1. Introduction
In the absence of a vaccine or treatment for SARS-CoV-2 after the COVID-19 pandemic, many studies have been conducted across the world to screen substances for inhibition of SARS-CoV-2 infection, replication, and propagation from marine bioactive compounds, such as polyphenols, carotenoids, and polysaccharides [
1,
2,
3,
4]. Polyphenols derived from brown algae, commonly known as phlorotannins, were predicted in silico as a potential inhibitor of SARS-CoV-2 3CLpro [
5]. Siphonaxanthin, a marine carotenoid extracted from
Codium fragile, inhibited SARS-CoV-2 pseudovirus cell entry in vitro [
6]. In the field of marine compounds, much work has been focused on polysaccharides for the development of therapeutics and prophylaxis for COVID-19.
Recently, it was reported that iota-carrageenan inhibits SARS-CoV-2 replication in various cell lines [
7,
8] and also inhibits replication of its variants, alpha, beta, gamma, and delta [
9]. Lambda-carrageenan has been reported to have antiviral effects against SARS-CoV-2 in vitro [
10]. In addition, an iota-carrageenan nasal spray exhibited a relative risk reduction of 79.8% in preventing SARS-CoV-2 in healthcare workers managing patients with COVID-19 disease [
11].
Furthermore, a number of marine sulfated polysaccharides (RPI-27, RPI-28, and sulfated galactofucan from
Saccharina japonica, sea cucumber sulfated polysaccharide (SCSP), and rhamnan sulfate from
Monostroma nitidum) effectively inhibited SARS-CoV-2 entry by interfering with the interaction of the spike protein with the ACE2 receptor of the host cell in vitro [
12,
13,
14,
15,
16]. Kwon et al. was reported that RPI-27 (Mw ~ 100 kDa) has a higher molecular weight than RPI-28 (Mw ~ 12 kDa) and is a branched polysaccharide with an EC
50 value of 8.3 ± 4.6 μg/mL against SARS-CoV-2 compared with RPI-28′s EC
50 of 16 ± 11 μg/mL [
13]. SCSP exhibited strong inhibitory activity in Vero E6 cells with an IC
50 of 9.10 μg mL
−1 [
15]. In addition, the neutralizing effect of rhamnan sulfate on wildtype and delta SARS-CoV-2 pseudovirus particles in vitro resulted in IC
50 values of 2.39 and 1.66 μg/mL, respectively [
16]. In our previous study [
17], the crude polysaccharides (CPs) from seaweed and abalone viscera effectively inhibited SARS-CoV-2 pseudovirus entry. Among them, CPSH (crude polysaccharide from
Sargassum horneri), CPAV (crude polysaccharide from abalone viscera), and CPHF (crude polysaccharide from
Hizikia fusiforme) inhibited virus infection with IC
50 values of 12 μg/mL, 33 μg/mL, and 47 μg/mL, respectively. These marine polysaccharides primarily inhibited SARS-CoV-2 infection in a noncompetitive manner.
In order to show various spectrums of the antiviral activity of marine polysaccharides for application in the development of therapeutics and prophylaxis, we evaluated the inhibitory effect on SARS-CoV-2 propagation in vitro using the three polysaccharides (CPAV, CPSH, and CPHF) that showed the highest inhibition of SARS-CoV-2 pseudovirus cell entry in our previous study [
17].
