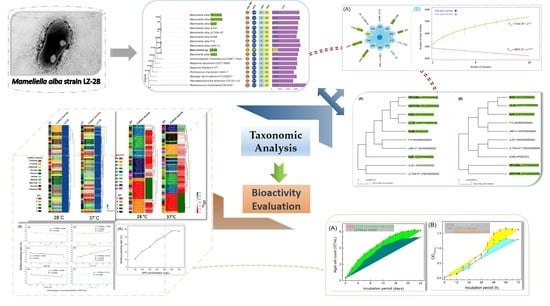
Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09
Abstract
:1. Introduction
2. Results and Discussion
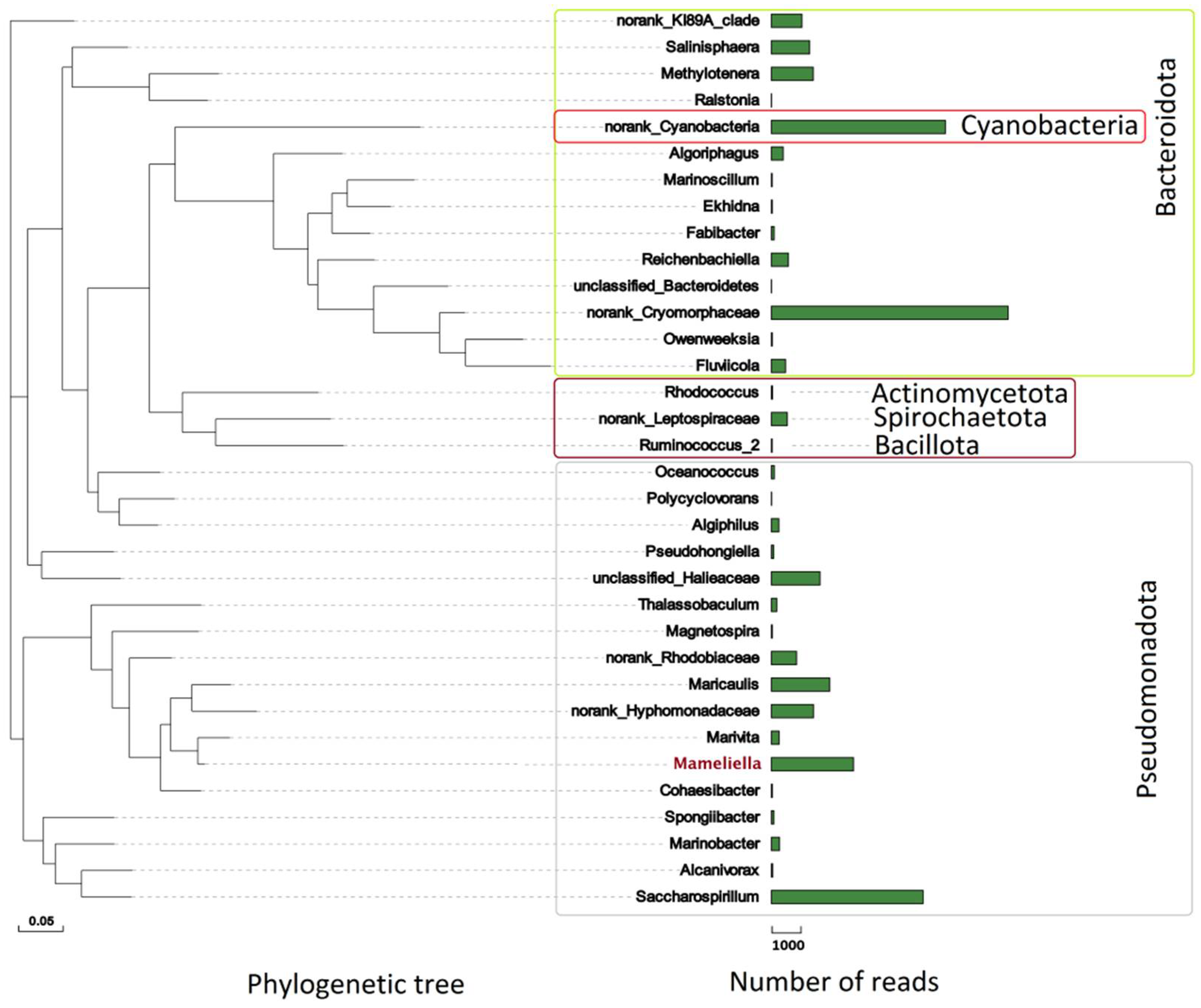
2.1. Characterization of the Composition of the Bacterial Community of Algal Strain LZT09
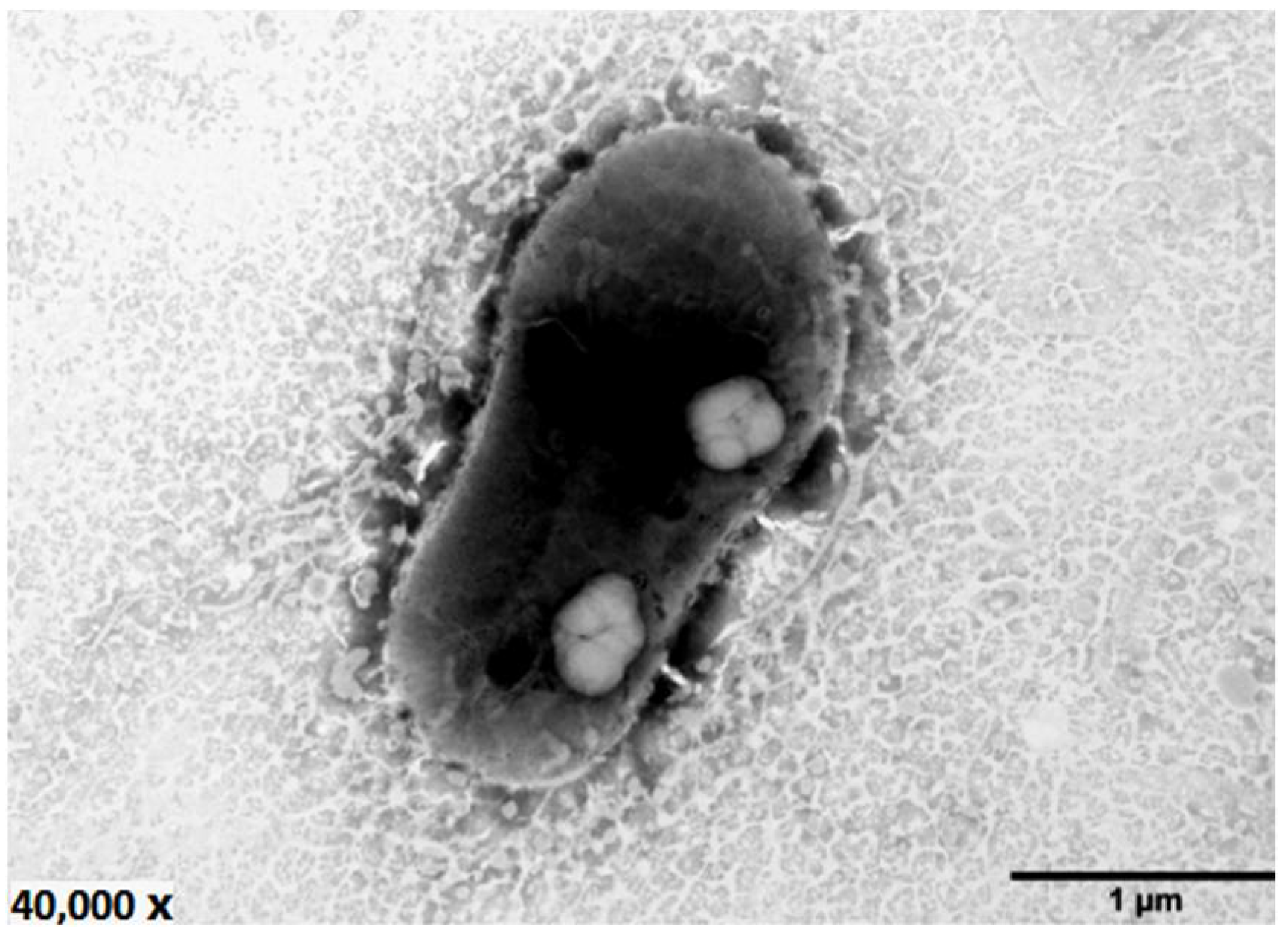
2.2. Phenotypic and Biochemical Characteristics of Bacterial Strain LZ-28
2.3. Phylogenetic Analysis Based on 16S rRNA Gene Sequences
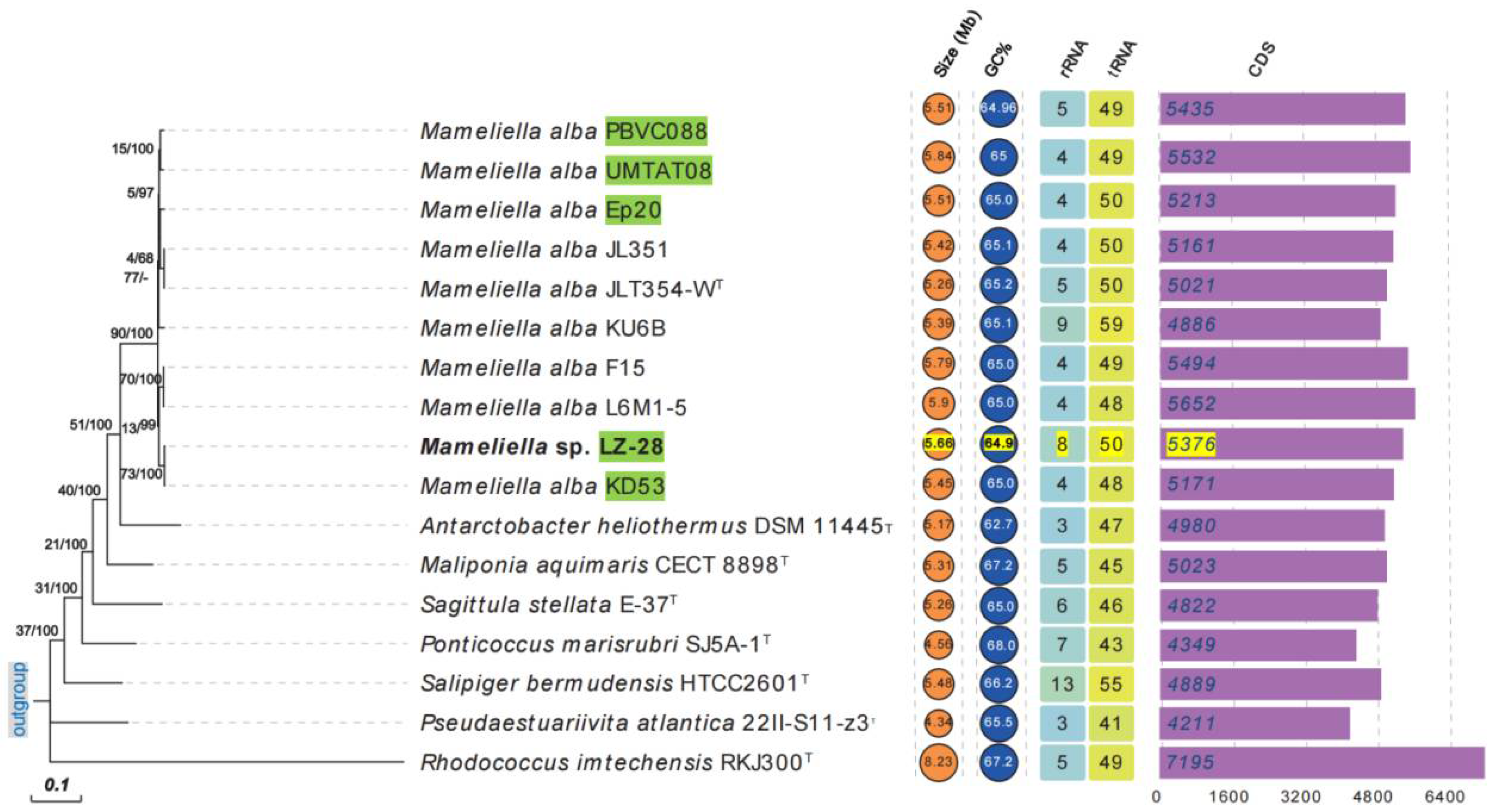
2.4. Genomic Features of the Selected M. alba Strains
2.5. Phylogenomic Characterization of Bacterial Strain LZ-28
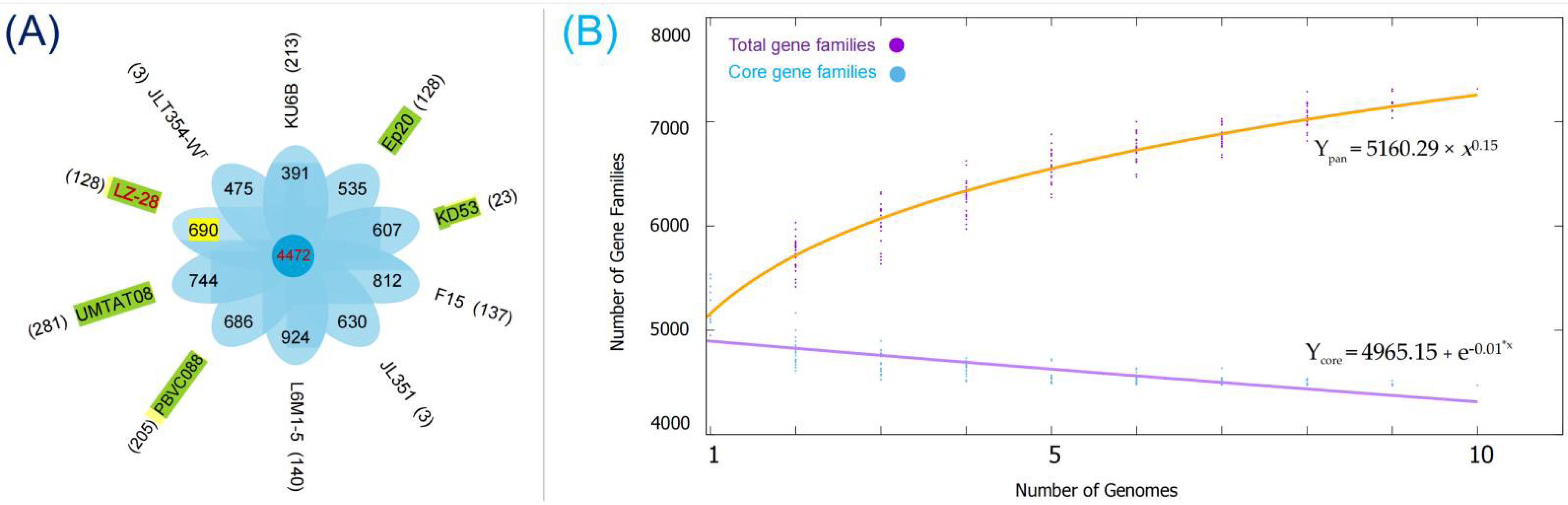
2.6. Comparison of the Core- and Pan-Genomic Profiles among the Selected M. alba Strains
2.7. Comparison of the Functional Classes of Predicted Genes among the Selected M. alba Strains
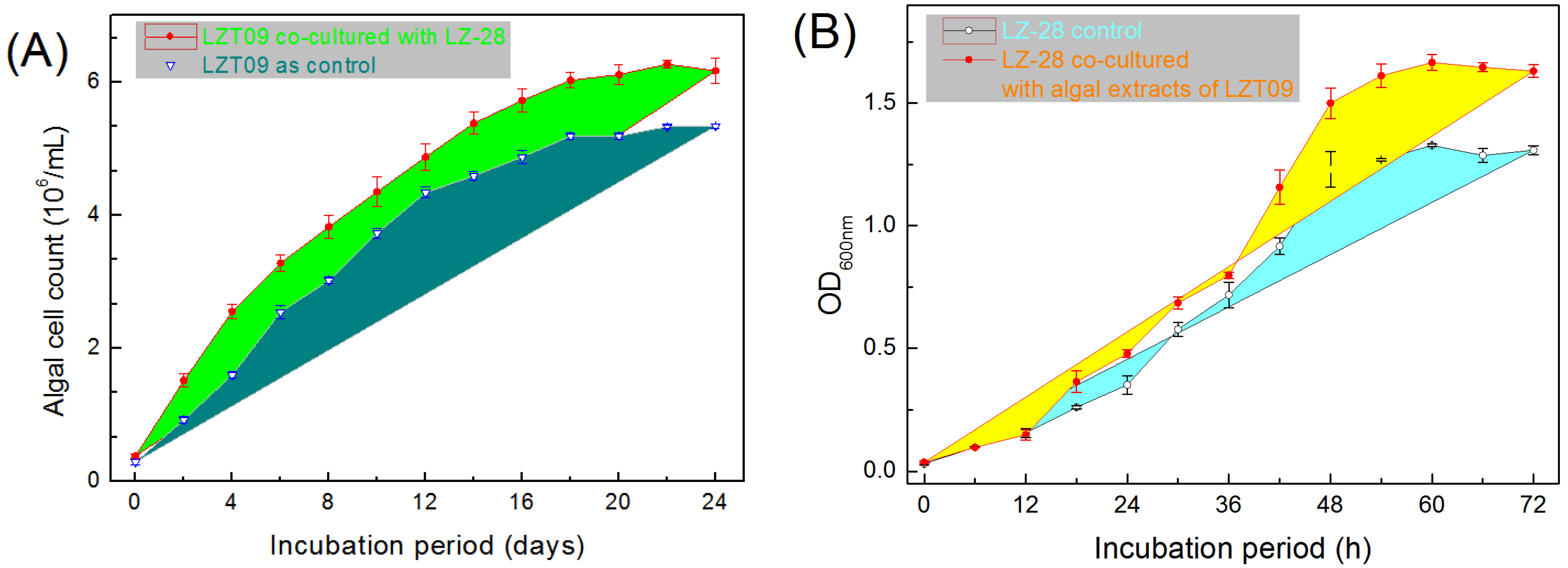
2.8. Growth-Promoting Effects of Bacterial Strain LZ-28 and Algal Strain LZT09
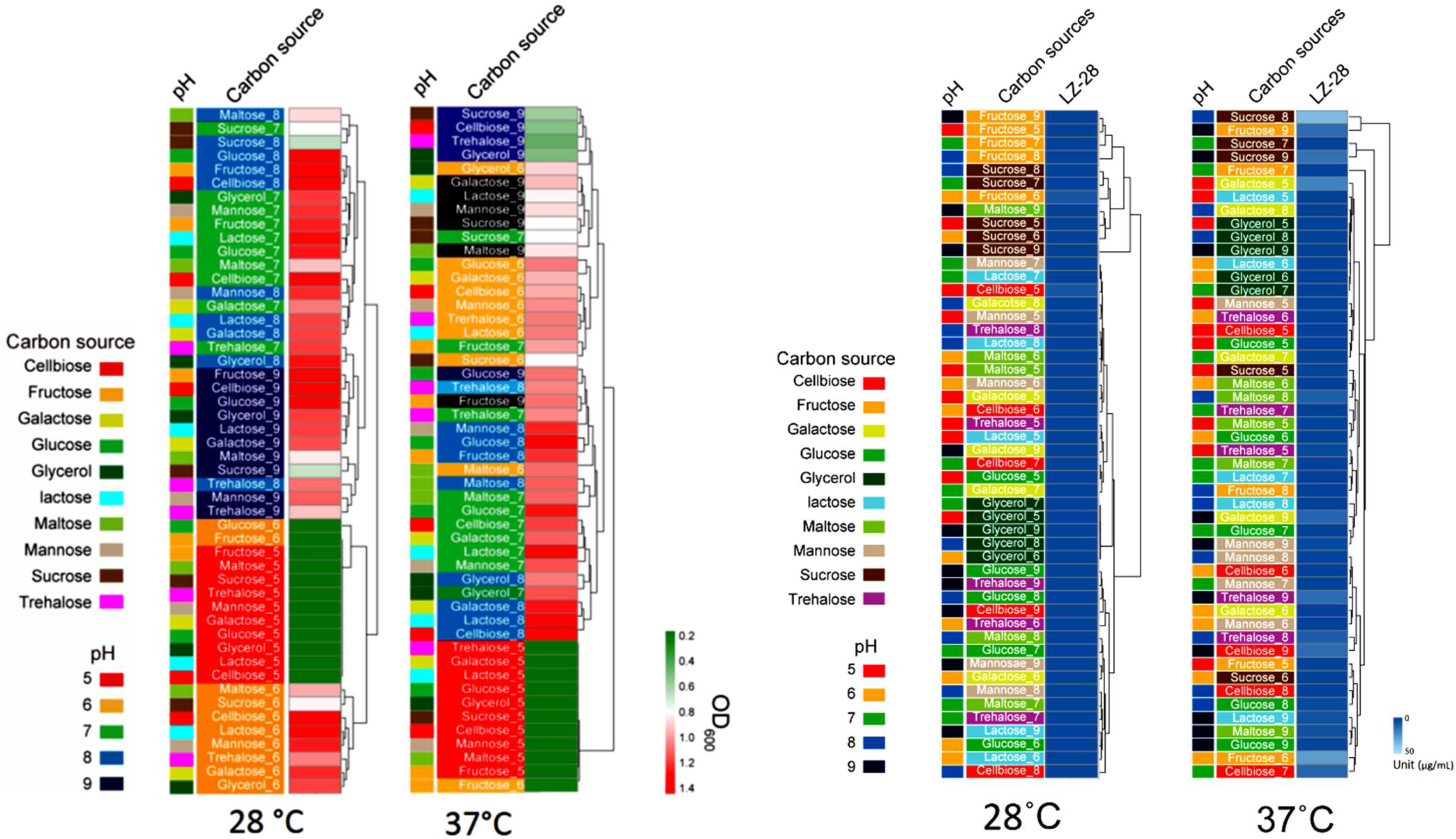
2.9. Optimization of Bacterial Growth and EPS Accumulation
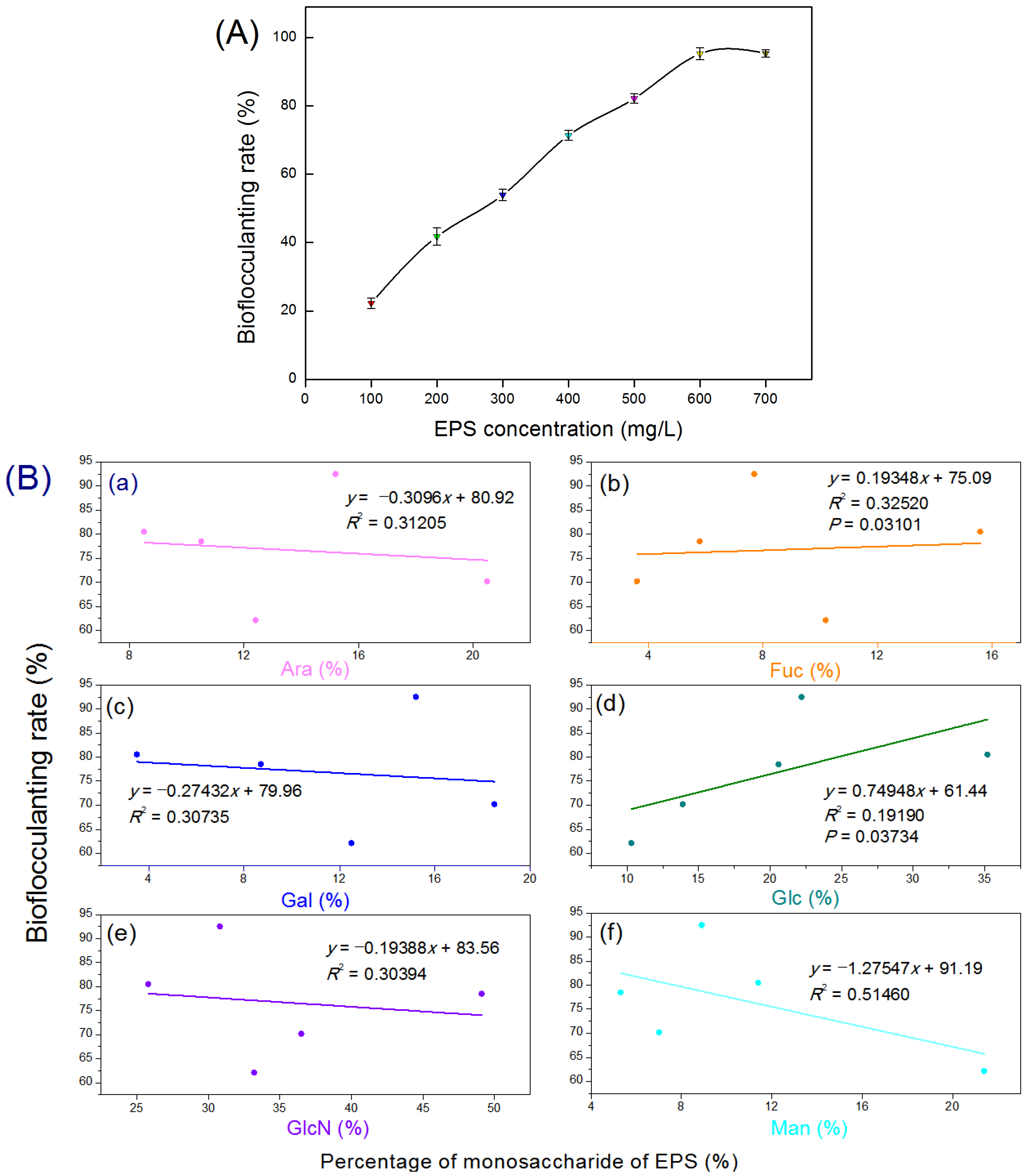
2.10. Bioflocculanting-Activity Evaluation and Correlation Analysis
3. Materials and Methods
3.1. Algal and Bacterial Strains and Culture
3.2. PCR Amplification, 16S rRNA Sequencing, and Data Analysis
3.3. Morpho-Physiological and Biochemical Characterizations
3.4. Analysis of Fatty Acid Profiles
3.5. Phylogenetic Analysis Based on the 16S rRNA Gene Sequences
3.6. Phylogenomic Calculations and UBCG Tree Construction
3.7. Pan-Genome Analysis
3.8. Comparative Analysis of Functional Genes
3.9. Bacterial Growth and EPS-Accumulation Analysis
3.10. Characterization of the Monosaccharides of EPSs
3.11. Evaluation of Bioflocculanting and MGP Bioactivities
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wayne, B.; Ralph, M. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 1972, 143, 265–277. [Google Scholar]
- Samo, T.J.; Kimbrel, J.A.; Nilson, D.J.; Pett-Ridge, J.; Weber, P.K.; Mayali, X. Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environ. Microbiol. 2018, 20, 4385–4400. [Google Scholar] [CrossRef] [PubMed]
- Seymour, J.R.; Amin, S.A.; Raina, J.B.; Stocker, R. Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, R.; Kang, Z.; Kim, B.H.; Cho, D.H.; Jin, L.; Oh, H.M.; Kim, H.S. Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. 2015, 8, 140–144. [Google Scholar] [CrossRef]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouzuma, A.; Watanabe, K. Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotechnol. 2015, 33, 125–129. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Nemergut, D. Microbes ride the current. Science 2014, 345, 1246. [Google Scholar] [CrossRef] [PubMed]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef] [Green Version]
