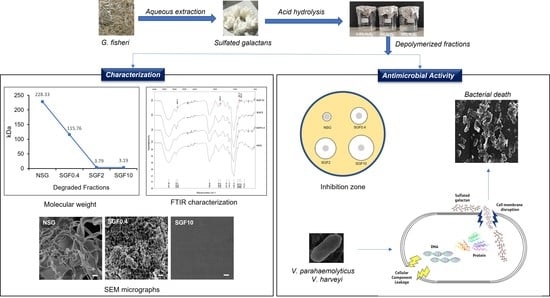
Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi
Abstract
:1. Introduction
2. Results
2.1. Physicochemical Properties of SGF
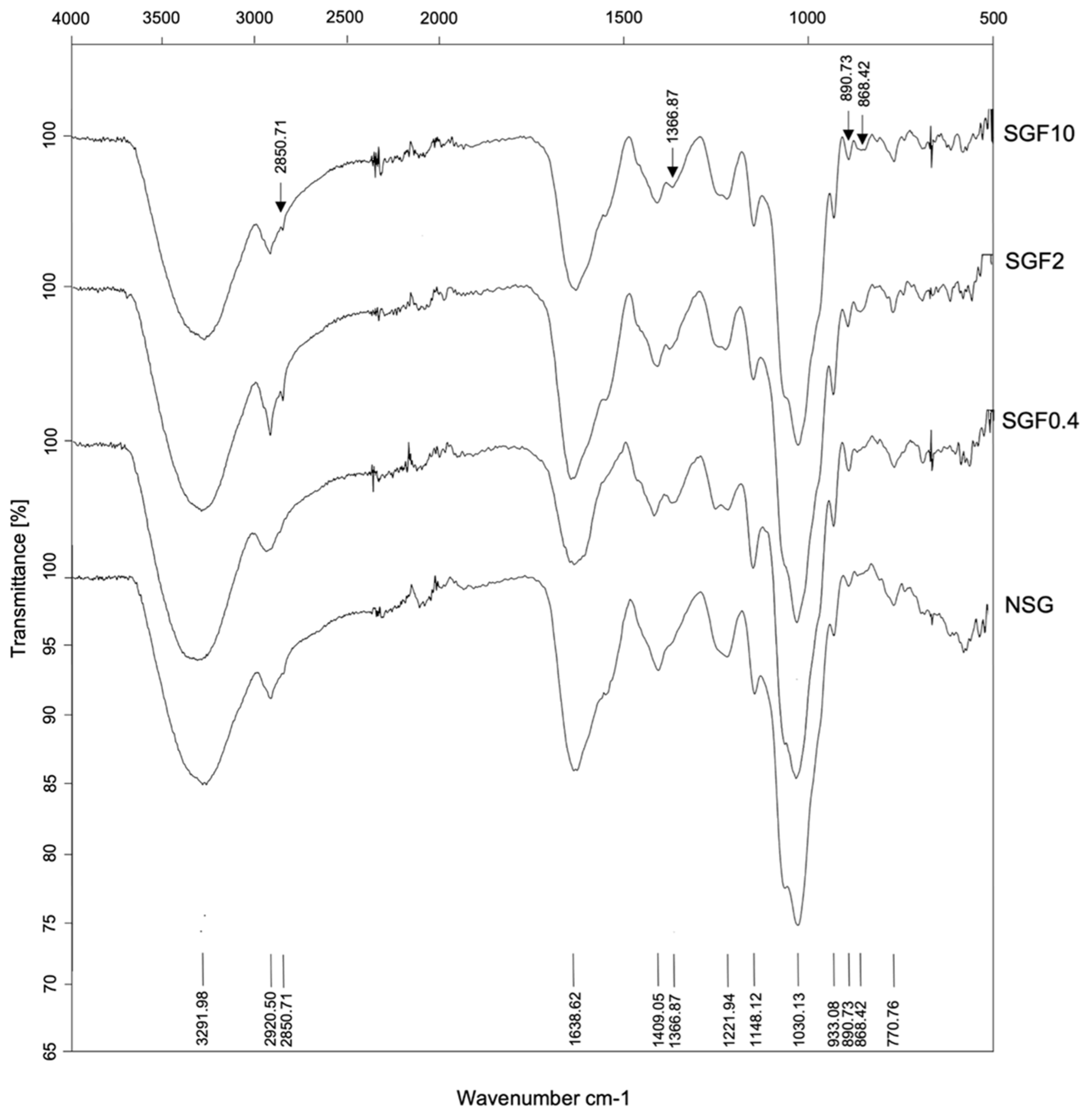
2.2. Structural Characterization and Morphology of SGFs
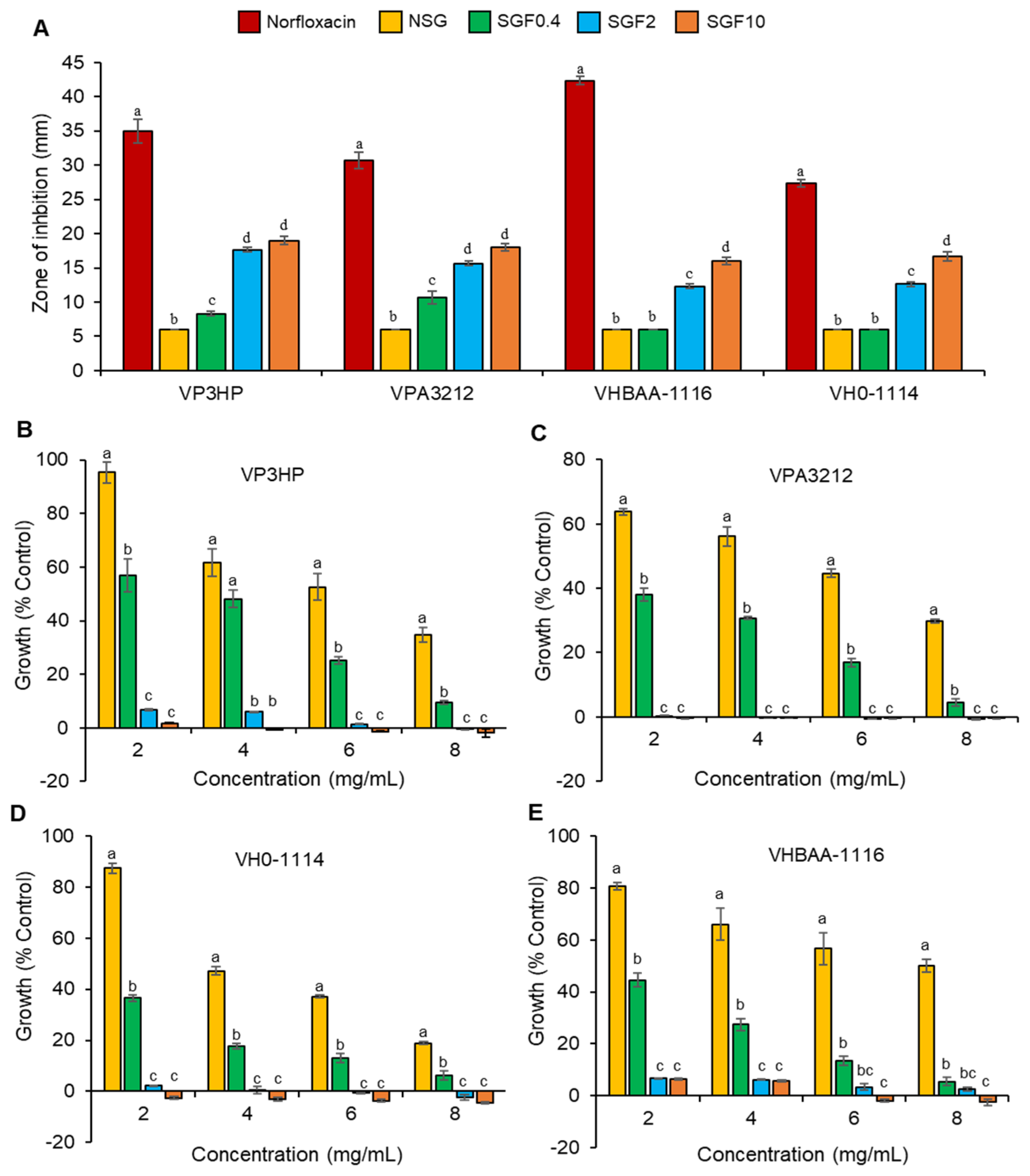
2.3. Degraded Fractions of SG Exhibited Greater Antimicrobial Activity
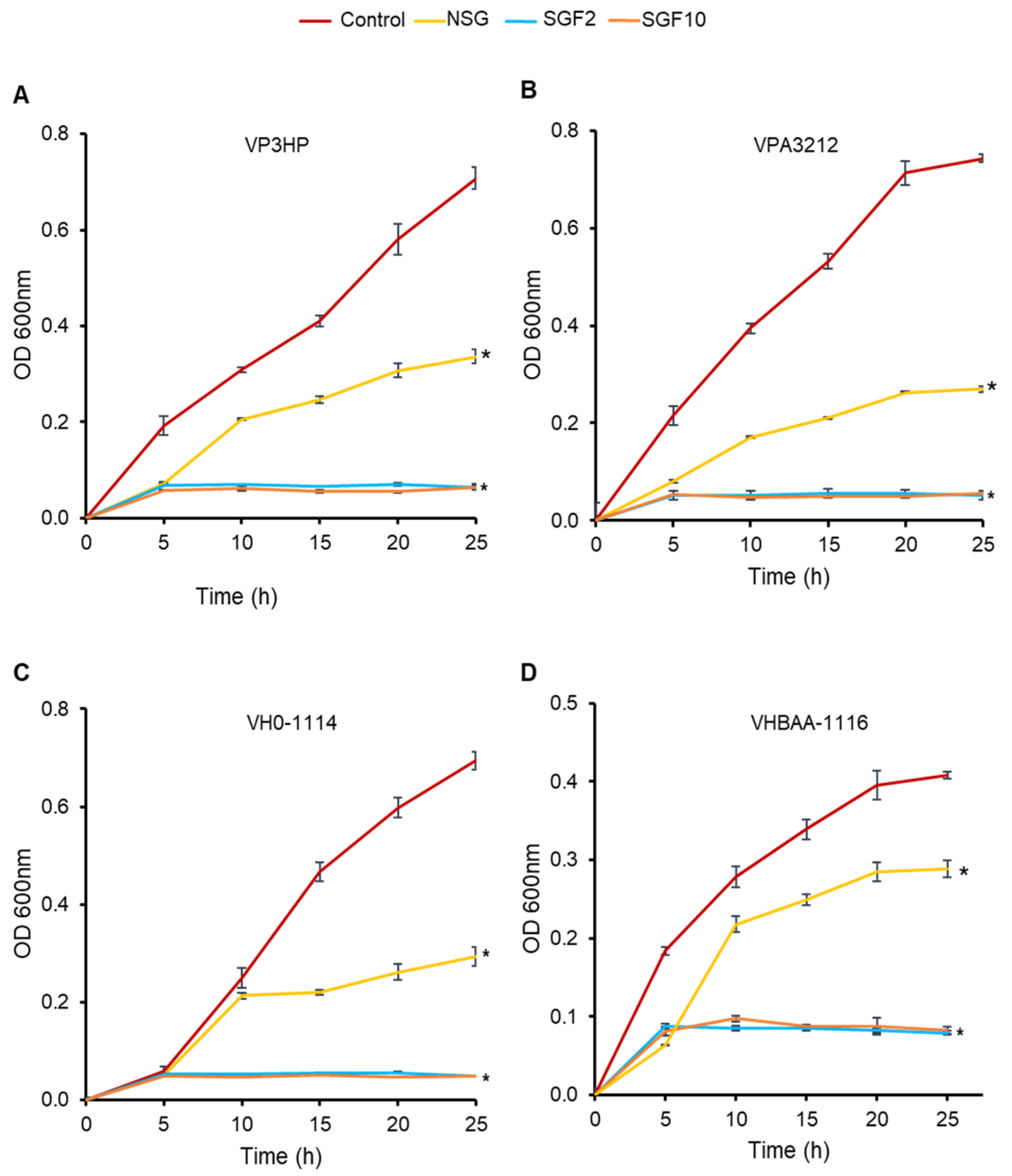
2.4. Degraded Fractions of SG Inhibited the Growth of V. parahaemolyticus and V. harveyi
2.5. Degraded Fractions of SG Disrupted the Cell Membrane of V. parahaemolyticus and V. harveyi
3. Discussion
4. Materials and Methods
4.1. Native Sulfated Galactans (NSG) and NSG Degradation
4.2. Sulfate and Carbohydrate Contents
4.3. Molecular Weight Determination
4.4. Fourier Transform Infrared (FTIR) Spectroscopic Analysis
4.5. Nuclear Magnetic Resonance (NMR) Spectroscopy Analysis
4.6. Bacterial Culture
4.7. Antibacterial Activity
4.8. MIC and MBC
4.9. Bacterial Growth Curve
4.10. Integrity of Cell Membrane
4.11. Scanning Electron Microscopy
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, B.K.; Deekshit, V.K.; Raj, J.R.M.; Rai, P.; Shivanagowda, B.M.; Karunasagar, I.; Karunasagar, I. Diversity of Vibrio parahaemolyticus associated with disease outbreak among cultured Litopenaeus vannamei (Pacific white shrimp) in India. Aquaculture 2014, 433, 247–251. [Google Scholar] [CrossRef]
- Joshi, J.; Srisala, J.; Truong, V.H.; Chen, I.-T.; Nuangsaeng, B.; Suthienkul, O.; Lo, C.F.; Flegel, T.W.; Sritunyalucksana, K.; Thitamadee, S. Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture 2014, 428, 297–302. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, F.; Wang, X.; Chen, H.; Wang, Q.; Guo, J.; Cao, X.; Wang, L. Antibacterial activity and action mechanism of questin from marine Aspergillus flavipes HN4-13 against aquatic pathogen Vibrio Harveyi. 3 Biotech 2019, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; Gregory, S.; Plummer, S.; Shields, R.; Rowley, A. An in vitro and in vivo assessment of the potential of Vibrio spp. as probiotics for the Pacific White shrimp, Litopenaeus vannamei. J. Appl. Microbiol. 2010, 109, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, R.; Balasundaram, C.; Heo, M.-S. Effect of probiotics enriched diet on Paralichthys olivaceus infected with lymphocystis disease virus (LCDV). Fish Shellfish Immunol. 2010, 29, 868–874. [Google Scholar] [CrossRef]
