Geographus. Pflügers Archiv. Eur. J. Physiol. 1986, 407, 241–243. [Google Scholar] [CrossRef]
- Noda, M.; Shimizu, S.; Tanabe, T.; Takai, T.; Kayano, T.; Ikeda, T.; Takahashi, H.; Nakayama, H.; Kanaoka, Y.; Minamino, N.; et al. Primary Structure of Electrophorus Electricus Sodium Channel Deduced from CDNA Sequence. Nature 1984, 312, 121–127. [Google Scholar] [CrossRef]
- Noda, M.; Ikeda, T.; Kayano, T.; Suzuki, H.; Takeshima, H.; Kurasaki, M.; Takahashi, H.; Numa, S. Existence of Distinct Sodium Channel Messenger RNAs in Rat Brain. Nature 1986, 320, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Kayano, T.; Noda, M.; Flockerzi, V.; Takahashi, H.; Numa, S. Primary Structure of Rat Brain Sodium Channel III Deduced from the CDNA Sequence. FEBS Lett. 1988, 228, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Rogart, R.B.; Cribbs, L.L.; Muglia, L.K.; Kephart, D.D.; Kaiser, M.W. Molecular Cloning of a Putative Tetrodotoxin-Resistant Rat Heart Na+ Channel Isoform. Proc. Natl. Acad. Sci. USA 1989, 86, 8170–8174. [Google Scholar] [CrossRef] [PubMed]
- Sills, M.N.; Xu, Y.C.; Baracchini, E.; Goodman, R.H.; Cooperman, S.S.; Mandel, G.; Chien, K.R. Expression of Diverse Na+ Channel Messenger RNAs in Rat Myocardium. Evidence for a Cardiac-Specific Na+ Channel. J. Clin. Investig. 1989, 84, 331–336. [Google Scholar] [CrossRef]
- Gellens, M.E.; George, A.L.; Chen, L.Q.; Chahine, M.; Horn, R.; Barchi, R.L.; Kallen, R.G. Primary Structure and Functional Expression of the Human Cardiac Tetrodotoxin-Insensitive Voltage-Dependent Sodium Channel. Proc. Natl. Acad. Sci. USA 1992, 89, 554–558. [Google Scholar] [CrossRef]
- White, M.M.; Chen, L.Q.; Kleinfield, R.; Kallen, R.G.; Barchi, R.L. SkM2, a Na+ Channel CDNA Clone from Denervated Skeletal Muscle, Encodes a Tetrodotoxin-Insensitive Na+ Channel. Mol. Pharmacol. 1991, 39, 604–608. [Google Scholar]
- Trimmer, J.S.; Cooperman, S.S.; Tomiko, S.A.; Zhou, J.; Crean, S.M.; Boyle, M.B.; Kalen, R.G.; Sheng, Z.; Barchi, R.L.; Sigworth, F.J.; et al. Primary Structure and Functional Expression of a Mammalian Skeletal Muscle Sodium Channel. Neuron 1989, 3, 33–49. [Google Scholar] [CrossRef]
- George, A.L.; Komisarof, J.; Kallen, R.G.; Barchi, R.L. Primary Structure of the Adult Human Skeletal Muscle Voltage-Dependent Sodium Channel. Ann. Neurol. 1992, 31, 131–137. [Google Scholar] [CrossRef]
- Goldin, A.L.; Snutch, T.; Lübbert, H.; Dowsett, A.; Marshall, J.; Auld, V.; Downey, W.; Fritz, L.C.; Lester, H.A.; Dunn, R. Messenger RNA Coding for Only the Alpha Subunit of the Rat Brain Na Channel Is Sufficient for Expression of Functional Channels in Xenopus Oocytes. Proc. Natl. Acad. Sci. USA 1986, 83, 7503–7507. [Google Scholar] [CrossRef]
- Stühmer, W.; Methfessel, C.; Sakmann, B.; Noda, M.; Numa, S. Patch Clamp Characterization of Sodium Channels Expressed from Rat Brain CDNA. Eur. Biophys. J. 1987, 14, 131–138. [Google Scholar] [CrossRef]
- Suzuki, H.; Beckh, S.; Kubo, H.; Yahagi, N.; Ishida, H.; Kayano, T.; Noda, M.; Numa, S. Functional Expression of Cloned CDNA Encoding Sodium Channel III. FEBS Lett. 1988, 228, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Chahine, M.; Bennett, P.B.; George, A.L.; Horn, R. Functional Expression and Properties of the Human Skeletal Muscle Sodium Channel. Pflügers Arch. 1994, 427, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Noda, M.; Ikeda, T.; Suzuki, H.; Takeshima, H.; Takahashi, T.; Kuno, M.; Numa, S. Expression of Functional Sodium Channels from Cloned CDNA. Nature 1986, 322, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.J.; Watson, M.; Adams, D.J.; Hammarström, A.K.; Gage, P.W.; Hill, J.M.; Craik, D.J.; Thomas, L.; Adams, D.; Alewood, P.F.; et al. Solution Structure of μ-Conotoxin PIIIA, a Preferential Inhibitor of Persistent Tetrodotoxin-Sensitive Sodium Channels. J. Biol. Chem. 2002, 277, 27247–27255. [Google Scholar] [CrossRef]
- Harvey, P.; Kurniawan, N.; Finol-Urdaneta, R.; McArthur, J.; Van Lysebetten, D.; Dash, T.; Hill, J.; Adams, D.; Durek, T.; Craik, D. NMR Structure of μ-Conotoxin GIIIC: Leucine 18 Induces Local Repacking of the N-Terminus Resulting in Reduced NaV Channel Potency. Molecules 2018, 23, 2715. [Google Scholar] [CrossRef]
- Sheets, M.F.; Hanck, D.A. Voltage-Dependent Open-State Inactivation of Cardiac Sodium Channels: Gating Current Studies with Anthopleurin—A Toxin. J. Gen. Physiol. 1995, 106, 617–640. [Google Scholar] [CrossRef]
- Sheets, M.F.; Hanck, D.A. Gating of Skeletal and Cardiac Muscle Sodium Channels in Mammalian Cells. J. Physiol. 1999, 514, 425–436. [Google Scholar] [CrossRef]
- Ohizumi, Y.; Nakamura, H.; Kobayashi, J.; Catterall, W.A. Specific Inhibition of [3H] Saxitoxin Binding to Skeletal Muscle Sodium Channels by Geographutoxin II, a Polypeptide Channel Blocker. J. Biol. Chem. 1986, 261, 6149–6152. [Google Scholar] [CrossRef]
- Moczydlowski, E.; Olivera, B.M.; Gray, W.R.; Strichartz, G.R. Discrimination of Muscle and Neuronal Na-Channel Subtypes by Binding Competition between [3H]Saxitoxin and Mu-Conotoxins. Proc. Natl. Acad. Sci. USA 1986, 83, 5321–5325. [Google Scholar] [CrossRef]
