A Glimpse at Siderophores Production by Anabaena flos-aquae UTEX 1444
Abstract
:1. Introduction
2. Results and Discussion
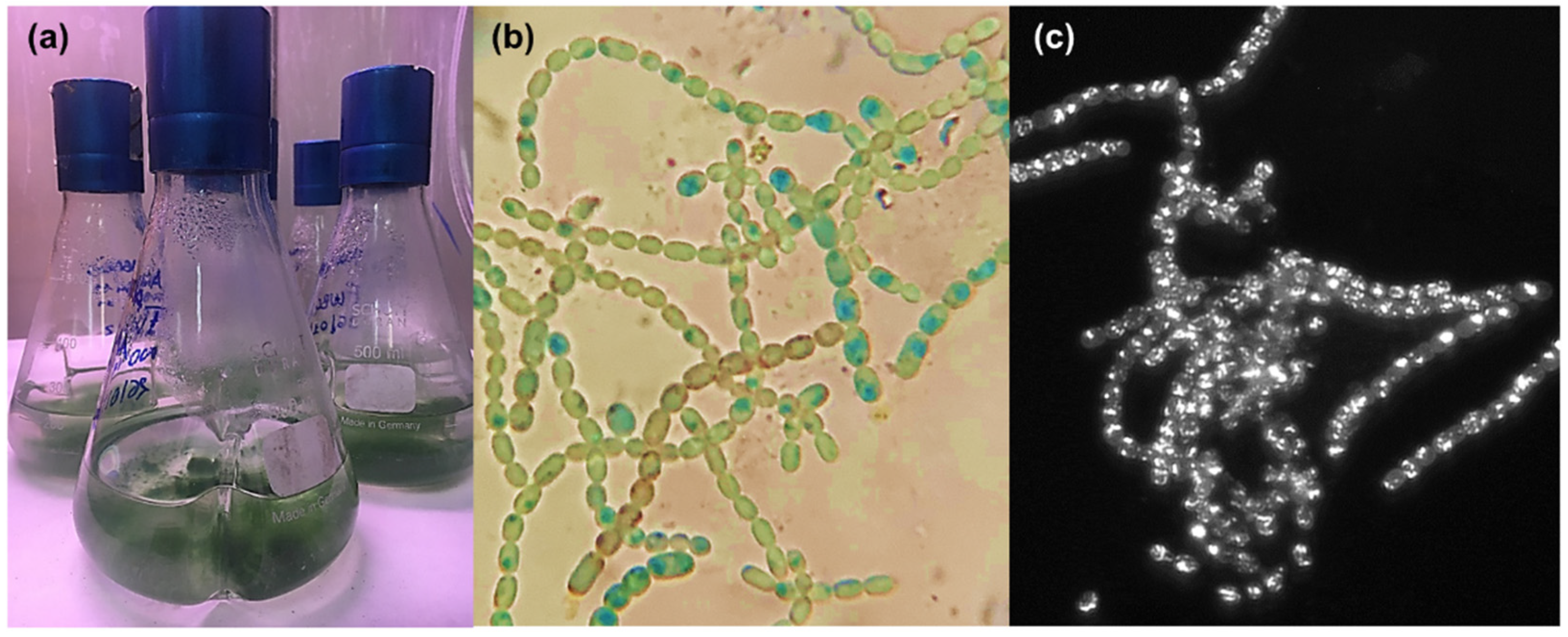
2.1. Cultivation and Extraction of Anabaena flos-aquae UTEX 1444
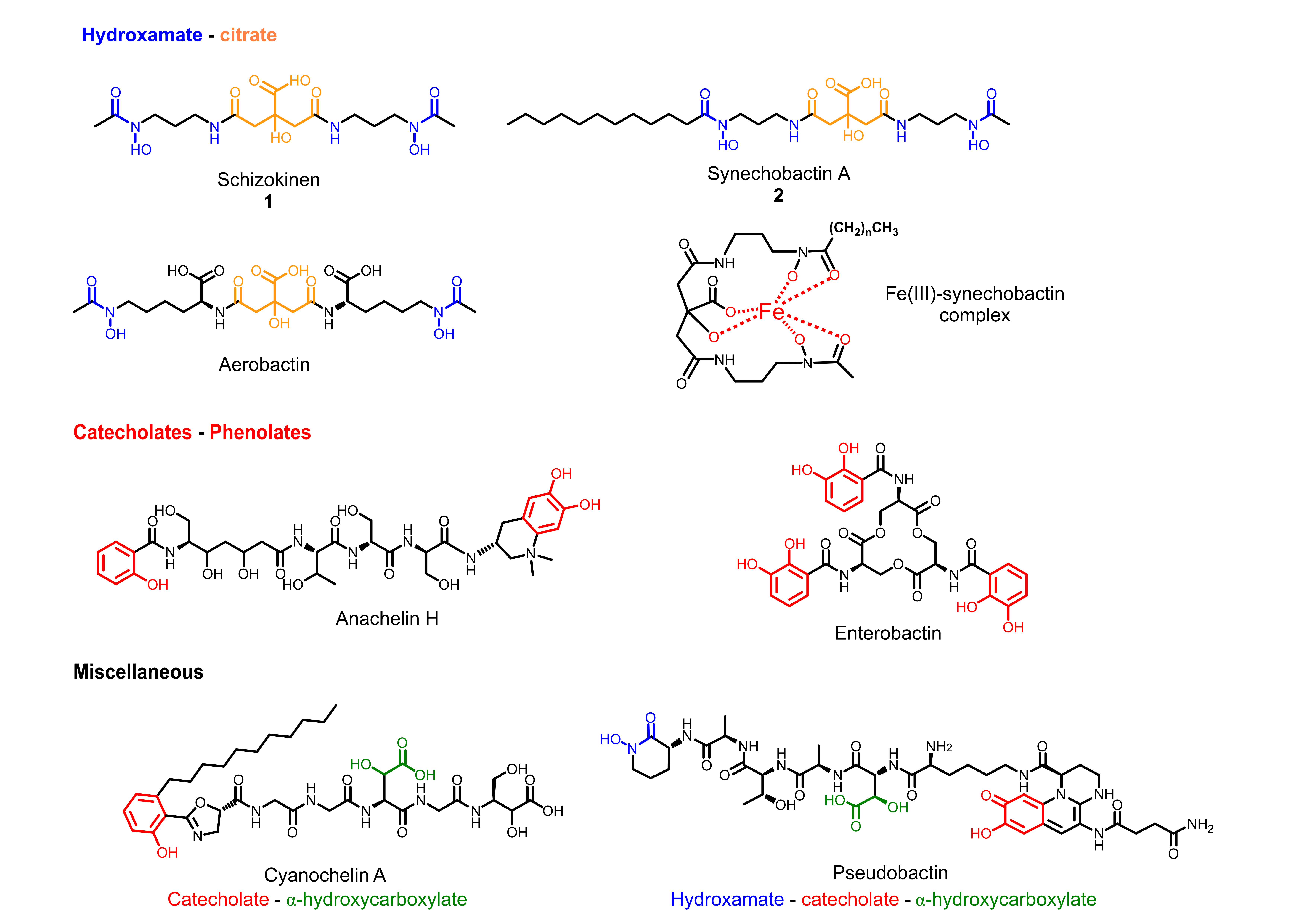
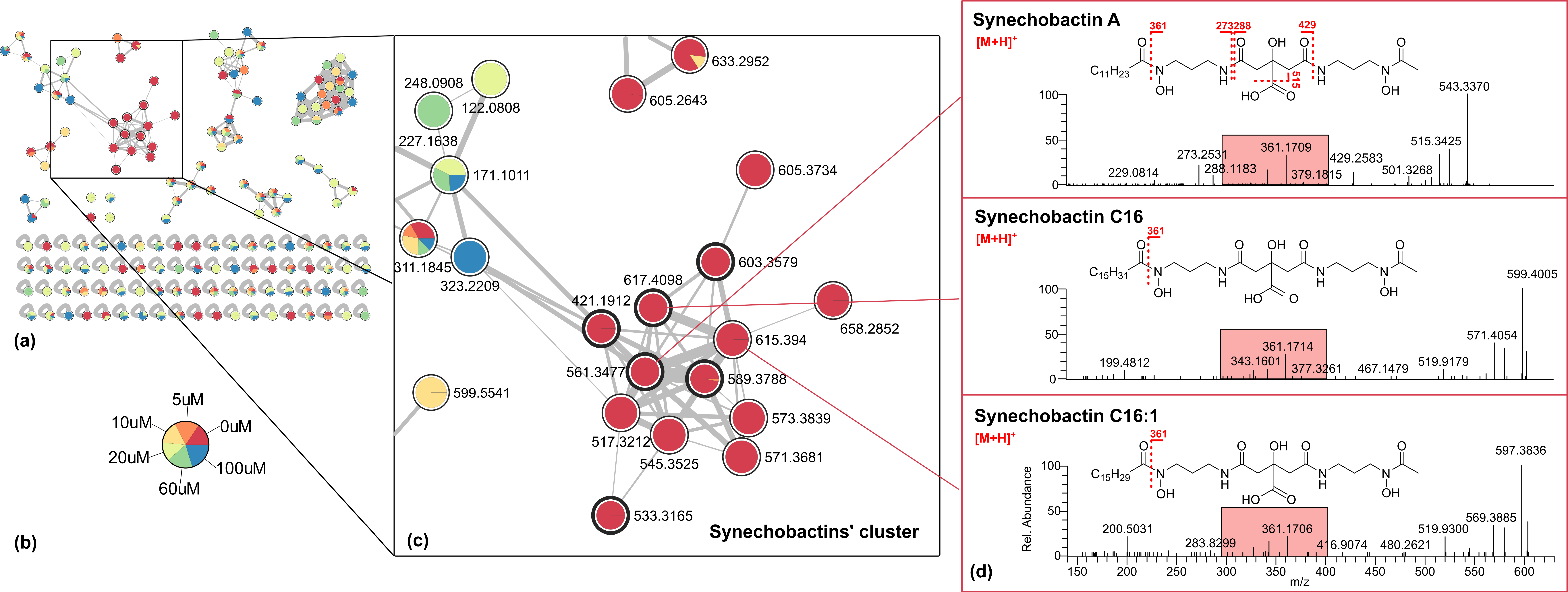
2.2. Molecular Networking and Synechobactins’ Identification
3. Materials and Methods
3.1. Anabaena flos-aquae UTEX 1444: Strain, Cultivation and Extraction
3.2. LC-HRMS Analyses and Molecular Networking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Oves-Costales, D.; Kadi, N.; Challis, G.L. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem. Commun. 2009, 43, 6530–6541. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Norton, I.; Salunkhe, A.S.; Goodluck, H.; Aly, W.S.M.; Mourad-Agha, H.; Cornelis, P. Control of Iron Metabolism in Bacteria. In Metallomics and the Cell; Springer: Berlin/Heidelberg, Germany, 2013; pp. 203–239. [Google Scholar]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Kurth, C.; Kage, H.; Nett, M. Siderophores as molecular tools in medical and environmental applications. Org. Biomol. Chem. 2016, 14, 8212–8227. [Google Scholar] [CrossRef]