- Kamble, M.T.; Yakupitiyage, A.; Salin, K.R.; Chavan, B.R. Effect of Psidium guajava and Phyllanthus acidus leaf extract on immunostimulant response of Nile tilapia against Streptococcus agalactiae infection. Isr. J. Aquac.—Bamidgeh 2018, 70, 1–9. [Google Scholar] [CrossRef]
- Kamble, M.T.; Chavan, B.R.; Ataguba, G.; Azpeitia, T.; Medhe, S.; Jain, S.; Jadhav, R. Application of Moringa oleifera for development of Sustainable and Biosecure Aquaculture. Aquac. Indones. 2015, 15, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Karnjana, K.; Soowannayan, C.; Wongprasert, K. Ethanolic extract of red seaweed Gracilaria fisheri and furanone eradicate Vibrio harveyi and Vibrio parahaemolyticus biofilms and ameliorate the bacterial infection in shrimp. Fish Shellfish Immunol. 2019, 88, 91–101. [Google Scholar] [CrossRef]
- Soowannayan, C.; Teja, D.N.C.; Yatip, P.; Mazumder, F.Y.; Krataitong, K.; Unagul, P.; Suetrong, S.; Preedanon, S.; Klaysuban, A.; Sakayaroj, J. Vibrio biofilm inhibitors screened from marine fungi protect shrimp against acute hepatopancreatic necrosis disease (AHPND). Aquaculture 2019, 499, 1–8. [Google Scholar] [CrossRef]
- FAO. FAO Global Fishery and Aquaculture Production Statistics (FishStatJ); FAO: Rome, Italy, 2021. [Google Scholar]
- FAO. The Global Status of Seaweed Production, Trade and Utilization; FAO: Rome, Italy, 2018; Volume 124, p. 120. [Google Scholar]
- Phi, T.N. Cultivation Characteristics and Biological Responses of Agarophytic Seaweed, Gracilaria fisheri (Rhodophyta), in Southern Thailand; Prince of Songkla University, Pattani Campus: Songkla, Thailand, 2014. [Google Scholar]
- Chirapart, A.; Munkit, J.; Lewmanomont, K. Changes in yield and quality of agar from the agarophytes, Gracilaria fisheri and G. tenuistipitata var. liui cultivated in earthen ponds. Agric. Nat. Resour. 2006, 40, 529–540. [Google Scholar]