- Landry, Z.C.; Vergin, K.; Mannenbach, C.; Block, S.; Yang, Q.; Blainey, P.; Carlson, C.; Giovannoni, S. Optofluidic Single-Cell Genome Amplification of Sub-micron Bacteria in the Ocean Subsurface. Front. Microbiol. 2018, 9, 1152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.L.; Yang, X.; Wang, S.J.; Jiang, Z.W.; Xie, Z.X.; Zhang, L. Draft Genome Sequences of Nine Cultivable Heterotrophic Proteobacteria Isolated from Phycosphere Microbiota of Toxic Alexandrium catenella LZT09. Microbiol. Resour. Announc. 2020, 9, e00281-20. [Google Scholar] [CrossRef]
- Zheng, Q.; Chen, C.; Yan, X.J.; Wang, Y.N.; Zeng, Y.H.; Hao, L.K.; He, W.H.; Jiao, N.Z. Mameliella alba gen. nov., sp. nov., a marine bacterium of the Roseobacter clade in the order Rhodobacterales. Int. J. Syst. Evol. Microbiol. 2010, 60, 953–957. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, J.; Lei, X.; Lai, Q.; Yang, L.; Zhang, H.; Li, Y.; Zheng, W.; Tian, Y.; Yu, Z.; et al. Mameliella phaeodactyli sp. nov., a member of the family Rhodobacteraceae isolated from the marine algae Phaeodactylum tricornutum. Int. J. Syst. Evol. Microbiol. 2015, 65, 1617–1621. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, X.; Lai, Q.; Shao, Z. Reclassification of Mameliella phaeodactyli, Mameliella atlantica, Ponticoccus lacteus and Alkalimicrobium pacificum as later heterotypic synonyms of Mameliella alba and an emended description of Mameliella alba. Int. J. Syst. Evol. Microbiol. 2018, 68, 1047–1051. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Andersen, A.J.; Andersen, N.G.; Nielsen, K.F.; Hansen, P.J.; Larsen, T.O. Chemical Diversity, Origin, and Analysis of Phycotoxins. J. Nat. Prod. 2016, 79, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Brosnahan, M.L.; Fischer, A.D.; Lopez, C.B.; Moore, S.K.; Anderson, D.M. Cyst-forming dinoflagellates in a warming climate. Harmful Algae 2020, 91, 101728. [Google Scholar] [CrossRef]
- McKenzie, C.H.; Bates, S.S.; Martin, J.L.; Haigh, N.; Howland, K.L.; Lewis, N.I.; Locke, A.; Peña, A.; Poulin, M.; Rochon, A.; et al. Three decades of Canadian marine harmful algal events: Phytoplankton and phycotoxins of concern to human and ecosystem health. Harmful Algae 2021, 102, 101852. [Google Scholar] [CrossRef]
- Lewis, A.M.; Coates, L.N.; Turner, A.D.; Percy, L.; Lewis, J. A review of the global distribution of Alexandrium minutum (Dinophyceae) and comments on ecology and associated paralytic shellfish toxin profiles, with a focus on Northern Europe. J. Phycol. 2018, 54, 581–598. [Google Scholar] [CrossRef]
- Thottumkara, A.P.; Parsons, W.H.; Du, B.J. Saxitoxin. Angew. Chem. Int. Ed. Engl. 2014, 53, 5760–5784. [Google Scholar] [CrossRef]