3. Discussion
A number of marine polysaccharides, comprised of carrageenan, laminarin, alginate, fucoidan, chitosan, ulvan, agar, and porphyran, have exhibited directly and indirectly antiviral effects against SARS-CoV-2 [
7,
8,
9,
10,
11,
12,
13,
14,
15,
16]. Iota-carrageenan inhibits the replication of SARS-CoV-2 and its variants alpha, beta, gamma, and delta [
9], and lambda-carrageenan also has demonstrated antiviral activity against influenza virus and SARS-CoV-2 [
7,
8]. Sulfated polysaccharides effectively inhibited SARS-CoV-2 entry by interfering with the interaction of the spike protein with the ACE2 receptor of the host cell in vitro [
13,
14,
15,
16]. In our previous study [
17], crude polysaccharides (CPs) from seaweed and abalone viscera inhibited SARS-CoV-2 pseudovirus cell entry with an IC
50 of 12~289 μg/mL. Among them, the total carbohydrate content was the highest in CPSH (99.1%), followed by CPHF (94.4%) and CPAV (62.7%), whereas the highest level of total protein content was found in CPAV (22.3%), followed by CPHF (10.9%) and CPSH (4.0%), respectively. In addition, CPHF had the highest sulfate ion content (20.4%), followed by CPSH (9.8%) and CPAV (0.5%), respectively. In monosaccharide composition, total fucose and galactose contents were CPSH (66.2% and 6.9%), CPHF (41.8% and 9.6%), and CPAV (37.3% and 12.9%), respectively [
17].
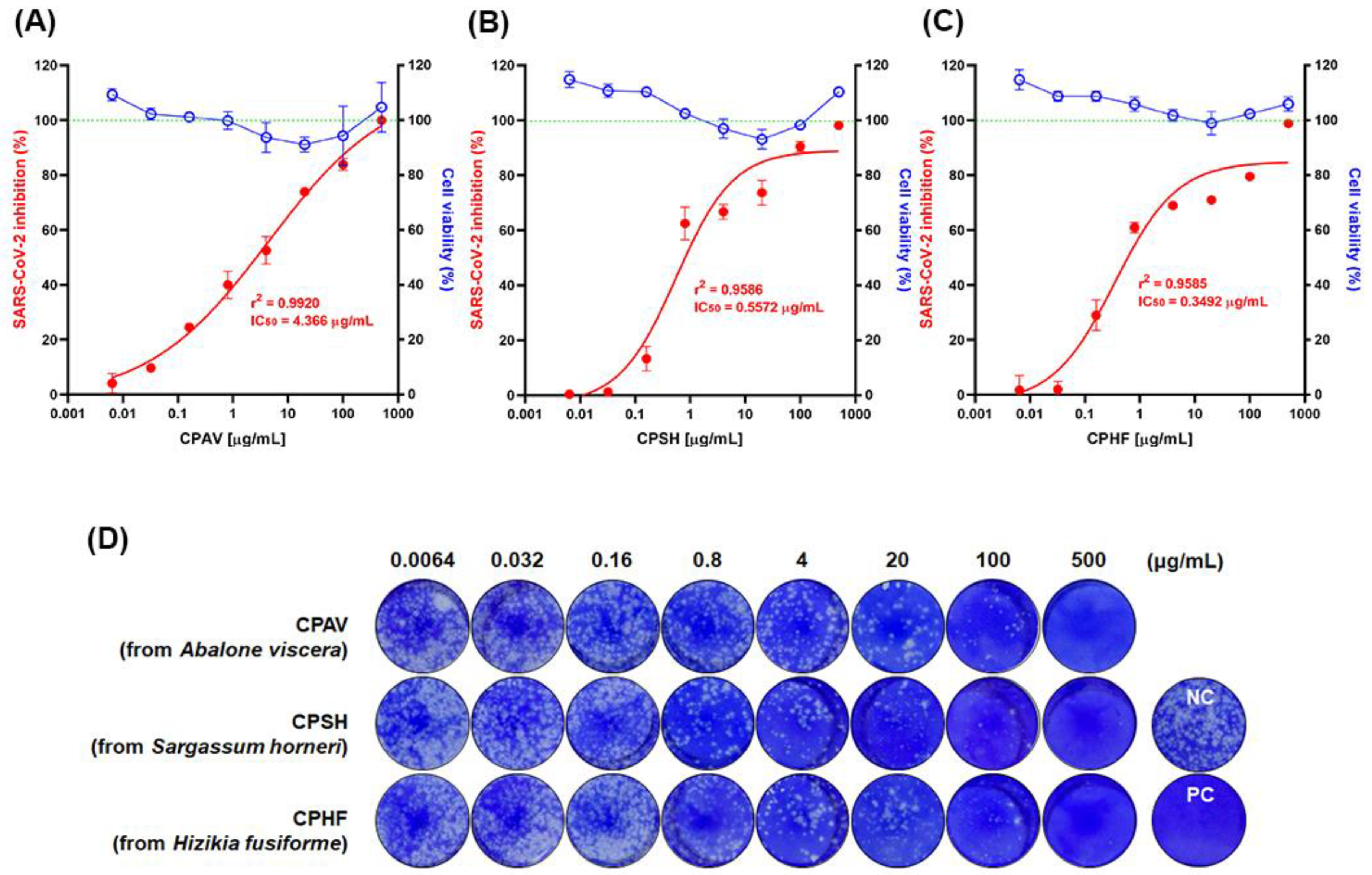
In this study, the same crude polysaccharides used in our previous study [
17], CPAV, CPSH, and CPHF, which effectively inhibited SARS-CoV-2 pseudovirus entry in that study, were evaluated for antiviral effects against SARS-CoV-2 (NCCP 43326/Wuhan) propagation in vitro. CPAV, CPSH, and CPHF drastically inhibited SARS-CoV-2 propagation in a dose-dependent manner (