- Wongprasert, K.; Rudtanatip, T.; Praiboon, J. Immunostimulatory activity of sulfated galactans isolated from the red seaweed Gracilaria fisheri and development of resistance against white spot syndrome virus (WSSV) in shrimp. Fish Shellfish. Immunol. 2014, 36, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.; Santos, J.P.; Chow, F.; dos Santos, D.Y. A comprehensive review of traditional uses, bioactivity potential, and chemical diversity of the genus Gracilaria (Gracilariales, Rhodophyta). Algal Res. 2019, 37, 288–306. [Google Scholar] [CrossRef]
- Hentati, F.; Tounsi, L.; Djomdi, D.; Pierre, G.; Delattre, C.; Ursu, A.V.; Fendri, I.; Abdelkafi, S.; Michaud, P. Bioactive polysaccharides from seaweeds. Molecules 2020, 25, 3152. [Google Scholar] [CrossRef]
- Sun, L.; Chu, J.; Sun, Z.; Chen, L. Physicochemical properties, immunomodulation and antitumor activities of polysaccharide from Pavlova viridis. Life Sci. 2016, 144, 156–161. [Google Scholar] [CrossRef]
- Wu, G.-J.; Shiu, S.-M.; Hsieh, M.-C.; Tsai, G.-J. Anti-inflammatory activity of a sulfated polysaccharide from the brown alga Sargassum cristaefolium. Food Hydrocoll. 2016, 53, 16–23. [Google Scholar] [CrossRef]
- Li, S.; Xiong, Q.; Lai, X.; Li, X.; Wan, M.; Zhang, J.; Yan, Y.; Cao, M.; Lu, L.; Guan, J. Molecular modification of polysaccharides and resulting bioactivities. Compr. Rev. Food Sci. Food Saf. 2016, 15, 237–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Wang, J.; Jin, W.; Zhang, H.; Zhang, Q. Degradation of Laminaria japonica fucoidan by hydrogen peroxide and antioxidant activities of the degradation products of different molecular weights. Carbohydr. Polym. 2012, 87, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ayanda, O.S.; Nelana, S.M.; Naidoo, E.B. Ultrasonic degradation of aqueous phenolsulfonphthalein (PSP) in the presence of nano-Fe/H2O2. Ultrason. Sonochemistry 2018, 47, 29–35. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, R.; Li, Y.; Li, X.; You, L.; Kulikouskaya, V.; Hileuskaya, K. Degradation of polysaccharides from Sargassum fusiforme using UV/H2O2 and its effects on structural characteristics. Carbohydr. Polym. 