- Yanagawa, Y.; Abe, T.; Satake, M. Blockade of [3H]Lysine-Tetrodotoxin Binding to Sodium Channel Proteins by Conotoxin GIII. Neurosci. Lett. 1986, 64, 7–12. [Google Scholar] [CrossRef]
- Cruz, L.J.; Kupryszewski, G.; LeCheminant, G.W.; Gray, W.R.; Olivera, B.M.; Rivier, J.M. Conotoxin GIIIA, a Peptide Ligand for Muscle Sodium Channels: Chemical Synthesis, Radiolabeling and Receptor Characterization. Biochemistry 1989, 28, 3437–3442. [Google Scholar] [CrossRef] [PubMed]
- French, R.J.; Horn, R. Shifts of Macroscopic Current Activation in Partially Blocked Sodium Channels. Interaction between the Voltage Sensor and a μ-Conotoxin. In From Ion Channels to Cell-to-Cell Conversations; Latorre, R., Sáez, J.C., Eds.; Springer: Boston, MA, USA, 1997; pp. 67–89. ISBN 978-1-4899-1797-3. [Google Scholar]
- French, R.J.; Yoshikami, D.; Sheets, M.F.; Olivera, B.M. The Tetrodotoxin Receptor of Voltage-Gated Sodium Channels—Perspectives from Interactions with μ-Conotoxins. Mar. Drugs 2010, 8, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Capes, D.L.; Arcisio-Miranda, M.; Jarecki, B.W.; French, R.J.; Chanda, B. Gating Transitions in the Selectivity Filter Region of a Sodium Channel Are Coupled to the Domain IV Voltage Sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Lancelin, J.M.; Kohda, D.; Tate, S.; Yanagawa, Y.; Abe, T.; Satake, M.; Inagaki, F. Tertiary Structure of Conotoxin GIIIA in Aqueous Solution. Biochemistry 1991, 30, 6908–6916. [Google Scholar] [CrossRef]
- Hill, J.M.; Alewood, P.F.; Craik, D.J. Three-Dimensional Solution Structure of μ-Conotoxin GIIIB, a Specific Blocker of Skeletal Muscle Sodium Channels. Biochemistry 1996, 35, 8824–8835. [Google Scholar] [CrossRef]
- Sato, K.; Ishida, Y.; Wakamatsu, K.; Kato, R.; Honda, H.; Ohizumi, Y.; Nakamura, H.; Ohya, M.; Lancelin, J.M.; Kohda, D. Active Site of Mu-Conotoxin GIIIA, a Peptide Blocker of Muscle Sodium Channels. J. Biol. Chem. 1991, 266, 16989–16991. [Google Scholar] [CrossRef]
- Becker, S.; Prusak-Sochaczewski, E.; Zamponi, G.; Beck-Sickinger, A.G.; Gordon, R.D.; French, R.J. Action of Derivatives of.Mu.-Conotoxin GIIIA on Sodium Channels. Single Amino Acid Substitutions in the Toxin Separately Affect Association and Dissociation Rates. Biochemistry 1992, 31, 8229–8238. [Google Scholar] [CrossRef]
- Chahine, M.; Chen, L.Q.; Fotouhi, N.; Walsky, R.; Fry, D.; Santarelli, V.; Horn, R.; Kallen, R.G. Characterizing the Mu-Conotoxin Binding Site on Voltage-Sensitive Sodium Channels with Toxin Analogs and Channel Mutations. Recept. Channels 1995, 3, 161–174. [Google Scholar]
- Han, P.; Wang, K.; Dai, X.; Cao, Y.; Liu, S.; Jiang, H.; Fan, C.; Wu, W.; Chen, J. The Role of Individual Disulfide Bonds of μ-Conotoxin GIIIA in the Inhibition of NaV1.4. Mar. Drugs 2016, 14, 213. [Google Scholar] [CrossRef]
- Khan, A.; Romantseva, L.; Lam, A.; Lipkind, G.; Fozzard, H.A. Role of Outer Ring Carboxylates of the Rat Skeletal Muscle Sodium Channel Pore in Proton Block. J. Physiol. 2002, 543, 71–84. [Google Scholar] [CrossRef]
- Lipkind, G.M.; Fozzard, H.A. Voltage-Gated Na Channel Selectivity: The Role of the Conserved Domain III Lysine Residue. J. Gen. Physiol. 2008, 131, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Lim, C. Ion Selectivity Strategies of Sodium Channel Selectivity Filters. Acc. Chem. Res. 2014, 47, 3580–3587. [Google Scholar] [CrossRef]
- Flood, E.; Boiteux, C.; Allen, T.W. Selective Ion Permeation Involves Complexation with Carboxylates and Lysine in a Model Human Sodium Channel. PLoS Comput. Biol. 2018, 14, e1006398. [Google Scholar] [CrossRef] [PubMed]
- Penzotti, J.L.; Fozzard, H.A.; Lipkind, G.M.; Dudley, S.C. Differences in Saxitoxin and Tetrodotoxin Binding Revealed by Mutagenesis of the Na+ Channel Outer Vestibule. Biophys. J. 1998, 75, 2647–2657. [Google Scholar] [CrossRef] [PubMed]
- Terlau, H.; Heinemann, S.H.; Stühmer, W.; Pusch, M.; Conti, F.; Imoto, K.; Numa, S. Mapping the Site of Block by Tetrodotoxin and Saxitoxin of Sodium Channel II. FEBS Lett. 1991, 293, 93–96. [Google Scholar] [CrossRef]
- Dudley, S.C.; Todt, H.; Lipkind, G.; Fozzard, H.A. A Mu-Conotoxin-Insensitive Na+ Channel Mutant: Possible Localization of a Binding Site at the Outer Vestibule. Biophys. J. 1995, 69, 1657–1665. [Google Scholar] [CrossRef]
- Li, R.A.; Tsushima, R.G.; Kallen, R.G.; Backx, P.H. Pore Residues Critical for Mu-CTX Binding to Rat Skeletal Muscle Na+ Channels Revealed by Cysteine Mutagenesis. Biophys. J. 1997, 73, 1874–1884. [Google Scholar] [CrossRef]
- Chahine, M.; Sirois, J.; Marcotte, P.; Chen, L.-Q.; Kallen, R.G. Extrapore Residues of the S5-S6 Loop of Domain 2 of the Voltage-Gated Skeletal Muscle Sodium Channel (RSkM1) Contribute to the μ-Conotoxin GIIIA Binding Site. Biophys. J. 1998, 75, 236–246. [Google Scholar] [CrossRef]
- Carbonneau, E.; Vijayaragavan, K.; Chahine, M. A Tryptophan Residue (W736) in the Amino-Terminus of the P-Segment of Domain II Is Involved in Pore Formation in Nav1.4 Voltage-Gated Sodium Channels. Pflügers Arch. Eur. J. Physiol. 2002, 445, 18–24. [Google Scholar] [CrossRef]