Figure 1). As described previously for crude polysaccharides [
17], these results suggest that the antiviral effect of CPs could mainly be explained by interference with SARS-CoV-2 entry because propagation of SARS-CoV-2 was strongly inhibited and the virus titer sharply reduced when CPs were used to pretreat before viral infection (
Figure 1 and
Figure 2), while the inhibitory effect was less significant when CPs were used for post-treatment after viral infection (
Figure 3 and
Figure 4). These results show that, although not purified polysaccharides, CPs can inhibit SARS-CoV-2 propagation as well as cell entry, and high molecular weight, high total carbohydrate, and high fucose content are important factors in the inhibitory effect. Kwon et al. [
13] reported that RPI-27(~100 kDa), a branched polysaccharide from
Saccharina japonica, was shown to have higher antiviral activity (EC
50 = 8.3 μg/mL) than RPI-28 (~12 kDa, EC
50 = 16 μg/mL). Jin et al. [
14] reported that sulfated galactofucan (195 kDa) and glucuronomannan (7.0 kDa) strongly inhibited interaction between the SARS-CoV-2 S-glycoprotein and heparin (IC
50s are 27 and 231 nM, respectively). In addition, CPAV has a higher total protein content compared with CPSH and CPHF, which is presumed to exhibit strong antiviral activity due to the synergistic effect of various bioactive molecules [
18,
19,
20,
21] and high molecular weight polysaccharides present in the abalone viscera.
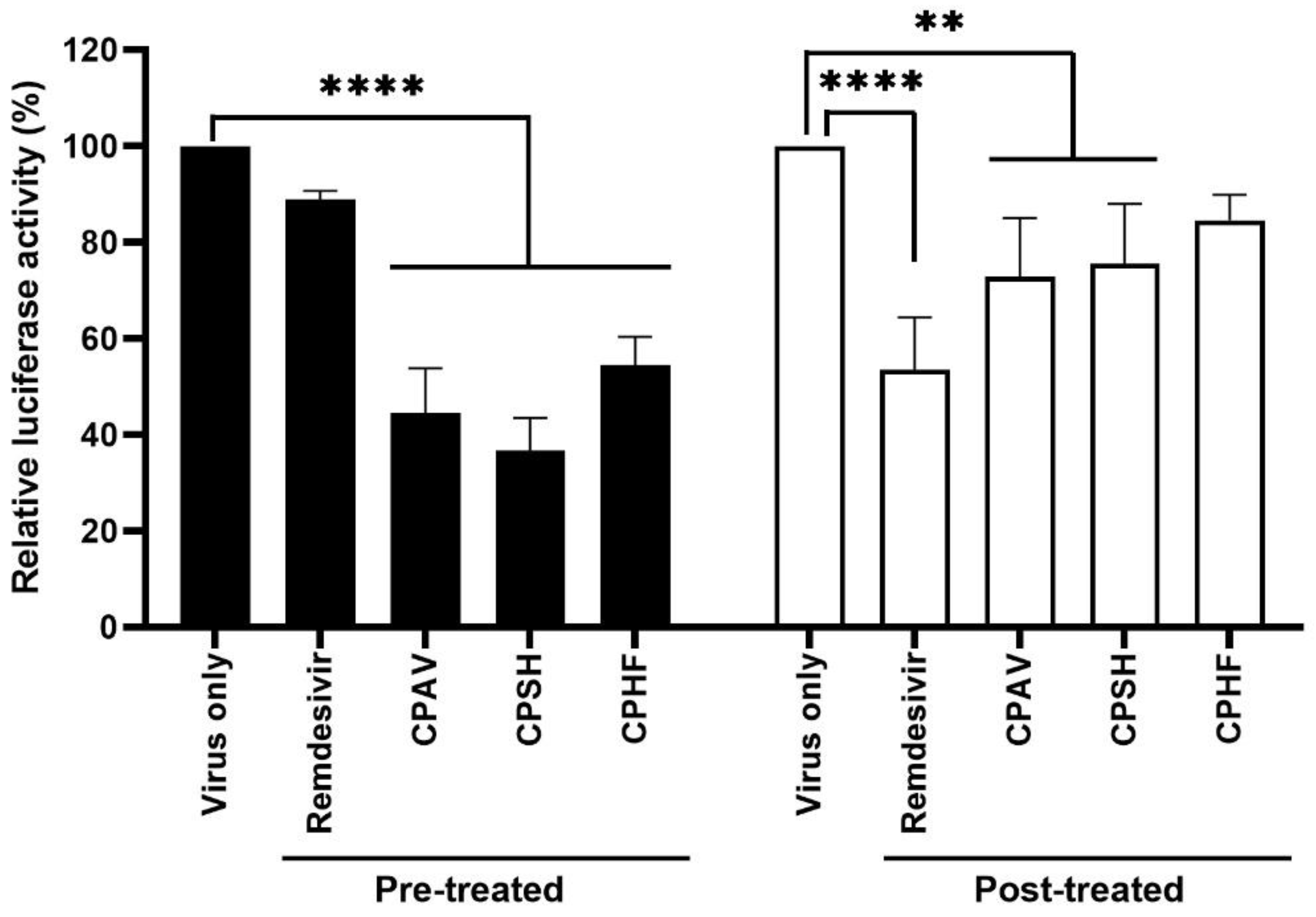