2020, 230, 115647. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Kuo, C.-H.; Lee, C.-H. Antibacterial and antioxidant capacities and attenuation of lipid accumulation in 3T3-L1 adipocytes by low-molecular-weight fucoidans prepared from compressional-puffing-pretreated Sargassum crassifolium. Mar. Drugs 2018, 16, 24. [Google Scholar] [CrossRef] [Green Version]
- Shao, L.-L.; Xu, J.; Shi, M.-J.; Wang, X.-L.; Li, Y.-T.; Kong, L.-M.; Hider, R.C.; Zhou, T. Preparation, antioxidant and antimicrobial evaluation of hydroxamated degraded polysaccharides from Enteromorpha prolifera. Food Chem. 2017, 237, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Turki, A.; El Oudiani, A.; Msahli, S.; Sakli, F. Infrared spectra for alfa fibers treated with thymol. J. Glycobiol. 2018, 7, 8. [Google Scholar] [CrossRef]
- Rudtanatip, T.; Pariwatthanakun, C.; Somintara, S.; Sakeaw, W.; Wongprasert, K. Structural characterization, antioxidant activity, and protective effect against hydrogen peroxide-induced oxidative stress of chemically degraded Gracilaria fisheri sulfated galactans. Int. J. Biol. Macromol. 2022, 206, 51–63. [Google Scholar] [CrossRef]
- Souza, B.W.; Cerqueira, M.A.; Bourbon, A.I.; Pinheiro, A.C.; Martins, J.T.; Teixeira, J.A.; Coimbra, M.A.; Vicente, A.A. Chemical characterization and antioxidant activity of sulfated polysaccharide from the red seaweed Gracilaria birdiae. Food Hydrocoll. 2012, 27, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Ordóñez, E.; Rupérez, P. FTIR-ATR spectroscopy as a tool for polysaccharide identification in edible brown and red seaweeds. Food Hydrocoll. 2011, 25, 1514–1520. [Google Scholar] [CrossRef]
- Yuan, F.; Gao, Z.; Liu, W.; Li, H.; Zhang, Y.; Feng, Y.; Song, X.; Wang, W.; Zhang, J.; Huang, C.; et al. Characterization, antioxidant, anti-aging and organ protective effects of sulfated polysaccharides from Flammulina velutipes. Molecules 2019, 24, 3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciel, J.S.; Chaves, L.S.; Souza, B.W.S.; Teixeira, D.I.A.; Freitas, A.L.P.; Feitosa, J.P.A.; Paula, R.C.M. Structural characterization of cold extracted fraction of soluble sulphated polysaccharide from red seaweed Gracilaria birdiae. Carbohydr. Polym. 2008, 71, 559–565. [Google Scholar] [CrossRef]