- Zhang, X.L.; Tian, X.Q.; Ma, L.Y.; Feng, B.; Liu, Q.H.; Yuan, L.D. Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. J. Biosci. Med. 2015, 3, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.L.; Ma, L.Y.; Tian, X.Q.; Huang, H.L.; Yang, Q. Biodiversity study of intracellular bacteria closely associated with paralytic shellfish poisoning dinoflagellates Alexandrium tamarense and A. minutum. Int. J. Environ. Resour. 2015, 4, 23–27. [Google Scholar] [CrossRef]
- Yang, Q.; Feng, Q.; Zhang, B.P.; Gao, J.J.; Sheng, Z.; Xue, Q.P.; Zhang, X.L. Marinobacter alexandrii sp. nov., a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie Leeuwenhoek 2021, 114, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Duan, Y.; Yang, X.; Yao, B.; Zeng, T.; Wang, X.; Feng, Q.; Qi, M.; Yang, Q.; Zhang, X.L. Nitratireductor alexandrii sp. nov., from phycosphere microbiota of toxic marine dinoflagellate Alexandrium tamarense. Int. J. Syst. Evol. Microbiol. 2020, 70, 4390–4397. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Xie, Z.; Zhang, S.; Wu, Y.; Ge, Y.; Zhang, X. Haliea alexandrii sp. nov., isolated from phycosphere microbiota of the toxin-producing dinoflagellate Alexandrium catenella. Int. J. Syst. Evol. Microbiol. 2020, 70, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jiang, Z.W.; Zhang, J.; Zhou, X.; Zhang, X.L.; Wang, L.; Yu, T.; Wang, Z.; Bei, J.; Dong, B. Mesorhizobium alexandrii sp. nov., isolated from phycosphere microbiota of PSTs-producing marine dinoflagellate Alexandrium minutum amtk4. Antonie Leeuwenhoek 2020, 113, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Z.W.; Huang, C.H.; Zhang, R.N.; Li, L.Z.; Yang, G.; Feng, L.J.; Yang, G.F.; Zhang, H.; Zhang, X.L. Hoeflea prorocentri sp. nov., isolated from a culture of the marine dinoflagellate Prorocentrum mexicanum PM01. Antonie Leeuwenhoek 2018, 111, 1845–1853. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Li, G.X.; Ge, Y.M.; Iqbal, N.M.; Yang, X.; Cui, Z.D.; Yang, Q. Sphingopyxis microcysteis sp. nov., a novel bioactive exopolysaccharides-bearing Sphingomonadaceae isolated from the Microcystis phycosphere. Antonie Leeuwenhoek 2021, 114, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.Z.; Ge, Y.M.; Dai, J.; Zhang, X.L.; Yang, Q. Alexandriicola marinus gen. nov., sp. nov., a new member of the family Rhodobacteraceae isolated from marine phycosphere. Antonie Leeuwenhoek 2022, 115, 473–486. [Google Scholar] [CrossRef]
- Duan, Y.; Jiang, Z.; Wu, Z.; Sheng, Z.; Yang, X.; Sun, J.; Zhang, X.; Yang, Q.; Yu, X.; Yan, J. Limnobacter alexandrii sp. nov., a thiosulfate-oxidizing, heterotrophic and EPS-bearing Burkholderiaceae isolated from cultivable phycosphere microbiota of toxic Alexandrium catenella LZT09. Antonie Leeuwenhoek 2020, 13, 1689–1698. [Google Scholar] [CrossRef]