Furthermore, the crude polysaccharides clearly inhibited SARS-CoV-2 replication and viral protein expression even as a post-treatment. These results are supported by IFA, western blot, and qRT-PCR (
Figure 3). The inhibition of virus replication and viral protein expression by CPs may be due to the post-treatment CPs inhibiting the re-infection of surrounding cells by released virions after replication. These results show a more significant inhibitory action with the prophylactic application. In the immunofluorescence assays (IFA), the CP pretreatment blocked about 90% of SARS-CoV-2 propagation, although CPAV displayed a lower inhibitory effect than CPSH and CPHF, and CP post-treatment blocked about 60% (
Figure 3A). Immunoblotting supported that the pretreatment inhibited the expression of SARS-CoV-2 spikes and nucleocapsid proteins, but the post-treatment did not completely inhibit it (
Figure 3C). In addition, pre and post-treatment with CPs dramatically reduced RdRp and nucleocapsid RNA copies similar to remdesivir as a positive control in immunofluorescence assays (
Figure 3D). Similar to these results, testing with a SARS-CoV-2 pseudovirus for cell entry showed a high inhibitory effect when CPs were administered before virus infection compared with after virus infection (
Figure 4). It is presumed that crude polysaccharides from seaweed and abalone viscera inhibit SARS-CoV-2 cell entry, resulting in the inhibition of viral RNA replication and protein expression. Fröba et al. and Varese et al. reported that iota-carrageenan inhibits the replication of SARS-CoV-2 in various cell lines [
8,
9], and iota-carrageenan also inhibited the variants alpha, beta, gamma, and delta [
9].