- Negash, K.H.; Norris, J.K.; Hodgkinson, J.T. Siderophore–Antibiotic Conjugate Design: New Drugs for Bad Bugs? Molecules 2019, 24, 3314. [Google Scholar] [CrossRef] [Green Version]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 2016, 23, 3984–3999. [Google Scholar] [CrossRef]
- Nagoba, B.; Vedpathak, D. Medical applications of siderophores. Eur. J. Gen. Med. 2011, 8, 229–235. [Google Scholar] [CrossRef]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.-L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Madamwar, D.; Incharoensakdi, A. Bloom Dynamics of Cyanobacteria and Their Toxins: Environmental Health Impacts and Mitigation Strategies. Front. Microbiol. 2015, 6, 1254. [Google Scholar] [CrossRef] [Green Version]
- Årstøl, E.; Hohmann-Marriott, M.F. Cyanobacterial Siderophores—Physiology, Structure, Biosynthesis, and Applications. Mar. Drugs 2019, 17, 281. [Google Scholar] [CrossRef] [Green Version]
- Řezanka, T.; Palyzová, A.; Sigler, K. Isolation and identification of siderophores produced by cyanobacteria. Folia Microbiol. 2018, 63, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.J.; Lammers, P.J.; Berman, M.S.; Sanders Loehr, J. Siderophore-mediated in iron uptake in different strains of Anabaena sp. J. Bacteriol. 