- Yang, Q.; Ge, Y.M.; Iqbal, N.M.; Yang, X.; Zhang, X.L. Sulfitobacter alexandrii sp. nov., a new microalgae growth-promoting bacterium with exopolysaccharides bioflocculanting potential isolated from marine phycosphere. Antonie Leeuwenhoek 2021, 114, 1091–1106. [Google Scholar] [CrossRef]
- Zhang, X.L.; Qi, M.; Li, Q.H.; Cui, Z.D.; Yang, Q. Maricaulis alexandrii sp. nov., a novel active bioflocculants-bearing and dimorphic prosthecate bacterium isolated from marine phycosphere. Antonie Leeuwenhoek 2021, 114, 1195–1203. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Wu, Y.; Lou, L.; Ma, Z.; Wang, D.; Ge, Y.; Zhang, X.; et al. Nioella ostreopsis sp. nov., isolated from toxic dinoflagellate, Ostreopsis lenticularis. Int. J. Syst. Evol. Microbiol. 2020, 70, 759–765. [Google Scholar] [CrossRef]
- Simon, M.; Scheuner, C.; Meier-Kolthoff, J.P.; Brinkhoff, T.; Wagner-Döbler, I.; Ulbrich, M.; Klenk, H.P.; Schomburg, D.; Petersen, J.; Göker, M. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017, 11, 1483–1499. [Google Scholar] [CrossRef]
- Attar, N. Marine microbiology. An interkingdom partnership. Nat. Rev. Microbiol. 2015, 13, 400. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, K.; Shen, L.; Chen, H.; Hou, F.; Zhou, X.; Zhang, D.; Zhu, X. Microbial Community Dynamics and Assembly Follow Trajectories of an Early-Spring Diatom Bloom in a Semienclosed Bay. Appl. Environ. Microbiol. 2018, 84, e01000-18. [Google Scholar] [CrossRef] [Green Version]
- Shibl, A.A.; Isaac, A.; Ochsenkühn, M.A.; Cárdenas, A.; Fei, C.; Behringer, G.; Arnoux, M.; Drou, N.; Santos, M.P.; Gunsalus, K.C.; et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. USA 2020, 117, 27445–27455. [Google Scholar] [CrossRef]
- Bowman, J.P. Out from the Shadows—Resolution of the Taxonomy of the Family Cryomorphaceae. Front. Microbiol. 2020, 11, 795. [Google Scholar] [CrossRef]
- Nakayama, T.; Nomura, M.; Takano, Y.; Tanifuji, G.; Shiba, K.; Inaba, K.; Inagaki, Y.; Kawata, M. Single-cell genomics unveiled a cryptic cyanobacterial lineage with a worldwide distribution hidden by a dinoflagellate host. Proc. Natl. Acad. Sci. USA 2019, 116, 15973–15978. [Google Scholar] [CrossRef] [Green Version]
- Giovannoni, S.J.; Stingl, U. The importance of culturing bacterioplankton in the ‘omics’ age. Nat. Rev. Microb. 2007, 5, 820–826. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Xu, H.; Jiang, L.; Li, S.; Zeng, X.; Shao, Z. Mameliella atlantica sp. nov., a marine bacterium of the Roseobacter clade isolated from deep-sea sediment of the South Atlantic Ocean. Int. J. Syst. Evol. Microbiol. 2015, 65, 2255–2259. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, J.; Tang, K.; Lin, D.; Li, C.; Lin, Y. Ponticoccus lacteus sp. nov. of the family Rhodobacteraceae, isolated from surface seawater. Int. J. Syst. Evol. Microbiol. 2015, 65, 1247–1250. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Y.; Wang, S.; Sun, Z.; Jiao, K. Alkalimicrobium pacificum gen. nov., sp. nov., a marine bacterium in the family Rhodobacteraceae. Int. J. Syst. Evol. Microbiol. 2015, 65, 2453–2458. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species defnition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Liu, Y.; Hasegawa, Y.; Iwaki, H. Complete Genome Sequence of Mameliella alba Strain KU6B, a Cyclohexylamine-Utilizing Marine Bacterium. Microbiol. Resour. Announc. 2020, 9, e00273-20. [Google Scholar] [CrossRef]
- Polz, M.F.; Alm, E.J.; Hanage, W.P. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013, 29, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Filipski, A.J.; Battistuzzi, F.U.; Kosakovsky Pond, S.L.; Tamura, K. Statistics and truth in phylogenomics. Mol. Biol. Evol. 2012, 29, 457–472. [Google Scholar] [CrossRef] [Green Version]
- Danish-Daniel, M.; Han, M.G.; Noor, M.E.; Yeong, Y.S.; Usup, G. Draft genome sequence of Mameliella alba strain UMTAT08 isolated from clonal culture of toxic dinoflagellate Alexandrium tamiyavanichii. Genom. Data 2016, 10, 12–14. [Google Scholar] [CrossRef] [Green Version]
- Geng, H.; Belas, R. Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr. Opin. Biotechnol. 2010, 21, 332–338. [Google Scholar] [CrossRef]
- Gao, H.M.; Xie, P.F.; Zhang, X.L.; Yang, Q. Isolation, Phylogenetic and Gephyromycin Metabolites Characterization of New Exopolysaccharides-Bearing Antarctic Actinobacterium from Feces of Emperor Penguin. Mar. Drugs 2021, 19, 458. [Google Scholar] [CrossRef]
- Gutleben, J.; Chaib De Mares, M.; van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: From sequence to microbial isolate. Crit. Rev. Microbiol. 2018, 44, 212–229. [Google Scholar] [CrossRef] [Green Version]
- Stingl, U.; Tripp, H.J.; Giovannoni, S.J. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 2007, 1, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Stingl, U.; Cho, J.C.; Foo, W.; Vergin, K.L.; Lanoil, B.; Giovannoni, S.J. Dilution-to-extinction culturing of psychrotolerant planktonic bacteria from permanently ice-covered lakes in the McMurdo Dry Valleys, Antarctica. Microb. Ecol. 2007, 55, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Hmelo, L.R.; van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.Z.; Ge, Y.M.; Gao, H.M.; Dai, J.; Zhang, X.L.; Yang, Q. Gephyromycinifex aptenodytis gen. nov., sp. nov., isolated from gut of Antarctic emperor penguin. Antonie Leeuwenhoek 2021, 114, 2003–2017. [Google Scholar] [CrossRef]