There was a slight correlation between molecular weight of the CPs and antiviral inhibitory effects. Regarding the IC
50 of CPs, the antiviral activity of CPAV increased in a dose-dependent manner, while CPSH and CPHF showed almost no inhibitory effect at concentrations below 0.032 μg/mL. However, these showed strong antiviral effects at concentrations above 0.16 μg/mL and the effects increased gently at concentrations above 0.8 μg/mL. When CPSH and CPHF were used at concentrations below 0.032 μg/mL, plaques formed 98.8 ± 1.1% and 97.9 ± 2.9%, respectively, and with CPAV they formed 90.2 ± 0.1%. On the other hand, when CPSH and CPHF were used at concentrations over 0.8 μg/mL, plaque formation was sharply reduced to 37.5 ± 5.1% and 39.0 ± 2.8% compared with CPAV (59.9 ± 3.7%). In addition, the plaque formation with CPs was similarly reduced to 26.1 ± 0.8% (CPAV), 26.3 ± 3.9% (CPSH), and 29.1 ± 1.7% (CPHF), respectively, when CPs were used at concentrations over 20 μg/mL. These differences seem to be related to the difference in molecular weight of the CPs. In our previous study, CPAV appeared to be constituted of whole polysaccharides and fragments (over 800 kDa and 400~800 kDa) due to degradation by marine microorganisms in the abalone viscera, whereas CPSH and CPHF were both over 800 kDa [
17]. We suspect that the small molecular weight of CPAV nonspecifically interferes with the binding of the SARS-CoV-2 spike with the ACE receptor at low concentrations (
Figure 1A), but large molecular weight CPs inhibit it more effectively at higher concentrations (
Figure 1B,C). These results show that crude polysaccharides of over 800 kDa required at least a certain concentration of CPs to effectively inhibit SARS-CoV-2 cell entry.
4. Materials and Methods
4.1. Chemicals and Reagents
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), phosphate buffered saline (PBS) pH 7.4, and geneticin (G-418 sulfate) were purchased from Thermo Fisher Scientific (Waltham, MA, USA), and the Cell Titer-Glo Luminescent cell viability assay kit was purchased from Promega (Madison, WI, USA). Dimethyl sulfoxide (DMSO), agarose, formaldehyde solution (37%), chloroform, 2-propanol, and crystal violet were purchased from Sigma-Aldrich (St. Louis, MO, USA).
CPAV (from abalone viscera), CPSH (from
Sargassum horneri), and CPHF (from
Hizikia fusiforme) were prepared for use in our previous study. The algae
Hizikia fusiforme (collected in May 2020),
Sargassum horneri (collected in August 2020), and abalone (
Haliotis discus hannai, Dashimachonbok) were cultured in Wando, Jeollanam-do, South Korea. The fresh seaweed and abalone viscera were immediately washed with tap water and then dried. A concentrated water extract was rendered from each by treating with 2% CaCl
2 and 1.2 L of 85% (
v/v) ethanol to remove alginic acid and dialyzed using an ultrafiltration hollow fiber cartridge (10,000 NMWC, 31.8 L × 3.2 cm O.D., Cytiva, Middlesex County, MA, USA). Finally, the CP was lyophilized and stored at −20 °C [
17].