- Li, R.A.; Hui, K.; French, R.J.; Sato, K.; Henrikson, C.A.; Tomaselli, G.F.; Marbán, E. Dependence of μ-Conotoxin Block of Sodium Channels on Ionic Strength but Not on the Permeating [Na+]. J. Biol. Chem. 2003, 278, 30912–30919. [Google Scholar] [CrossRef]
- Nakamura, M.; Ishida, Y.; Kohno, T.; Sato, K.; Oba, Y.; Nakamura, H. Effects of Modification at the Fifth Residue of Mu-Conotoxin GIIIA with Bulky Tags on the Electrically Stimulated Contraction of the Rat Diaphragm. J. Pept. Res. Off. J. Am. Pept. Soc. 2004, 64, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, G.; Aliste, M.P.; Tieleman, D.P.; French, R.J.; Dudley, S.C., Jr. Docking of μ-Conotoxin GIIIA in the Sodium Channel Outer Vestibule. Channels 2007, 1, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.; Lipkind, G.; Fozzard, H.A.; French, R.J. Electrostatic and Steric Contributions to Block of the Skeletal Muscle Sodium Channel by μ-Conotoxin. J. Gen. Physiol. 2002, 119, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.S.; French, R.J.; Lipkind, G.M.; Fozzard, H.A.; Dudley, S. Predominant Interactions between μ-Conotoxin Arg-13 and the Skeletal Muscle Na+ Channel Localized by Mutant Cycle Analysis. Biochemistry 1998, 37, 4407–4419. [Google Scholar] [CrossRef]
- Li, R.A.; Sato, K.; Kodama, K.; Kohno, T.; Xue, T.; Tomaselli, G.F.; Marbán, E. Charge Conversion Enables Quantification of the Proximity between a Normally-Neutral μ-Conotoxin (GIIIA) Site and the Na+ Channel Pore. FEBS Lett. 2002, 511, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Li, R.A.; Ennis, I.L.; Tomaselli, G.F.; French, R.J.; Marbán, E. Latent Specificity of Molecular Recognition in Sodium Channels Engineered To Discriminate between Two “Indistinguishable” μ-Conotoxins. Biochemistry 2001, 40, 6002–6008. [Google Scholar] [CrossRef] [PubMed]
- Cummins, T.R.; Aglieco, F.; Dib-Hajj, S.D. Critical Molecular Determinants of Voltage-Gated Sodium Channel Sensitivity to μ-Conotoxins GIIIA/B. Mol. Pharmacol. 2002, 61, 1192–1201. [Google Scholar] [CrossRef]
- Pan, X.; Li, Z.; Huang, X.; Huang, G.; Gao, S.; Shen, H.; Liu, L.; Lei, J.; Yan, N. Molecular Basis for Pore Blockade of Human Na+ Channel Nav 1.2 by the μ-Conotoxin KIIIA. Science 2019, 363, 1309–1313. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Li, R.A.; Ennis, I.L.; Vélez, P.; Tomaselli, G.F.; Marbán, E. Novel Structural Determinants of Mu-Conotoxin (GIIIB) Block in Rat Skeletal Muscle (Mu1) Na+ Channels. J. Biol. Chem. 2000, 275, 27551–27558. [Google Scholar] [CrossRef]
- Li, R.A.; Vélez, P.; Chiamvimonvat, N.; Tomaselli, G.F.; Marbán, E. Charged Residues between the Selectivity Filter and S6 Segments Contribute to the Permeation Phenotype of the Sodium Channel. J. Gen. Physiol. 2000, 115, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Ennis, I.L.; Sato, K.; French, R.J.; Li, R.A. Novel Interactions Identified between μ-Conotoxin and the Na+ Channel Domain I P-Loop: Implications for Toxin-Pore Binding Geometry. Biophys. J. 2003, 85, 2299–2310. [Google Scholar] [CrossRef] [PubMed]
- Todt, H.; Dudley, S.C., Jr.; Kyle, J.W.; French, R.J.; Fozzard, H.A. Ultra-Slow Inactivation in Μ1 Na+ Channels Is Produced by a Structural Rearrangement of the Outer Vestibule. Biophys. J. 1999, 76, 1335–1345. [Google Scholar] [CrossRef]
- Li, R.A.; Ennis, I.L.; French, R.J.; Dudley, S.C.; Tomaselli, G.F.; Marbán, E. Clockwise Domain Arrangement of the Sodium Channel Revealed by ¼-Conotoxin (GIIIA) Docking Orientation. J. Biol. Chem. 2001, 276, 11072–11077. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.M.; Hasson, A.; Spira, M.E.; Gray, W.R.; Li, W.; Marsh, M.; Hillyard, D.R.; Olivera, B.M. A New Family of Conotoxins That Blocks Voltage-Gated Sodium Channels. J. Biol. Chem. 1995, 270, 16796–16802. [Google Scholar] [CrossRef] [PubMed]
- Shon, K.J.; Olivera, B.M.; Watkins, M.; Jacobsen, R.B.; Gray, W.R.; Floresca, C.Z.; Cruz, L.J.; Hillyard, D.R.; Brink, A.; Terlau, H.; et al. Mu-Conotoxin PIIIA, a New Peptide for Discriminating among Tetrodotoxin-Sensitive Na Channel Subtypes. J. Neurosci. Off. J. Soc. Neurosci. 1998, 18, 4473–4481. [Google Scholar] [CrossRef]
- Lewis, R.J.; Schroeder, C.I.; Ekberg, J.; Nielsen, K.J.; Loughnan, M.; Thomas, L.; Adams, D.A.; Drinkwater, R.; Adams, D.J.; Alewood, P.F. Isolation and Structure-Activity of μ-Conotoxin TIIIA, A Potent Inhibitor of Tetrodotoxin-Sensitive Voltage-Gated Sodium Channels. Mol. Pharmacol. 2007, 71, 676–685. [Google Scholar] [CrossRef]
- Walewska, A.; Skalicky, J.J.; Davis, D.R.; Zhang, M.-M.; Lopez-Vera, E.; Watkins, M.; Han, T.S.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. NMR-Based Mapping of Disulfide Bridges in Cysteine-Rich Peptides: Application to the μ-Conotoxin SxIIIA. J. Am. Chem. Soc. 2008, 130, 14280–14286. [Google Scholar] [CrossRef]
- Yang, M.; Zhao, S.; Min, X.; Shao, M.; Chen, Y.; Chen, Z.; Zhou, M. A Novel μ-Conotoxin from Worm-Hunting Conus Tessulatus That Selectively Inhibit Rat TTX-Resistant Sodium Currents. Toxicon 2017, 130, 11–18. [Google Scholar] [CrossRef]