- Sudharsan, S.S.; Giji, P.; Seedevi, S.; Vairamani, A. Shanmugam, Isolation, characterization and bioactive potential of sulfated galactans from Spyridia hypnoides (Bory) papenfuss. Int. J. Biol. Macromol. 2018, 109, 589–597. [Google Scholar] [CrossRef]
- Li, X.; Xu, A.; Xie, H.; Yu, W.; Xie, W.; Ma, X. Preparation of low molecular weight alginate by hydrogen peroxide depolymerization for tissue engineering. Carbohydr. Polym. 2010, 79, 660–664. [Google Scholar] [CrossRef]
- Najafabadi, S.A.A.; Honarkar, H.; Moghadam, M.; Mirkhani, V.; Tahriri, M.; Tayebi, L. UV irradiation-H2O2 system as an effective combined depolymerization technique to produce oligosaccharides from chitosan. Bio-Des. Manuf. 2018, 1, 62–68. [Google Scholar] [CrossRef]
- Li, B.; Liu, S.; Xing, R.; Li, K.; Li, R.; Qin, Y.; Wang, X.; Wei, Z.; Li, P. Degradation of sulfated polysaccharides from Enteromorpha prolifera and their antioxidant activities. Carbohydr. Polym. 2013, 92, 1991–1996. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Yu, K.; Sun, X.-Y.; Ouyang, J.-M. Structural characterization and repair mechanism of Gracilaria lemaneiformis sulfated polysaccharides of different molecular weights on damaged renal epithelial cells. Oxidative Med. Cell. Longev. 2018, 2018, 7410389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Chen, X.; Cheng, Z.; Liu, S.; Yu, H.; Wang, X.; Li, P. Degradation of polysaccharides from Grateloupia filicina and their antiviral activity to avian leucosis virus subgroup . J. Mar. Drugs 2017, 15, 345. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Liu, Y.; Hu, K.; Zhao, B. The depolymerization mechanism of chitosan by hydrogen peroxide. J. Mater. Sci. 2003, 38, 4709–4712. [Google Scholar] [CrossRef]
- Trabelsi, L.; M’sakni, N.H.; Ouada, H.B.; Bacha, H.; Roudesli, S. Partial characterization of extracellular polysaccharides produced by cyanobacterium Arthrospira platensis. Biotechnol. Bioprocess Eng. 2009, 14, 27–31. [Google Scholar] [CrossRef]
