Insights into the Influence of Signal Peptide on the Enzymatic Properties of Alginate Lyase AlyI1 with Removal Effect on Pseudomonas aeruginosa Biofilm
Abstract
1. Introduction
2. Results and Discussion
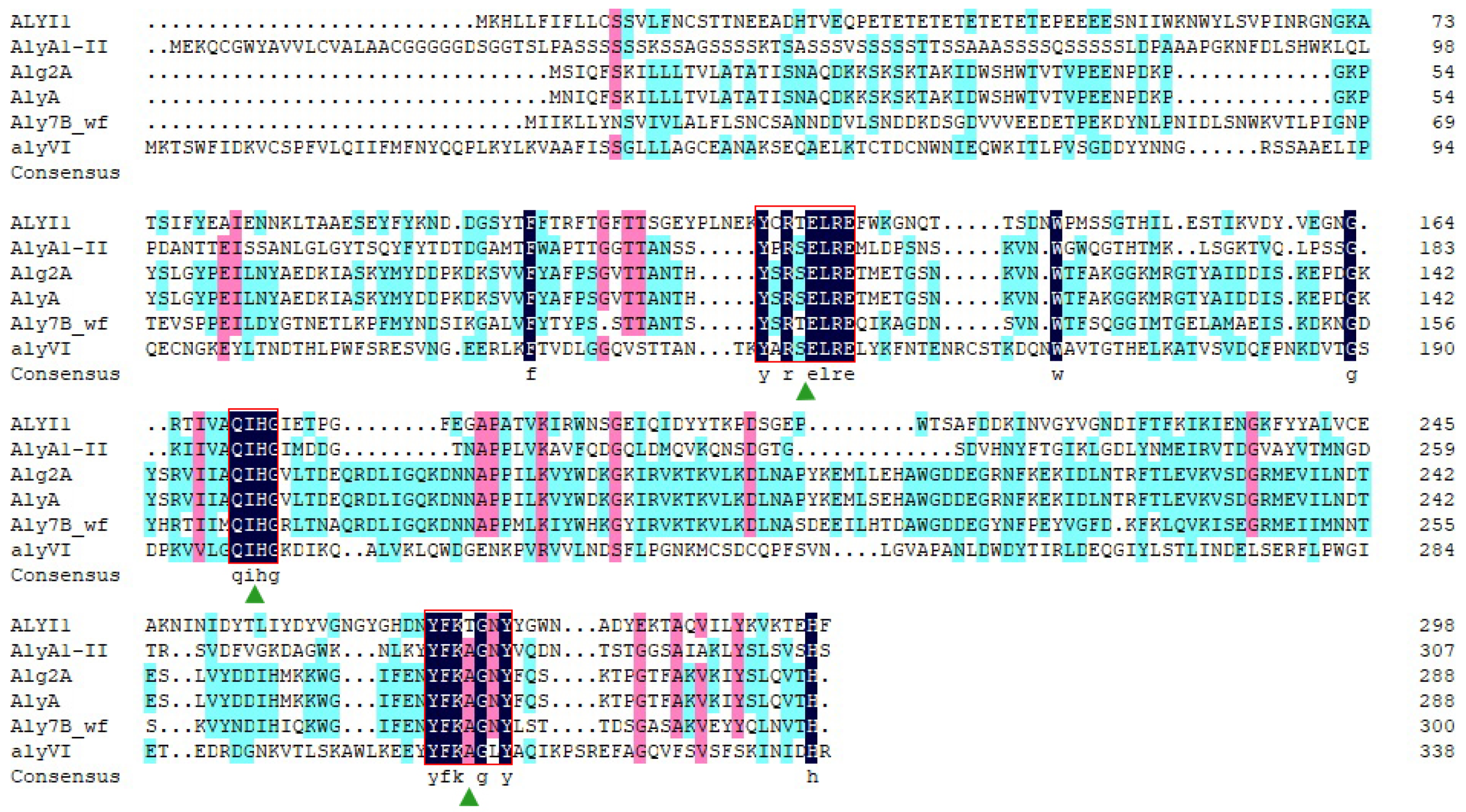
2.1. Sequence Analysis of Alyi1/alyI1-1