- West, P.J.; Bulaj, G.; Garrett, J.E.; Olivera, B.M.; Yoshikami, D. μ-Conotoxin SmIIIA, a Potent Inhibitor of Tetrodotoxin-Resistant Sodium Channels in Amphibian Sympathetic and Sensory Neurons. Biochemistry 2002, 41, 15388–15393. [Google Scholar] [CrossRef]
- Bulaj, G.; West, P.J.; Garrett, J.E.; Watkins, M.; Zhang, M.-M.; Norton, R.S.; Smith, B.J.; Yoshikami, D.; Olivera, B.M. Novel Conotoxins from Conus Striatus and Conus Kinoshitai Selectively Block TTX-Resistant Sodium Channels. Biochemistry 2005, 44, 7259–7265. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Z.; Zhang, H.; Jiang, H.; Lu, W.; Zhao, Z.-Q.; Chi, C.-W. A Novel Conotoxin from Conus Striatus, μ-SIIIA, Selectively Blocking Rat Tetrodotoxin-Resistant Sodium Channels. Toxicon 2006, 47, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-M.; Fiedler, B.; Green, B.R.; Catlin, P.; Watkins, M.; Garrett, J.E.; Smith, B.J.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Structural and Functional Diversities among μ-Conotoxins Targeting TTX-Resistant Sodium Channels. Biochemistry 2006, 45, 3723–3732. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.I.; Ekberg, J.; Nielsen, K.J.; Adams, D.; Loughnan, M.L.; Thomas, L.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Neuronally Selective μ-Conotoxins from Conus Striatus Utilize an α-Helical Motif to Target Mammalian Sodium Channels. J. Biol. Chem. 2008, 283, 21621–21628. [Google Scholar] [CrossRef]
- Holford, M.; Zhang, M.-M.; Gowd, K.H.; Azam, L.; Green, B.R.; Watkins, M.; Ownby, J.-P.; Yoshikami, D.; Bulaj, G.; Olivera, B.M. Pruning Nature: Biodiversity-Derived Discovery of Novel Sodium Channel Blocking Conotoxins from Conus Bullatus. Toxicon 2009, 53, 90–98. [Google Scholar] [CrossRef]
- Khoo, K.K.; Gupta, K.; Green, B.R.; Zhang, M.-M.; Watkins, M.; Olivera, B.M.; Balaram, P.; Yoshikami, D.; Bulaj, G.; Norton, R.S. Distinct Disulfide Isomers of μ-Conotoxins KIIIA and KIIIB Block Voltage-Gated Sodium Channels. Biochemistry 2012, 51, 9826–9835. [Google Scholar] [CrossRef]
- Favreau, P.; Benoit, E.; Hocking, H.G.; Carlier, L.; D’hoedt, D.; Leipold, E.; Markgraf, R.; Schlumberger, S.; Córdova, M.A.; Gaertner, H.; et al. A Novel Μ-Conopeptide, CnIIIC, Exerts Potent and Preferential Inhibition of NaV1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors: A µ-Conopeptide with Novel Neuropharmacology. Br. J. Pharmacol. 2012, 166, 1654–1668. [Google Scholar] [CrossRef]
- McMahon, K.L.; Tran, H.N.T.; Deuis, J.R.; Lewis, R.J.; Vetter, I.; Schroeder, C.I. Discovery, Pharmacological Characterisation and NMR Structure of the Novel µ-Conotoxin SxIIIC, a Potent and Irreversible NaV Channel Inhibitor. Biomedicines 2020, 8, 391. [Google Scholar] [CrossRef]
- Floresca, C.Z. A Comparison of the μ-Conotoxins by [3H]Saxitoxin Binding Assays in Neuronal and Skeletal Muscle Sodium Channel. Toxicol. Appl. Pharmacol. 2003, 190, 95–101. [Google Scholar] [CrossRef]
- McArthur, J.R.; Singh, G.; O’Mara, M.L.; McMaster, D.; Ostroumov, V.; Tieleman, D.P.; French, R.J. Orientation of μ-Conotoxin PIIIA in a Sodium Channel Vestibule, Based on Voltage Dependence of Its Binding. Mol. Pharmacol. 2011, 80, 219–227. [Google Scholar] [CrossRef]
- McArthur, J.R.; Ostroumov, V.; Al-Sabi, A.; McMaster, D.; French, R.J. Multiple, Distributed Interactions of μ-Conotoxin PIIIA Associated with Broad Targeting among Voltage-Gated Sodium Channels. Biochemistry 2011, 50, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Safo, P.; Rosenbaum, T.; Shcherbatko, A.; Choi, D.-Y.; Han, E.; Toledo-Aral, J.J.; Olivera, B.M.; Brehm, P.; Mandel, G. Distinction among Neuronal Subtypes of Voltage-Activated Sodium Channels by μ-Conotoxin PIIIA. J. Neurosci. 2000, 20, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Keizer, D.W.; West, P.J.; Lee, E.F.; Yoshikami, D.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structural Basis for Tetrodotoxin-Resistant Sodium Channel Binding by μ-Conotoxin SmIIIA. J. Biol. Chem. 2003, 278, 46805–46813. [Google Scholar] [CrossRef]
- Zhang, M.-M.; Gruszczynski, P.; Walewska, A.; Bulaj, G.; Olivera, B.M.; Yoshikami, D. Cooccupancy of the Outer Vestibule of Voltage-Gated Sodium Channels by μ-Conotoxin KIIIA and Saxitoxin or Tetrodotoxin. J. Neurophysiol. 2010, 104, 88–97. [Google Scholar] [CrossRef]
- Tran, H.N.T.; McMahon, K.L.; Deuis, J.R.; Vetter, I.; Schroeder, C.I. Structural and Functional Insights into the Inhibition of Human Voltage-Gated Sodium Channels by μ-Conotoxin KIIIA Disulfide Isomers. J. Biol. Chem. 2022, 298, 101728. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A.; Goldin, A.L.; Waxman, S.G. International Union of Pharmacology. XLVII. Nomenclature and Structure-Function Relationships of Voltage-Gated Sodium Channels. Pharmacol. Rev. 2005, 57, 397–409. [Google Scholar] [CrossRef]
- Vetter, I.; Mozar, C.A.; Durek, T.; Wingerd, J.S.; Alewood, P.F.; Christie, M.J.; Lewis, R.J. Characterisation of Nav Types Endogenously Expressed in Human SH-SY5Y Neuroblastoma Cells. Biochem. Pharmacol. 2012, 83, 1562–1571. [Google Scholar] [CrossRef]