- Prasetyaningrum, A.; Purwati, D.; Dharmawan, Y.; Ratnawati, R.; Jos, B. Effect of H2O2 concentration on molecular weight and functional properties of sulfated polysaccharides from red seaweed (Kappaphycus alvarezii). In Proceedings of 2nd International Conference on Chemical Process and Product Engineering (ICCPPE) 2019. AIP Conf. Proc. 2020, 2197, 070002. [Google Scholar]
- Duarte, M.E.R.; Cauduro, J.P.; Noseda, D.G.; Noseda, M.D.; Goncalves, A.G.; Pujol, C.A.; Damonte, E.B.; Cerezo, A.S. The structure of the agaran sulfate from Acanthophora spicifera (Rhodomelaceae, Ceramiales) and its antiviral activity. Relation between structure and antiviral activity in agarans. Carbohydr. Polym. 2004, 339, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Rangel, A.; Sánchez-Polo, M.; Polo, A.; Rivera-Utrilla, J.; Berber-Mendoza, M. Sulfonamides degradation assisted by UV, UV/H2O2 and UV/K2S2O8: Efficiency, mechanism and byproducts cytotoxicity. J. Environ. Manag. 2018, 225, 224–231. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Cao, M.-J.; Liu, G.-M.; Chen, Q.; Sun, L.; Chen, H. Antibacterial activity and mechanisms of depolymerized fucoidans isolated from Laminaria japonica. Carbohydr. Polym. 2017, 172, 294–305. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, B.; Tang, C.; Gou, Y.; Chen, H.; Wang, Y.; Jin, C.; Liu, J.; Niu, F.; Kan, J. Characterization, antioxidant activity and hepatoprotective effect of purple sweet potato polysaccharides. Int. J. Biol. Macromol. 2018, 115, 69–76. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, W.; Wang, Y.; Ma, Y.; Huang, L.; Zou, C.; Li, D.; Cao, M.-J.; Liu, G.-M. Inhibitory effect of depolymerized sulfated galactans from marine red algae on the growth and adhesion of diarrheagenic Escherichia Coli. Mar. Drugs 2019, 17, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palanisamy, S.; Vinosha, M.; Rajasekar, P.; Anjali, R.; Sathiyaraj, G.; Marudhupandi, T.; Selvam, S.; Prabhu, N.M.; You, S. Antibacterial efficacy of a fucoidan fraction (Fu-F2) extracted from Sargassum polycystum. Int. J. Biol. Macromol. 