- Zhu, W.Z.; Wang, S.H.; Gao, H.M.; Ge, Y.M.; Dai, J.; Zhang, X.L.; Yang, Q. Characterization of Bioactivities and Biosynthesis of Angucycline/angucyclinone Derivatives Derived from Gephyromycinifex aptenodytis gen. nov., sp. nov. Mar. Drugs 2022, 20, 34. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Xie, Z.; Wang, Y.; Wang, D.; Feng, L.; Yang, G.; Ge, Y.; Zhang, X. Saccharospirillum alexandrii sp. nov., isolated from the toxigenic marine dinoflagellate Alexandrium catenella LZT09. Int. J. Syst. Evol. Microbiol. 2020, 70, 820–826. [Google Scholar] [CrossRef]
- Yang, X.; Xiang, R.; Iqbal, N.M.; Duan, Y.H.; Zhang, X.A.; Wang, L.; Yu, L.Z.; Li, J.Z.; Sun, M.F.; Yang, Q. Marinobacter shengliensis subsp. alexandrii Subsp. Nov.; Isolated from Cultivable Phycosphere Microbiota of Highly Toxic Dinoflagellate Alexandrium catenella LZT09 and Description of Marinobacter shengliensis Subsp. shengliensis Subsp. Nov. Curr. Microbiol. 2021, 78, 1648–1655. [Google Scholar] [CrossRef]
- Wang, X.; Ye, Y.; Xu, F.F.; Duan, Y.H.; Xie, P.F.; Yang, Q.; Zhang, X. Maritimibacter alexandrii sp. nov.; a New Member of Rhodobacteraceae Isolated from Marine Phycosphere. Curr. Microbiol. 2021, 78, 3996–4003. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X.; Jiang, Z.; Yang, X.; Zhang, X.; Yang, Q. Combined characterization of a new member of Marivita cryptomonadis, strain LZ-15-2 isolated from cultivable phycosphere microbiota of toxic HAB dinoflagellate Alexandrium catenella LZT09. Braz. J. Microbiol. 2021, 52, 739–748. [Google Scholar] [CrossRef]
- Siddharth, T.; Sridhar, P.; Vinila, V.; Tyagi, R.D. Environmental applications of microbial extracellular polymeric substance (EPS): A review. J. Environ. Manag. 2021, 287, 112307. [Google Scholar] [CrossRef]
- Lai, H.; Fang, H.; Huang, L.; He, G.; Reible, D. A review on sediment bioflocculation: Dynamics, influencing factors and modeling. Sci. Total Environ. 2018, 642, 1184–1200. [Google Scholar] [CrossRef]
- Su, J.; Yang, X.; Zheng, T.; Hong, H. An efficient method to obtain axenic cultures of Alexandrium tamarense-a PSP-producing dinoflagellate. J. Microbiol. Methods 2007, 69, 425–430. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA Gene Analysis with QIIME2. Methods Mol. Biol. 2018, 1849, 113–129. [Google Scholar]
- Prasad, D.V.; Madhusudanan, S.; Jaganathan, S. uCLUST-A new algorithm for clustering unstructured data. ARPN J. Eng. Appl. Sci. 2015, 10, 2108–2117. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note 101; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Varasteh, T.; Moreira, A.P.B.; Silva Lima, A.W.; Leomil, L.; Otsuki, K.; Tschoeke, D.; Garcia, G.; Thompson, C.; Thompson, F. Genomic repertoire of Mameliella alba Ep20 associated with Symbiodinium from the endemic coral Mussismilia braziliensis. Symbiosis 2020, 80, 53–60. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Jeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998, 23, 403–405. [Google Scholar] [CrossRef]