- Wilson, M.J.; Yoshikami, D.; Azam, L.; Gajewiak, J.; Olivera, B.M.; Bulaj, G.; Zhang, M.-M. μ-Conotoxins That Differentially Block Sodium Channels NaV1.1 through 1.8 Identify Those Responsible for Action Potentials in Sciatic Nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 10302–10307. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, M. μ-Conotoxin TsIIIA, a Peptide Inhibitor of Human Voltage-Gated Sodium Channel HNav1.8. Toxicon 2020, 186, 29–34. [Google Scholar] [CrossRef]
- Zhang, M.-M.; Green, B.R.; Catlin, P.; Fiedler, B.; Azam, L.; Chadwick, A.; Terlau, H.; McArthur, J.R.; French, R.J.; Gulyas, J.; et al. Structure/Function Characterization of μ-Conotoxin KIIIA, an Analgesic, Nearly Irreversible Blocker of Mammalian Neuronal Sodium Channels. J. Biol. Chem. 2007, 282, 30699–30706. [Google Scholar] [CrossRef]
- Khoo, K.K.; Feng, Z.-P.; Smith, B.J.; Zhang, M.-M.; Yoshikami, D.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structure of the Analgesic μ-Conotoxin KIIIA and Effects on the Structure and Function of Disulfide Deletion. Biochemistry 2009, 48, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Zhang, M.-M.; Yoshikami, D.; Azam, L.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structure, Dynamics, and Selectivity of the Sodium Channel Blocker μ-Conotoxin SIIIA. Biochemistry 2008, 47, 10940–10949. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.I.; Adams, D.; Thomas, L.; Alewood, P.F.; Lewis, R.J. N- and c-Terminal Extensions of μ-Conotoxins Increase Potency and Selectivity for Neuronal Sodium Channels. Biopolymers 2012, 98, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Leipold, E.; Markgraf, R.; Miloslavina, A.; Kijas, M.; Schirmeyer, J.; Imhof, D.; Heinemann, S.H. Molecular Determinants for the Subtype Specificity of μ-Conotoxin SIIIA Targeting Neuronal Voltage-Gated Sodium Channels. Neuropharmacology 2011, 61, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Markgraf, R.; Leipold, E.; Schirmeyer, J.; Paolini-Bertrand, M.; Hartley, O.; Heinemann, S.H. Mechanism and Molecular Basis for the Sodium Channel Subtype Specificity of Μ-Conopeptide CnIIIC: Subtype Specificity of µ-CnIIIC. Br. J. Pharmacol. 2012, 167, 576–586. [Google Scholar] [CrossRef]
- Stevens, M.; Peigneur, S.; Dyubankova, N.; Lescrinier, E.; Herdewijn, P.; Tytgat, J. Design of Bioactive Peptides from Naturally Occurring μ-Conotoxin Structures. J. Biol. Chem. 2012, 287, 31382–31392. [Google Scholar] [CrossRef]
- Van Der Haegen, A.; Peigneur, S.; Tytgat, J. Importance of Position 8 in μ-Conotoxin KIIIA for Voltage-Gated Sodium Channel Selectivity: Residue 8 in KIIIA as Key Residue for Selectivity. FEBS J. 2011, 278, 3408–3418. [Google Scholar] [CrossRef]
- McMahon, K.L.; Tran, H.N.T.; Deuis, J.R.; Craik, D.J.; Vetter, I.; Schroeder, C.I. Μ-Conotoxins Targeting the Human Voltage-Gated Sodium Channel Subtype NaV1.7. Toxins 2022, 14, 600. [Google Scholar] [CrossRef]
- Pallaghy, P.K.; Norton, R.S.; Nielsen, K.J.; Craik, D.J. A Common Structural Motif Incorporating a Cystine Knot and a Triple-Stranded β-Sheet in Toxic and Inhibitory Polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef]
- Norton, R.S.; Pallaghy, P.K. The Cystine Knot Structure of Ion Channel Toxins and Related Polypeptides. Toxicon 1998, 36, 1573–1583. [Google Scholar] [CrossRef]
- Craik, D.J.; Daly, N.L.; Waine, C. The Cystine Knot Motif in Toxins and Implications for Drug Design. Toxicon 2001, 39, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.; Dekan, Z.; Knapp, O.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Isolation, Characterization and Total Regioselective Synthesis of the Novel ΜO-Conotoxin MfVIA from Conus Magnificus That Targets Voltage-Gated Sodium Channels. Biochem. Pharmacol. 2012, 84, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Hillyard, D.R.; Olivera, B.M.; Woodward, S.; Corpuz, G.P.; Gray, W.R.; Ramilo, C.A.; Cruz, L.J. A Molluskivorous Conus Toxin: Conserved Frameworks in Conotoxins. Biochemistry 1989, 28, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Fainzilber, M.; Gordon, D.; Hasson, A.; Spira, M.E.; Zlotkin, E. Mollusc-Specific Toxins from the Venom of Conus Textile Neovicarius. Eur. J. Biochem. 1991, 202, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Sasaki, T.; Kobayashi, K.; Fainzilber, M.; Sato, K. Three-Dimensional Solution Structure of the Sodium Channel Agonist/Antagonist δ-Conotoxin TxVIA. J. Biol. Chem. 2002, 277, 36387–36391. [Google Scholar] [CrossRef]
- Shon, K.-J.; Hasson, A.; Spira, M.E.; Cruz, L.J.; Gray, W.R.; Olivera, B.M. Delta.-Conotoxin GmVIA, a Novel Peptide from the Venom of Conus Gloriamaris. Biochemistry 1994, 33, 11420–11425. [Google Scholar] [CrossRef]
- Fainzilber, M.; van der Schors, R.; Lodder, J.C.; Li, K.W.; Geraerts, W.P.M.; Kits, K.S. New Sodium Channel-Blocking Conotoxins Also Affect Calcium Currents in Lymnaea Neurons. Biochemistry 1995, 34, 5364–5371. [Google Scholar] [CrossRef]