4.2. Cells and Viruses
African green monkey kidney Vero E6 cell culture (ATCC CRL-1586TM) was purchased from the American Type Culture Collection (Manassas, VA, USA). Vero E6 cells were maintained at 37 °C, 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) supplemented with 10% FBS. SARS-CoV-2 (NCCP 43326/Wuhan) virus was provided by the Korea Disease Control and Prevention Agency and was amplified in Vero E6 cells at 37 °C for 3 days. After centrifugation at 1000× g for 5 min, viral stocks were stored at −80 °C and viral titers were determined in a plaque reduction assay. Vero E6 cells were seeded into 96-well microplates and grown overnight at 37 °C under 5% CO2. Ten-fold serial dilutions of SARS-CoV-2 virus from Vero E6 cells were added to monolayers of 80% confluent Vero E6 cells at 37 °C for 1 h in serum-free DMEM. For titer determination of SARS-CoV-2 virus stocks, Vero E6 cells were infected with serial dilutions of the virus stock over 72 h.
4.3. Cytotoxicity Assay
The cytotoxicity evaluation of CPHF, CPSH, and CPAV were performed using a Cell Titer-Glo Luminescent cell viability assay kit. Vero E6 cells were seeded into 96-well plates at a density of 2 × 10
4 cells per well for 24 h at 37 °C under 5% CO
2. The supernatants were then removed and washed one time with PBS. After the Vero E6 cells were treated with 20 μL of CPHF, CPSH or CPAV at various concentrations (5-fold serial dilutions in the range of 0.006 μg/mL to 500 μg/mL, final D.W 5%) for 72 h, respectively, 100 μL of Cell Titer-Glo 2.0 reagent was added per well and the plate was shaken for 2 min at 700 rpm. After 10 min, the mixture was read via luminometer and cell viability was calculated as follows:
where the RLU sample is the luminescence of the experimental sample and RLU conc. is the luminescence of the control. Cytotoxicity was calculated as follows:
The 50% cytotoxic concentration for CPHF, CPSH, and CPAV were not determined because there is no cytotoxicity. Each sample was analyzed in triplicate wells and repeated three times.
4.4. Plaque Reduction Assay
The plaque reduction assay was performed as described previously, with some modification [
22]. Various concentrations of CPHF, CPSH, and CPAV were prepared as 5-fold serial dilutions in the range from 0.006 μg/mL to 500 μg/mL using distilled water. Remdesivir was prepared using DMSO at various concentrations in the range from 1.56 μM to 50 μM as a positive control. Vero E6 cells were seeded into 12-well plates at a density of 3 × 10
5 cells per well for 24 h at 37 °C under 5% CO
2. Cells were treated with CPs for 1 h and infected with 1.5 × 10
4 TCID
50 of SARS-CoV-2 virus at an MOI of 0.02 for 1 h. Then, cells were overlaid with a final 0.6% agarose solution and incubated at 37 °C. At 3 days post-infection; cells were fixed with 3.65% formaldehyde for 1 h at room temperature and washed three times with PBS. Finally, the number of plaques was counted by 0.5% crystal violet staining. Each sample was analyzed in duplicate.
4.5. TCID50 Assay
For determination of 50% tissue culture infectious dose (TCID
50), Vero E6 cells were seeded into 96-well plates at a density of 2.0 × 10
4 cells per well for 24 h at 37 °C under 5% CO
2. Cells were treated with 500 μg/mL of CPs and 10 μM remdesivir for 1 h and infected with 10-fold serially diluted 1.0 × 10
5 TCID
50 of SARS-CoV-2 virus for 3 days post infection (dpi) and 4 dpi. TCID
50s were determined based on whether or not a cytopathic effect occurred. The infective dose 50 was then calculated by the method of Reed and Muench [
23]. Each sample was analyzed in triplicate wells and repeated three times.