2019, 125, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-C.; Lee, H.; Choi, H.-D.; Jeon, Y.-J. Antioxidant properties of a sulfated polysaccharide isolated from an enzymatic digest of Sargassum thunbergii. Int. J. Biol. Macromol. 2019, 132, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.B.; Nagai, A.; Jara, C.E.P.; Santos, J.P.; Chow, F.; Santos, D.Y.A.C.d. Determination of sulfate in algal polysaccharide samples: A step-by-step protocol using microplate reader. Ocean Coast. Res. 2021, 69, 4. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, Q.; Qi, H.; Zhang, H.; Niu, X.; Xu, Z.; Li, Z. Degradation of porphyran from Porphyra haitanensis and the antioxidant activities of the degraded porphyrans with different molecular weight. Int. J. Biol. Macromol. 2006, 38, 45–50. [Google Scholar] [CrossRef]
- Kamble, M.T.; Gallardo, W.; Yakupitiyage, A.; Chavan, B.R.; Rusydi, R.; Rahma, A. Antimicrobial activity of bioactive herbal extracts against Streptococcus agalactiae biotype 2. Int. J. Basic Appl. Biol. 2014, 2, 152–155. [Google Scholar]
- Kolsi, R.B.A.; Gargouri, B.; Sassi, S.; Frikha, D.; Lassoued, S.; Belghith, K. In vitro biological properties and health benefits of a novel sulfated polysaccharide isolated from Cymodocea nodosa. Lipids Health Dis. 2017, 16, 252. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Baek, K.-H. Antibacterial activity and action mechanism of the essential oil from Enteromorpha linza L. against foodborne pathogenic bacteria. Molecules 2016, 21, 388. [Google Scholar] [CrossRef] [Green Version]





| Treatment | H2O2 Concentration (%) | MW (kDa) | PD | pH | Carbohydrate Content (%) | Sulfate Content (%) | DS |
|---|---|---|---|---|---|---|---|
| NSG | 0 | 228.33 a | 1.29 | 6 | 66.95 ± 0.72 a | 9.95 ± 0.19 a | 0.74 ± 0.02 a |
| SGF0.4 | 0.4 | 115.76 b | 1.08 | 5 | 62.88 ± 1.17 a | 12.02 ± 0.12 b | 0.99 ± 0.02 b |
| SGF2 | 2 | 3.79 b | 1.04 | 4 | 58.97 ± 0.61 a | 12.28 ± 0.92 b | 1.03 ± 0.06 b |
| SGF10 | 10 | 3.19 b | 1.19 | 4 | 58.40±3.38 a | 12.33 ± 0.51 b | 1.03 ± 0.06 b |
| Functional Group Characteristic Vibration (Wavenumber cm−1) | Fraction | |||