- Shon, K.-J.; Grilley, M.M.; Marsh, M.; Yoshikami, D.; Hall, A.R.; Kurz, B.; Gray, W.R.; Imperial, J.S.; Hillyard, D.R.; Olivera, B.M. Purification, Characterization, Synthesis, and Cloning of the Lockjaw Peptide from Conus Purpurascens Venom. Biochemistry 1995, 34, 4913–4918. [Google Scholar] [CrossRef]
- Bulaj, G.; DeLaCruz, R.; Azimi-Zonooz, A.; West, P.; Watkins, M.; Yoshikami, D.; Olivera, B.M. δ-Conotoxin Structure/Function through a Cladistic Analysis. Biochemistry 2001, 40, 13201–13208. [Google Scholar] [CrossRef]
- Sudarslal, S.; Majumdar, S.; Ramasamy, P.; Dhawan, R.; Pal, P.P.; Ramaswami, M.; Lala, A.K.; Sikdar, S.K.; Sarma, S.P.; Krishnan, K.S.; et al. Sodium Channel Modulating Activity in a δ-Conotoxin from an Indian Marine Snail. FEBS Lett. 2003, 553, 209–212. [Google Scholar] [CrossRef]
- Barbier, J.; Lamthanh, H.; Le Gall, F.; Favreau, P.; Benoit, E.; Chen, H.; Gilles, N.; Ilan, N.; Heinemann, S.H.; Gordon, D.; et al. A δ-Conotoxin from Conus Ermineus Venom Inhibits Inactivation in Vertebrate Neuronal Na+ Channels but Not in Skeletal and Cardiac Muscles. J. Biol. Chem. 2004, 279, 4680–4685. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; Paolini-Bertrand, M.; Gaertner, H.; Biass, D.; Violette, A.; Stöcklin, R.; Favreau, P.; Tytgat, J.; Hartley, O. δ-Conotoxins Synthesized Using an Acid-Cleavable Solubility Tag Approach Reveal Key Structural Determinants for NaV Subtype Selectivity. J. Biol. Chem. 2014, 289, 35341–35350. [Google Scholar] [CrossRef] [PubMed]
- Aman, J.W.; Imperial, J.S.; Ueberheide, B.; Zhang, M.-M.; Aguilar, M.; Taylor, D.; Watkins, M.; Yoshikami, D.; Showers-Corneli, P.; Safavi-Hemami, H.; et al. Insights into the Origins of Fish Hunting in Venomous Cone Snails from Studies of Conus tessulatus. Proc. Natl. Acad. Sci. USA 2015, 112, 5087–5092. [Google Scholar] [CrossRef] [PubMed]
- Terlau, H.; Stocker, M.; Shon, K.J.; McIntosh, J.M.; Olivera, B.M. MicroO-Conotoxin MrVIA Inhibits Mammalian Sodium Channels, but Not through Site I. J. Neurophysiol. 1996, 76, 1423–1429. [Google Scholar] [CrossRef]
- Leipold, E.; Hansel, A.; Borges, A.; Heinemann, S.H. Subtype Specificity of Scorpion β-Toxin Tz1 Interaction with Voltage-Gated Sodium Channels Is Determined by the Pore Loop of Domain 3. Mol. Pharmacol. 2006, 70, 340–347. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Yarov-Yarovoy, V.; Scheuer, T.; Karbat, I.; Cohen, L.; Gordon, D.; Gurevitz, M.; Catterall, W.A. Mapping the Interaction Site for a β-Scorpion Toxin in the Pore Module of Domain III of Voltage-Gated Na+ Channels. J. Biol. Chem. 2012, 287, 30719–30728. [Google Scholar] [CrossRef]
- Sokolov, S.; Kraus, R.L.; Scheuer, T.; Catterall, W.A. Inhibition of Sodium Channel Gating by Trapping the Domain II Voltage Sensor with Protoxin II. Mol. Pharmacol. 2008, 73, 1020–1028. [Google Scholar] [CrossRef]
- Cestèle, S.; Qu, Y.; Rogers, J.C.; Rochat, H.; Scheuer, T.; Catterall, W.A. Voltage Sensor–Trapping. Neuron 1998, 21, 919–931. [Google Scholar] [CrossRef]
- Cestèle, S.; Yarov-Yarovoy, V.; Qu, Y.; Sampieri, F.; Scheuer, T.; Catterall, W.A. Structure and Function of the Voltage Sensor of Sodium Channels Probed by a β-Scorpion Toxin. J. Biol. Chem. 2006, 281, 21332–21344. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Yarov-Yarovoy, V.; Scheuer, T.; Karbat, I.; Cohen, L.; Gordon, D.; Gurevitz, M.; Catterall, W.A. Structure-Function Map of the Receptor Site for β-Scorpion Toxins in Domain II of Voltage-Gated Sodium Channels. J. Biol. Chem. 2011, 286, 33641–33651. [Google Scholar] [CrossRef]
- Leipold, E.; DeBie, H.; Zorn, S.; Adolfo, B.; Olivera, B.M.; Terlau, H.; Heinemann, S.H. ΜO-Conotoxins Inhibit NaV Channels by Interfering with Their Voltage Sensors in Domain-2. Channels 2007, 1, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Zorn, S.; Leipold, E.; Hansel, A.; Bulaj, G.; Olivera, B.M.; Terlau, H.; Heinemann, S.H. The ΜO-Conotoxin MrVIA Inhibits Voltage-Gated Sodium Channels by Associating with Domain-3. FEBS Lett. 2006, 580, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Stürzebecher, A.S.; Hu, J.; Smith, E.S.J.; Frahm, S.; Santos-Torres, J.; Kampfrath, B.; Auer, S.; Lewin, G.R.; Ibañez-Tallon, I. RAPID REPORT: An in Vivo Tethered Toxin Approach for the Cell-Autonomous Inactivation of Voltage-Gated Sodium Channel Currents in Nociceptors: Pain Modulation by Tethered Toxins. J. Physiol. 2010, 588, 1695–1707. [Google Scholar] [CrossRef]
- Bulaj, G.; Zhang, M.-M.; Green, B.R.; Fiedler, B.; Layer, R.T.; Wei, S.; Nielsen, J.S.; Low, S.J.; Klein, B.D.; Wagstaff, J.D.; et al. Synthetic ΜO-Conotoxin MrVIB Blocks TTX-Resistant Sodium Channel NaV1.8 and Has a Long-Lasting Analgesic Activity. Biochemistry 2006, 45, 7404–7414. [Google Scholar] [CrossRef]
- Daly, N.L.; Ekberg, J.A.; Thomas, L.; Adams, D.J.; Lewis, R.J.; Craik, D.J. Structures of ΜO-Conotoxins from Conus Marmoreus. J. Biol. Chem. 2004, 279, 25774–25782. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.J.; Zhang, M.-M.; Azam, L.; Olivera, B.M.; Bulaj, G.; Yoshikami, D. NaV β Subunits Modulate the Inhibition of NaV 1.8 by the Analgesic Gating Modifier ΜO-Conotoxin MrVIB. J. Pharmacol. Exp. Ther. 2011, 338, 687–693. [Google Scholar] [CrossRef]