4.6. Immunofluorescence Assay
To assess the propagation of SARS-CoV-2 when Vero E6 cells were treated with 500 μg/mL of CPs before or after virus infection, immunofluorescence assays (IFA) were performed using a SARS-CoV-2 spike antibody. Vero E6 cells were seeded into 12-well plates at a density of 3 × 105 cells per well for 24 h at 37 °C under 5% CO2. Cells were treated with CPs for 1 h before or after infection with 6.0 × 104 TCID50 of SARS-CoV-2 virus at an MOI of 0.1 for 1 h, respectively. Remdesivir was used at 10 μM as a positive control. At 24 h post infection, the supernatants were removed and washed three times with PBS. Washed cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Then cells were washed three times with PBS and blocked with 5% bovine serum albumin (BSA, Bovogen Biologicals, Keilor East, VIC, Australia) for 3 h. Cells were incubated with a SARS-CoV-2 spike polyclonal antibody diluted 1:5000 (Sino Biological Inc., Beijing, China) for 90 min. After washing three times with PBS, the anti-spike antibody bound cells were subsequently labeled for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-mouse IgG diluted 1:2000 (Cell Signaling Technology, Boston, MA, USA), and the nuclei were stained with 4′,6-diamidino-2-phenylindole diluted 1:2000 (DAPI, Thermo Fisher Scientific, Waltham, MA, USA) for 30 min. Confocal laser scanning microscopy was performed with a CELENA® S Digital Imaging System (Logos Biosystems, Anyang, South Korea). Each sample was analyzed in triplicate.
4.7. Western Blotting
To examine the expression of viral spike and nucleocapsid proteins, immunoblotting was performed using anti-SARS-CoV-2 spike (S) and nucleocapsid (N) antibodies, both when Vero E6 cells were treated with 500 μg/mL of CPs for 1 h before or after cells were infected with 6.0 × 104 TCID50 of SARS-CoV-2 virus at an MOI of 0.1 for 1 h, respectively. Remdesivir was used at 10 μM as a positive control. At 24 h post infection, the supernatants were removed and washed three times with PBS. Washed cells were lysed by treatment with RIPA lysis buffer (Seongnam-si, Gyeonggi-do, South Korea), and the lysed cells were then centrifuged at 4 °C, 15,000× g, for 15 min. Equal amounts of each virus-infected cell lysate or each virus lysate were resolved in 4 × Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) and boiled at 100 °C for 10 min. After gel electrophoresis, the proteins were transferred onto a PVDF membrane which was blocked with PBS containing 5% BSA for 2 h at room temperature. Then, the membrane was incubated with primary antibodies specific to SARS-CoV-2 spike (1 μg) and nucleocapsid proteins (1 μg, Sino Biological Inc., Beijing, China) in PBST at 4 °C for 16 h, followed by washing six times with TBS-T. The washed membrane was incubated in PBST containing horseradish peroxidase-conjugated secondary antibody against rabbit IgG (1 μg, Cell Signaling Technology, Boston, MA, USA) for 1 h at room temperature and then visualized with ECL solution (ELPIS-BIOTECH, Daejeon, South Korea).
4.8. RNA Isolation and Quantitative RT-PCR
To quantify SARS-CoV-2 RNA copies, quantitative RT-PCR (qRT-PCR) was performed using RNA-dependent RNA polymerase (RdRp) and nucleocapsid (N) primers (
Table 1), respectively. Vero E6 cells were seeded into 12-well plates at a density of 3 × 10
5 cells per well for 24 h at 37 °C under 5% CO
2 and treated with 500 μg/mL of CPs for 1 h before or after cells were infected with 6.0 × 10
4 TCID
50 of SARS-CoV-2 virus at an MOI of 0.1 for 1 h, respectively. Remdesivir was used at 10 μM as a positive control. At 24 h post infection, the supernatants were removed and washed three times with PBS. The cells were then lysed by treatment with 0.5 mL of TriZol (Invitrogen, Waltham, MA, USA), followed by adding 0.1 mL of chloroform. After centrifugation at 13,000 rpm for 10 min, the supernatant was treated with 0.25 mL of 2-propanol at 4 °C for 10 min, and RNA was then precipitated by centrifugation at 13,000 rpm for 15 min. The precipitated RNA was treated with 0.5 mL of 75% ethanol and centrifuged at 7000 rpm for 5 min to remove ethanol and then dried for 10 min. RNA was suspended by treatment with RNase-free water at 55 °C for 10 min, then RNA was quantified at 100 ng/μL and cDNA was synthesized using All-in-One 5X cDNA Master Mix (Cellsafe, Yongin, South Korea). The PCR reaction was performed at 25 °C for 5 min, 42 °C for 1 h, and 85 °C for 5 s according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using the synthesized cDNA as a template at 95 °C for 10 s and at 55 °C for 30 s for 40 cycles with iQ SYBR Green (Bio-Rad, Hercules, CA, USA). PCR primers were used as follows: Actin_fwd: 5′-TGA-CAG-CAG-TCG-GTT-GGA-GCG-3′, Actin_rev: 5′-GAC-TTC-CTG-TAA-CAA-CGC-ATC-TCA-TA-3′, Nucleocapsid_fwd: 5′-TAA-TCA-GAC-AAG-GAA-CTG-ATT-A-3′, Nucleocapsid_rev: 5′-CGA-AGG-TGT-GAC-TTC-CAT-G-3′, RdRp_fwd: 5′-AGA-ATA-GAG-CTC-GCA-CCG-TAG -3′, and RdRp_rev: 5′- CTCCTCTAGTGGCGGCTATT-3′. Each sample was analyzed in triplicate.