|---|---|---|---|---|
| NSG | SGF0.4 | SGF2 | SGF10 | |
| OH group (3291.98) | ✓ | ✓ | ✓ | ✓ |
| Asymmetric CH group (2920.05) | ✓ | ✓ | (+) | ✓ |
| Symmetric CH2 group (2850.71) | Non | Non | ✓ | ✓ |
| COOH of carboxylate group (1638.62) | ✓ | (-) | ✓ | (-) |
| C=O of carboxylate group (1409.05) | ✓ | (-) | (-) | (-) |
| Sulfate ester (1366.87) | Non | ✓ | ✓ | ✓ |
| S=O of sulfate group (1221.49) | ✓ | (-) | (-) | (-) |
| C-O of pyranose ring (1148.12) | ✓ | (-) | (-) | (-) |
| C-C of pyranose ring (1030.13) | ✓ | ✓ | ✓ | (-) |
| C-O-C of 3,6-anhydro-L-galactose (933.08) | ✓ | (+) | (+) | (+) |
| L-galactose-6 sulfate (890.73) | ✓ | (-) | (+) | (+) |
| D-galactose-4 sulfate (857.54) | ✓ | (-) | (+) | (+) |
| C-O-S of sulfate group (770.76) | ✓ | ✓ | ✓ | ✓ |
| Bacteria | VP3HP | VPA3212 | VH0-1114 | VHBAA-1116 | ||||
|---|---|---|---|---|---|---|---|---|
| Fraction | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MIC |
| NSG | 20 | 25 | 20 | 25 | 20 | 25 | 20 | 20 |
| SGF0.4 | 15 | 25 | 15 | 25 | 15 | 25 | 15 | 15 |
| SGF2 | 5 | 5 | 2.5 | 10 | 2.5 | 2.5 | 10 | 10 |
| SGF10 | 2.5 | 2.5 | 1.25 | 1.25 | 1.25 | 1.25 | 5 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamble, M.T.; Rudtanatip, T.; Soowannayan, C.; Nambunruang, B.; Medhe, S.V.; Wongprasert, K. Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi. Mar. Drugs 2022, 20, 469. https://doi.org/10.3390/md20080469
Kamble MT, Rudtanatip T, Soowannayan C, Nambunruang B, Medhe SV, Wongprasert K. Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi. Marine Drugs. 2022; 20(8):469. https://doi.org/10.3390/md20080469
Chicago/Turabian StyleKamble, Manoj Tukaram, Tawut Rudtanatip, Chumporn Soowannayan, Boottoh Nambunruang, Seema Vijay Medhe, and Kanokpan Wongprasert. 2022. "Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi" Marine Drugs 20, no. 8: 469. https://doi.org/10.3390/md20080469
APA StyleKamble, M. T., Rudtanatip, T., Soowannayan, C., Nambunruang, B., Medhe, S. V., & Wongprasert, K. (2022). Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi. Marine Drugs, 20(8), 469. https://doi.org/10.3390/md20080469