- Zhang, M.-M.; Wilson, M.J.; Azam, L.; Gajewiak, J.; Rivier, J.E.; Bulaj, G.; Olivera, B.M.; Yoshikami, D. Co-Expression of NaV β Subunits Alters the Kinetics of Inhibition of Voltage-Gated Sodium Channels by Pore-Blocking μ-Conotoxins: NaV β Co-Expression Alters μ-Conotoxin Kinetics. Br. J. Pharmacol. 2013, 168, 1597–1610. [Google Scholar] [CrossRef]
- Pi, C.; Liu, J.; Wang, L.; Jiang, X.; Liu, Y.; Peng, C.; Chen, S.; Xu, A. Soluble Expression, Purification and Functional Identification of a Disulfide-Rich Conotoxin Derived from Conus Litteratus. J. Biotechnol. 2007, 128, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pi, C.; Liu, J.; Chen, S.; Peng, C.; Sun, D.; Zhou, M.; Xiang, H.; Ren, Z.; Xu, A. Identification and Characterization of a Novel O-Superfamily Conotoxin from Conus Litteratus: Discovery of a novel o-Superfamily Conotoxin. J. Pept. Sci. 2008, 14, 1077–1083. [Google Scholar] [CrossRef]
- Spira, M.E.; Hasson, A.; Fainzilber, M.; Gordon, D.; Zlotkin, E. Chemical and Electrophysiological Characterization of New Peptide Neurotoxins from the Venom of the Molluscivorous Snail Conus Textile Neovicarius: A Review. Isr. J. Med. Sci. 1993, 29, 530–543. [Google Scholar]
- Possani, L.D.; Becerril, B.; Delepierre, M.; Tytgat, J. Scorpion Toxins Specific for Na+-Channels. Eur. J. Biochem. 1999, 264, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Capes, D.L.; Goldschen-Ohm, M.P.; Arcisio-Miranda, M.; Bezanilla, F.; Chanda, B. Domain IV Voltage-Sensor Movement Is Both Sufficient and Rate Limiting for Fast Inactivation in Sodium Channels. J. Gen. Physiol. 2013, 142, 101–112. [Google Scholar] [CrossRef]
- Campos, F.V.; Chanda, B.; Beirão, P.S.L.; Bezanilla, F. α-Scorpion Toxin Impairs a Conformational Change That Leads to Fast Inactivation of Muscle Sodium Channels. J. Gen. Physiol. 2008, 132, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Groome, J.; Lehmann-Horn, F.; Holzherr, B. Open- and Closed-State Fast Inactivation in Sodium Channels: Differential Effects of a Site-3 Anemone Toxin. Channels 2011, 5, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Hasson, A.; Fainzilber, M.; Zlotkin, E.; Spira, M. Electrophysiological Characterization of a Novel Conotoxin That Blocks Molluscan Sodium Channels. Eur. J. Neurosci. 1995, 7, 815–818. [Google Scholar] [CrossRef]
- Hasson, A.; Shon, K.J.; Olivera, B.M.; Spira, M.E. Alterations of Voltage-Activated Sodium Current by a Novel Conotoxin from the Venom of Conus Gloriamaris. J. Neurophysiol. 1995, 73, 1295–1301. [Google Scholar] [CrossRef]
- Sarma, S.; Kumar, G.S.; Sudarslal, S.; Iengar, P.; Ramasamy, P.; Sikdar, S.; Krishnan, K.S.; Balaram, P. Solution Structure of Am2766: A Highly Hydrophobic d-Conotoxin from Conus Amadis That Inhibits Inactivation of Neuronal Voltage-Gated Sodium Channels. Chem. Biodivers. 2005, 2, 535–556. [Google Scholar] [CrossRef]
- West, P.J.; Bulaj, G.; Yoshikami, D. Effects of δ-Conotoxins PVIA and SVIE on Sodium Channels in the Amphibian Sympathetic Nervous System. J. Neurophysiol. 2005, 94, 3916–3924. [Google Scholar] [CrossRef]
- Leipold, E.; Lu, S.; Gordon, D.; Hansel, A.; Heinemann, S.H. Combinatorial Interaction of Scorpion Toxins Lqh-2, Lqh-3, and LqhαIT with Sodium Channel Receptor Sites-3. Mol. Pharmacol. 2004, 65, 685–691. [Google Scholar] [CrossRef]
- Jimenez, E.C.; Shetty, R.P.; Lirazan, M.; Rivier, J.; Walker, C.; Abogadie, F.C.; Yoshikami, D.; Cruz, L.J.; Olivera, B.M. Novel Excitatory Conus Peptides Define a New Conotoxin Superfamily: Peripherally Active Excitatory Conotoxins. J. Neurochem. 2003, 85, 610–621. [Google Scholar] [CrossRef]
- Buczek, O.; Yoshikami, D.; Watkins, M.; Bulaj, G.; Jimenez, E.C.; Olivera, B.M. Characterization of D-Amino-Acid-Containing Excitatory Conotoxins and Redefinition of the I-Conotoxin Superfamily: Defining the I-Conotoxin Superfamily. FEBS J. 2005, 272, 4178–4188. [Google Scholar] [CrossRef] [PubMed]
- Buczek, O.; Yoshikami, D.; Bulaj, G.; Jimenez, E.C.; Olivera, B.M. Post-Translational Amino Acid Isomerization. J. Biol. Chem. 2005, 280, 4247–4253. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, B.; Zhang, M.-M.; Buczek, O.; Azam, L.; Bulaj, G.; Norton, R.S.; Olivera, B.M.; Yoshikami, D. Specificity, Affinity and Efficacy of Iota-Conotoxin RXIA, an Agonist of Voltage-Gated Sodium Channels NaV1.2, 1.6 and 1.7. Biochem. Pharmacol. 2008, 75, 2334–2344. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Abe, T.; Satake, M.; Odani, S.; Suzuki, J.; Ishikawa, K. A Novel Sodium Channel Inhibitor from Conus Geographus: Purification, Structure, and Pharmacological Properties. Biochemistry 1988, 27, 6256–6262. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Atkins, A.R.; Loughnan, M.L.; Jones, A.; Adams, D.A.; Martin, R.C.; Lewis, R.J.; Craik, D.J.; Alewood, P.F. Conotoxin TVIIA, a Novel Peptide from the Venom of Conus Tulipa: 1. Isolation, Characterization and Chemical Synthesis. Eur. J. Biochem. 2000, 267, 4642–4648. [Google Scholar] [CrossRef]