4.9. Reporter Assay Using SARS-CoV-2 Pseudovirus
For preparing the SARS-CoV-2 pseudovirus, HEK293T cells were treated with 2 μg of pLV-Luc, psPAX2, and pAGCN-SARS-CoV-2, respectively, and 12 μL of lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA) and incubated at 37 °C under 5% CO2 for 3 days. After centrifugation at 1000 rpm, at 4 °C for 5 min, the supernatant was treated with Lenti-X concentrator (TaKaRa Bio inc., San Jose, CA, USA) in a ratio of 1:3, and left at 4 °C for 16 h. The supernatant was removed after centrifugation at 1500× g, 4 °C for 45 min and the pellet was suspended in serum-free DMEM and stored at −80 °C until use. For the pseudovirus efficiency test, HEK293T cells were infected with the prepared SARS-CoV-2 pseudovirus, and 24 h later the cells were lysed with reporter lysis buffer (Promega, Madison, WI, USA). The lysate was dispensed in a 96-well white plate, the same amount of Bright-Glo reagent (Promega, Madison, WI, USA) was added, and the luminescence was measured with a luminometer (GM3000, Promega, Madison, WI, USA) to confirm the infection efficacy of the pseudovirus. To investigate the inhibitory effect of crude polysaccharides before viral infection, HEK293T cells were treated with 500 μg/mL each of CPHF, CPSH, and CPAV, incubated at 37 °C for 1 h, and then infected with 130 μL of SARS-CoV-2 pseudovirus (about 10,000 luminescence) for 4 h. At the same time, to investigate the inhibitory effect of crude polysaccharides after viral infection, HEK293T cells were infected with 130 μL of SARS-CoV-2 pseudovirus (about 10,000 luminescence) for 4 h, and then treated with 500 μg/mL each of CPHF, CPSH, and CPAV for 1 h at 37 °C. The medium was replaced with DMEM containing 5% FBS, and after 24 h the cells were lysed by treatment with reporter lysis buffer. Lysates were dispensed in a 96-well white plate, the same amount of Bright-Glo reagent was added, and then luminescence was measured. Each sample was analyzed in duplicate wells and repeated two times.
4.10. Statistical Analysis
The half maximal inhibitory concentrations (IC
50) were calculated using nonlinear regression analysis of GraphPad Prism version 8 by plotting (inhibitor) versus response (variable slope, four parameters). The equation corresponds to:
where Y is the response, X is the concentrations or doses, Top and Bottom are plateaus in the units of the Y axis, and HillSlope describes the steepness of the family of curves. Statistical analyses were performed by ordinary one-way ANOVA
t-test according to the Dunnett’s multiple comparison method using GraphPad Prism version 8.
p values lower than 0.05 were considered statistically significant. *
p < 0.05; **
p < 0.01; ***
p < 0.001; ****
p < 0.0001.