1983, 156, 1144–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galica, T.; Borbone, N.; Mareš, J.; Kust, A.; Caso, A.; Esposito, G.; Saurav, K.; Hájek, J.; Řeháková, K.; Urajová, P.; et al. Cyanochelins, an Overlooked Class of Widely Distributed Cyanobacterial Siderophores, Discovered by Silent Gene Cluster Awakening. Appl. Environ. Microbiol. 2021, 87, e03128-20. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [Green Version]
- Berberoğlu, H.; Jay, J.; Pilon, L. Effect of nutrient media on photobiological hydrogen production by Anabaena variabilis ATCC 29413. Int. J. Hydrogen Energy 2008, 33, 1172–1184. [Google Scholar] [CrossRef] [Green Version]
- Teta, R.; Sala, G.D.; Esposito, G.; Stornaiuolo, M.; Scarpato, S.; Casazza, M.; Anastasio, A.; Lega, M.; Costantino, V. Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus. Toxins 2021, 13, 471. [Google Scholar] [CrossRef]
- Esposito, G.; Teta, R.; Marrone, R.; De Sterlich, C.; Casazza, M.; Anastasio, A.; Lega, M.; Costantino, V. A Fast Detection Strategy for Cyanobacterial blooms and associated cyanotoxins (FDSCC) reveals the occurrence of lyngbyatoxin A in campania (South Italy). Chemosphere 2019, 225, 342–351. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Global Natural Product Social. Available online: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp (accessed on 20 May 2021).