- Aguilar, M.B.; Lezama-Monfil, L.; Maillo, M.; Pedraza-Lara, H.; López-Vera, E.; Heimer de la Cotera, E.P. A Biologically Active Hydrophobic T-1-Conotoxin from the Venom of Conus Spurius. Peptides 2006, 27, 500–505. [Google Scholar] [CrossRef]
- Ruelas-Callejas, A.; Aguilar, M.B.; Arteaga-Tlecuitl, R.; Gomora, J.C.; López-Vera, E. The T-1 Conotoxin μ-SrVA from the Worm Hunting Marine Snail Conus Spurius Preferentially Blocks the Human NaV1.5 Channel. Peptides 2022, 156, 170859. [Google Scholar] [CrossRef]
- Mahdavi, S.; Kuyucak, S. Molecular Dynamics Study of Binding of Μ-Conotoxin GIIIA to the Voltage-Gated Sodium Channel Nav1.4. PLoS ONE 2014, 9, e105300. [Google Scholar] [CrossRef]
- Zhao, L.; Barber, L.M.; Hung, A. Structural and Dynamical Effects of Targeted Mutations on ΜO-Conotoxin MfVIA: Molecular Simulation Studies. J. Mol. Graph. Model. 2021, 102, 107777. [Google Scholar] [CrossRef]
- Rashid, M.; Mahdavi, S.; Kuyucak, S. Computational Studies of Marine Toxins Targeting Ion Channels. Mar. Drugs 2013, 11, 848–869. [Google Scholar] [CrossRef]
- Deplazes, E. Molecular Simulations of Disulfide-Rich Venom Peptides with Ion Channels and Membranes. Molecules 2017, 22, 362. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chung, S.-H. Binding Modes of μ-Conotoxin to the Bacterial Sodium Channel (NaVAb). Biophys. J. 2012, 102, 483–488. [Google Scholar] [CrossRef]
- Korkosh, V.S.; Zhorov, B.S.; Tikhonov, D.B. Folding Similarity of the Outer Pore Region in Prokaryotic and Eukaryotic Sodium Channels Revealed by Docking of Conotoxins GIIIA, PIIIA, and KIIIA in a NavAb-Based Model of Nav1.4. J. Gen. Physiol. 2014, 144, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Mahdavi, S.; Kuyucak, S. Computational Study of Binding of μ-Conotoxin GIIIA to Bacterial Sodium Channels NaV Ab and NaV Rh. Biochemistry 2016, 55, 1929–1938. [Google Scholar] [CrossRef]
- Finol-Urdaneta, R.K.; McArthur, J.R.; Korkosh, V.S.; Huang, S.; McMaster, D.; Glavica, R.; Tikhonov, D.B.; Zhorov, B.S.; French, R.J. Extremely Potent Block of Bacterial Voltage-Gated Sodium Channels by µ-Conotoxin PIIIA. Mar. Drugs 2019, 17, 510. [Google Scholar] [CrossRef]
- Chen, R.; Robinson, A.; Chung, S.-H. Mechanism of μ-Conotoxin PIIIA Binding to the Voltage-Gated Na+ Channel NaV1.4. PLoS ONE 2014, 9, e93267. [Google Scholar] [CrossRef]
- Tietze, A.A.; Tietze, D.; Ohlenschläger, O.; Leipold, E.; Ullrich, F.; Kühl, T.; Mischo, A.; Buntkowsky, G.; Görlach, M.; Heinemann, S.H.; et al. Structurally Diverse μ-Conotoxin PIIIA Isomers Block Sodium Channel NaV 1.4. Angew. Chem. Int. Ed. 2012, 51, 4058–4061. [Google Scholar] [CrossRef] [PubMed]
- Paul George, A.A.; Heimer, P.; Leipold, E.; Schmitz, T.; Kaufmann, D.; Tietze, D.; Heinemann, S.H.; Imhof, D. Effect of Conformational Diversity on the Bioactivity of Μ-Conotoxin PIIIA Disulfide Isomers. Mar. Drugs 2019, 17, 390. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep Venomics Reveals the Mechanism for Expanded Peptide Diversity in Cone Snail Venom. Mol. Cell. Proteomics 2013, 12, 312–329. [Google Scholar] [CrossRef]
- Gorson, J.; Holford, M. Small Packages, Big Returns: Uncovering the Venom Diversity of Small Invertebrate Conoidean Snails. Integr. Comp. Biol. 2016, 56, 962–972. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone Snails: A Big Store of Conotoxins for Novel Drug Discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef]
- Himaya, S.; Lewis, R. Venomics-Accelerated Cone Snail Venom Peptide Discovery. Int. J. Mol. Sci. 2018, 19, 788. [Google Scholar] [CrossRef] [PubMed]
- Koua, D.; Ebou, A.; Dutertre, S. Improved Prediction of Conopeptide Superfamilies with ConoDictor 2.0. Bioinform. Adv. 2021, 1, vbab011. [Google Scholar] [CrossRef] [PubMed]
- Espiritu, M.J.; Cabalteja, C.C.; Sugai, C.K.; Bingham, J.-P. Incorporation of Post-Translational Modified Amino Acids as an Approach to Increase Both Chemical and Biological Diversity of Conotoxins and Conopeptides. Amino Acids 2014, 46, 125–151. [Google Scholar] [CrossRef]
- Fu, Y.; Li, C.; Dong, S.; Wu, Y.; Zhangsun, D.; Luo, S. Discovery Methodology of Novel Conotoxins from Conus Species. Mar. Drugs 2018, 16, 417. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S. Enhancing the Therapeutic Potential of Peptide Toxins. Expert Opin. Drug Discov. 2017, 12, 611–623. [Google Scholar] [CrossRef]
- Bernáldez-Sarabia, J.; Figueroa-Montiel, A.; Dueñas, S.; Cervantes-Luévano, K.; Beltrán, J.; Ortiz, E.; Jiménez, S.; Possani, L.; Paniagua-Solís, J.; Gonzalez-Canudas, J.; et al. The Diversified O-Superfamily in Californiconus Californicus Presents a Conotoxin with Antimycobacterial Activity. Toxins 2019, 11, 128. [Google Scholar] [CrossRef]
- Ebou, A.; Koua, D.; Addablah, A.; Kakou-Ngazoa, S.; Dutertre, S. Combined Proteotranscriptomic-Based Strategy to Discover Novel Antimicrobial Peptides from Cone Snails. Biomedicines 2021, 9, 344. [Google Scholar] [CrossRef]
- Primon-Barros, M.; José Macedo, A. Animal Venom Peptides: Potential for New Antimicrobial Agents. Curr. Top. Med. Chem. 2017, 17, 1119–1156. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).