- Thorne, J.L.; Kishino, H.; Felsenstein, J. An evolutionary model for maximum likelihood alignment of DNA sequences. J. Mol. Evol. 1991, 33, 114–124. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Sardà Carbasse, J.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Blin, K.; Pascal Andreu, V.; de Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annottion through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Dai, J.; Wu, Y.; Chen, S.W.; Zhu, S.; Yin, H.P.; Wang, M.; Tang, J. Sugar compositional determination of polysaccharides from Dunaliella salina by modified RP-HPLC method of precolumn derivatization with 1-phenyl-3-methyl-5-pyrazolone. Carbohydr. Polym. 2010, 82, 629–635. [Google Scholar] [CrossRef]
- Gonzalez, L.E.; Bashan, Y. Increased growth of the microalga Chlorella vulgaris when coimmobilized and cocultured in alginate beads with the plant-growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 2000, 6, 1527–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Characteristic | LZ-28 | JLT354-WT | KD53 | L6M1-5 | JL351 | F15 |
|---|---|---|---|---|---|---|
| Isolation source | Marine dinoflagellate | Seawater | Marine diatom | Deep-sea sediment | Surface seawater | Deep-sea sediment |
| Colony color | Light-Yellow | White | Yellow | Yellow–white | Yellow | Cream–yellow |
| Cell size (µm) | (0.7–1.0) × (2.0–2.9) | (0.7–0.9) × (2.0–3.2) | (0.5–1.0) × (2.2–2.9) | (0.7–0.8) × (1.0–2.0) | (0.5–0.8) × (0.7–1.5) | (0.7–1.2) × (1.6–3.4) |
| NaCl range (optimum, w/v, %) | 1.0–10.0 (2.5) | 1.0–10.0 (1.0–3.0) | 1.0–6.0 (3.0) | 0.5–15.0 (3.0–5.0) | 0.5–9.0 (1.0–4.0) | 0–10.0 (3.5) |
| pH range (optimum) | 5.0–10.0 (7.0) | 6.0–9.0 (8.0) | 6.0–9.5 (7.5–8.0) | 5.0–10.5 (7.0) | 7.0–10.0 (8.0) | 6.0–11.0 (7.0–8.0) |
| Temperature range (optimum, °C) | 15–40 (25–28) | 10–30 (25) | 16–37 (28) | 10–41 (28–30) | 15–40 (30) | 4–50 (35–37) |
| Oxidase activity | + | + | + | − | + | + |
| Catalase activity | + | + | − | + | + | + |
| Poly-β-hydroxybutyrate | + | + | − | − | + | − |
| API 20E test | ||||||
| Citrate utilization | − | − | − | w | w | + |
| Gelatinase | + | + | − | + | + | + |
| API 20NE test | ||||||
| Reduction of nitrite | + | + | + | + | − | + |
| Gelatin hydrolysis | + | + | − | + | + | + |
| Phenylacetic acid utilization | − | + | + | + | + | − |
| Polar lipid profile a | PC, DPG, PE | AL | PC, PE, AL | PE, AL | PC, DPG, PE, AL, GL | PC, PE, GL |
| Fatty acid profile b | ||||||
| C16:0 | 3.2 | 5.7 | 5.3 | 4.8 | 5.3 | 4.5 |
| C17:0 | 1.1 | tr | 1.0 | tr | tr | ND |
| C18:0 | 7 | 9.2 | 11.1 | 10 | 8.6 | 8.5 |
| C12:1 3OH | 4.7 | 3.3 | 2.8 | 3.1 | 3.2 | 3.1 |
| C17:1ω8c | − | tr | tr | tr | tr | 1 |
| C18:1ω7c 11-methyl | 10.8 | 6.6 | 7.9 | 7.5 | 6.1 | 5.4 |
| C19:0 cyclo ω8c | 3.2 | 5.7 | 5.3 | 4.8 | 5.3 | 4.5 |