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ito, Y.; Butler, A. Structure of synechobactins, new siderophores of the marine cyanobacterium. Limnol. Oceanogr. 2005, 50, 1918–1923. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Repeta, D.J. An extended siderophore suite from Synechococcus sp. PCC 7002 revealed by LC-ICPMS-ESIMS. Metallomics 2015, 7, 877–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persmark, M.; Pittman, P.; Buyer, J.S.; Schwyn, B.; Gill, P.R.; Neilands, J.B. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J. Am. Chem. Soc. 1993, 115, 3950–3956. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Modern approach to the classification system of cyanophytes 4. nostocales. Arch. Fuer Hydrobiol. Suppl. 1989, 82, 247–345. [Google Scholar]
- Zapomělová, E.; Řeháková, K.; Jezberová, J.; Komárková, J. Polyphasic characterization of eight planktonic Anabaena strains (Cyanobacteria) with reference to the variability of 61 Anabaena populations observed in the field. Hydrobiologia 2010, 639, 99–113. [Google Scholar] [CrossRef]
- Li, R.; Watanabe, M.; Watanabe, M.M. Taxonomic studies of planktic species of Anabaena based on morphological characteristics in cultured strains. Hydrobiologia 2000, 438, 117–138. [Google Scholar] [CrossRef]
- Moreira, C.; Vasconcelos, V.; Antunes, A. Cyanobacterial Blooms: Current Knowledge and New Perspectives. Earth 2022, 3, 127–135. [Google Scholar] [CrossRef]
- Zapomělová, E.; Řeháková, K.; Znachor, P.; Komárková, J. Morphological diversity of coiled planktonic types of the genus Anabaena (cyanobacteria) in natural populations—Taxonomic consequences. Cryptogam. Algol. 2007, 28, 353–371. [Google Scholar]
- Wacklin, P.; Hoffmann, L.; Komárek, J. Nomenclatural validation of the genetically revised cyanobacterial genus Dolichospermum (RALFS ex BORNET et FLAHAULT) comb. nova. Fottea 2009, 9, 59–64. [Google Scholar] [CrossRef] [Green Version]
| No. | Name | apo [M + H]+ | Molecular Formula | tR (min) |
|---|---|---|---|---|
| 1 | Skizokinen | 421.1934 | C16H29O9N4+ | 4.7 |
| 2 | Synechobactin A | 561.3477 | C26H49O9N4+ | 30.7 |
| 3 | Synechobactin B | 533.3165 | C24H45O9N4+ | 27.7 |
| 4 | Synechobactin C14 | 589.3788 | C28H53O9N4+ | 33.0 |
| 5 | Synechobactin C16 | 617.4098 | C30H57O9N4+ | 35.1 |
| 6 | Synechobactin C16:1 | 615.3940 | C30H55O9N4+ | 33.6 |
| 7 | Synechobactin oxyC14 | 605.3734 | C28H53O10N4+ | 30.9 |
| 8 | Synechobactin oxyC14:1 | 603.3579 | C28H51O10N4+ | 30.7 |
| 9 | Desacetyl-synechobactin A | 517.3212 | C24H45O8N4+ | 30.3 |
| 10 | Desacetyl-synechobactin C14 | 545.3525 | C26H49O8N4+ | 32.7 |
| 11 | Deoxysynechobactin C14 | 573.3839 | C28H53O8N4+ | 32.4 |
| 12 | Deoxysynechobactin C14:1 | 571.3681 | C28H51O8N4+ | 33.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teta, R.; Esposito, G.; Kundu, K.; Stornaiuolo, M.; Scarpato, S.; Pollio, A.; Costantino, V. A Glimpse at Siderophores Production by Anabaena flos-aquae UTEX 1444. Mar. Drugs 2022, 20, 256. https://doi.org/10.3390/md20040256
Teta R, Esposito G, Kundu K, Stornaiuolo M, Scarpato S, Pollio A, Costantino V. A Glimpse at Siderophores Production by Anabaena flos-aquae UTEX 1444. Marine Drugs. 2022; 20(4):256. https://doi.org/10.3390/md20040256
Chicago/Turabian StyleTeta, Roberta, Germana Esposito, Karishma Kundu, Mariano Stornaiuolo, Silvia Scarpato, Antonino Pollio, and Valeria Costantino. 2022. "A Glimpse at Siderophores Production by Anabaena flos-aquae UTEX 1444" Marine Drugs 20, no. 4: 256. https://doi.org/10.3390/md20040256
APA StyleTeta, R., Esposito, G., Kundu, K., Stornaiuolo, M., Scarpato, S., Pollio, A., & Costantino, V. (2022). A Glimpse at Siderophores Production by Anabaena flos-aquae UTEX 1444. Marine Drugs, 20(4), 256. https://doi.org/10.3390/md20040256