| Summed feature 8 | 65.5 | 70.8 | 69.5 | 70.7 | 72.5 | 74.3 |
| 16S rRNA gene similarity (%) | − | 99.70 | 99.77 | 99.92 | 99.62 | 99.40 |
| DNA G+C content (mol%) c | 64.9 | 65.2 | 65.0 | 65.0 | 65.1 | 65.0 |
| ANI value of LZ-28 (%) | − | 98.0 | 98.2 | 98.0 | 98.0 | 97.9 |
| AAI value of LZ-28 (%) | − | 98.4 | 98.1 | 98.1 | 98.5 | 98.2 |
| dDDH value of LZ-28 (%) | − | 84.3 | 83.5 | 83.9 | 84.3 | 83.1 |
| Strain | Isolation Source and Year | Genome Size (Mb) | G+C Content (mol%) a | Protein | CDS b | GenBank Accession No. |
|---|---|---|---|---|---|---|
| LZ-28 | Toxic marine dinoflagellate Alexandrium catenella LZT09, East China Sea, 2018 | 5.66 | 64.94 | 5502 | 5497 | JAANYX000000000 |
| KD53 | Marine diatom Phaeodactylum tricornutum, Xiamen, China, 2013 | 5.42 | 65.10 | 5278 | 5223 | NIWC00000000 |
| Ep20 | Marine dinoflagellate, Symbiodinium sp., 2015 | 5.51 | 65.04 | 5345 | 5343 | QAEF00000000 |
| PBVC088 | Toxic marine dinoflagellate, Pyrodinium bahamense var. Compressum, 2012 | 5.51 | 64.96 | 5689 | 5632 | LZNT00000000 |
| UMTAT08 | Marine dinoflagellate Alexandrium tamiyavanichii AcMS01, 1997 | 5.84 | 65.01 | 5761 | 5719 | JSUQ00000000 |
| JLT354-WT | Seawater, South China Sea, 2006 | 5.26 | 65.21 | 5126 | 5132 | FMZI00000000 |
| JL351 | Surface seawater (111°00′ E, 20°59′ N), South China Sea, 2006 | 5.42 | 65.15 | 5290 | 5275 | NIWA00000000 |
| KU6B c | Surface seawater, Boso Peninsula, Japan (34.9° N, 134.89° E), 2011 | 5.83 | 64.95 | 6009 | 4886 | AP022337-022340 |
| L6M1-5 | Deep-sea sediment (2835 m), South Atlantic Ocean (15.18° S, 13.88° W), 2011 | 5.90 | 64.96 | 5821 | 5783 | NIVZ00000000 |
| F15 | Deep-sea sediment (7118 m, 141°59.7′ E, 10°59.7′ N), Western Pacific Ocean, 2015 | 5.79 | 64.95 | 5694 | 5589 | NIWB00000000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, C.-Z.; Gao, H.-M.; Dai, J.; Zhu, W.-Z.; Xu, F.-F.; Ye, Y.; Zhang, X.-L.; Yang, Q. Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09. Mar. Drugs 2022, 20, 321. https://doi.org/10.3390/md20050321
Ren C-Z, Gao H-M, Dai J, Zhu W-Z, Xu F-F, Ye Y, Zhang X-L, Yang Q. Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09. Marine Drugs. 2022; 20(5):321. https://doi.org/10.3390/md20050321
Chicago/Turabian StyleRen, Cheng-Zhe, Hui-Min Gao, Jun Dai, Wen-Zhuo Zhu, Fei-Fei Xu, Yun Ye, Xiao-Ling Zhang, and Qiao Yang. 2022. "Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09" Marine Drugs 20, no. 5: 321. https://doi.org/10.3390/md20050321
APA StyleRen, C.-Z., Gao, H.-M., Dai, J., Zhu, W.-Z., Xu, F.-F., Ye, Y., Zhang, X.-L., & Yang, Q. (2022). Taxonomic and Bioactivity Characterizations of Mameliella alba Strain LZ-28 Isolated from Highly Toxic Marine Dinoflagellate Alexandrium catenella LZT09. Marine Drugs, 20(5), 321. https://doi.org/10.3390/md20050321