2.2. Expression, Purification, and Characterization of rALYI1/rALYI1-1
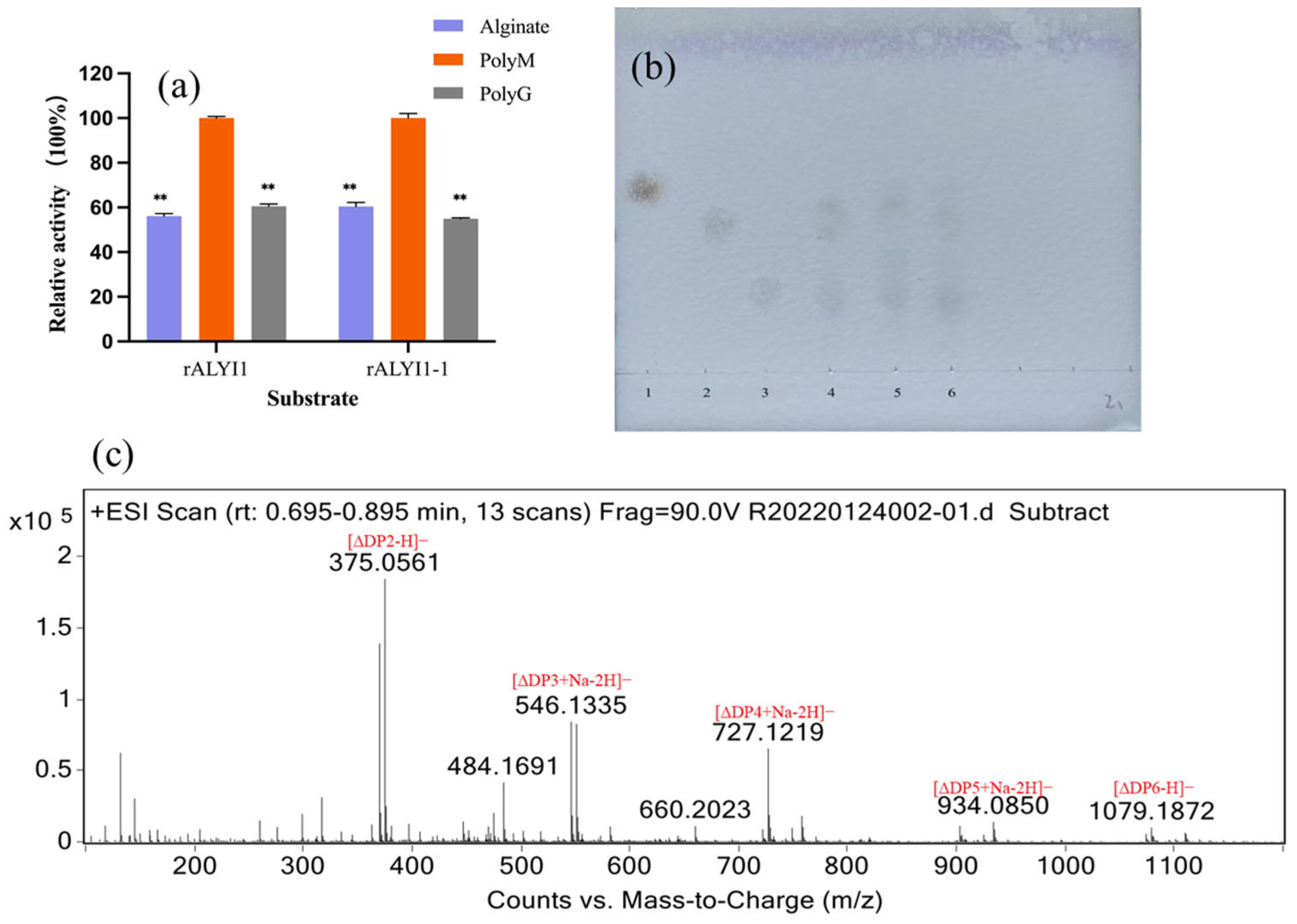
2.3. Analysis of Substrate Specificity and Final Products
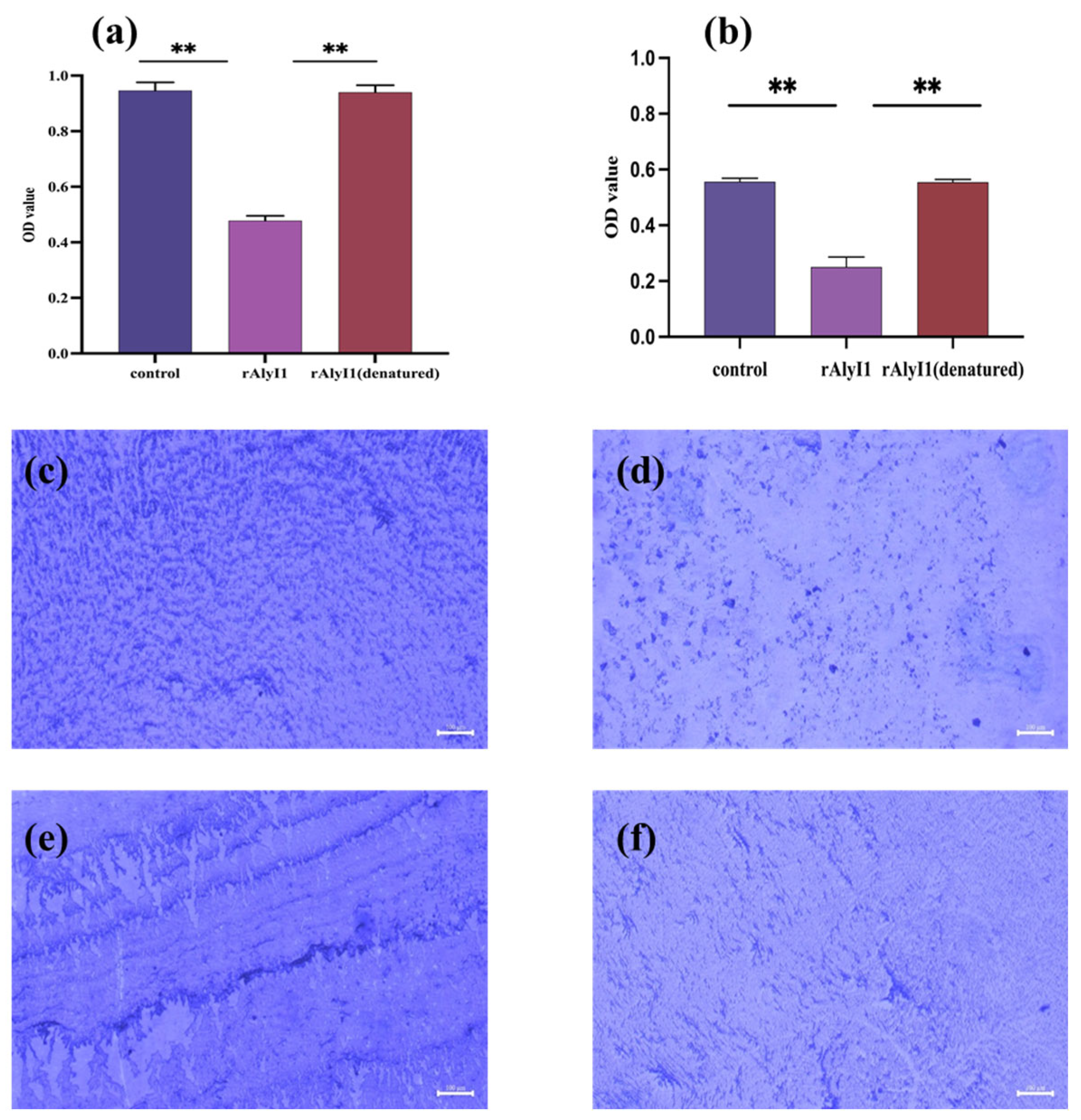
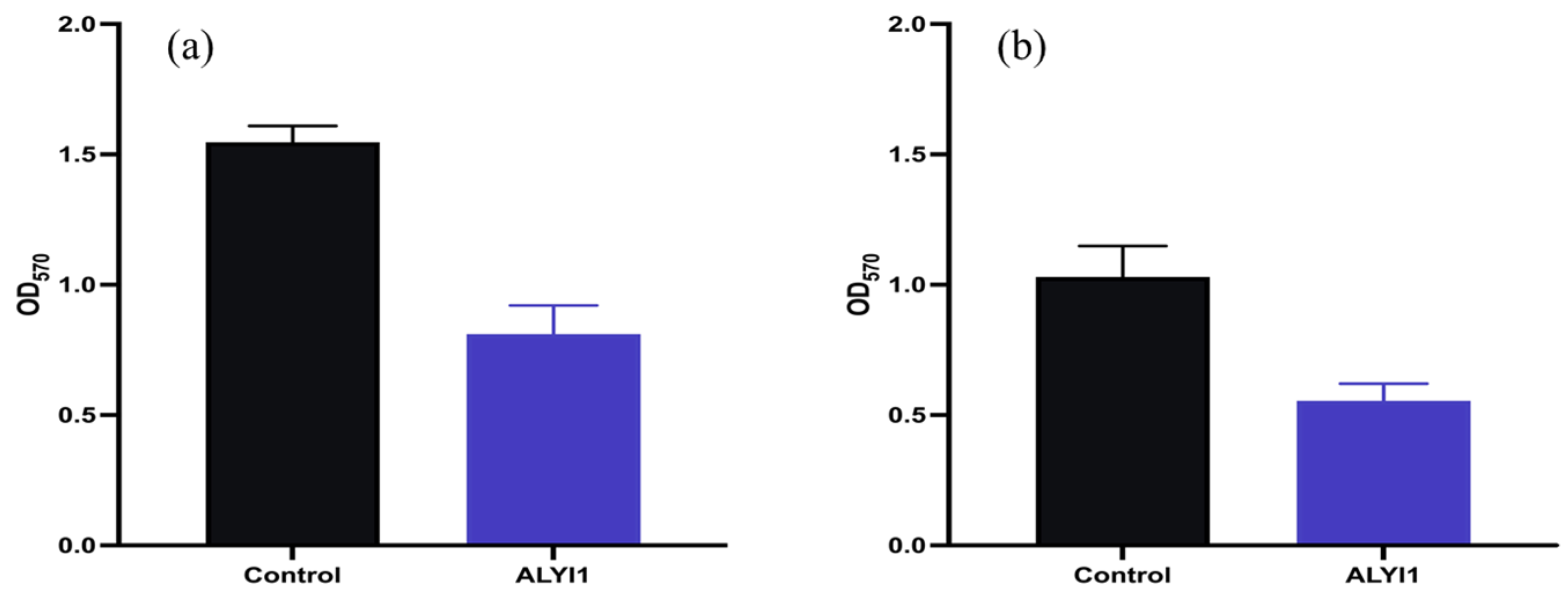
2.4. Effect of Alginate Lyase rALYI1 on Biofilm Produced by P. aeruginosa
3. Materials and Methods
3.1. Strains, Plasmids, and Reagents
3.2. Cloning, Expression, and Purification of rALYI1/rALYI1-1
3.3. Sequence Analysis of ALYI1
3.4. Enzyme Activity Assay
3.5. Characterization of rALYI1/rALYI1-1
3.5.1. Effect of Temperature on Enzyme Activity
3.5.2. Effect of pH on Enzyme Activity
3.5.3. Effects of Metal ions, Surfactants, and NaCl on Enzyme Activity
3.6. Substrate Specificity, Degradation Products, and Kinetic Parameters of rALYI1/rALYI1-1
3.7. Biofilm Formation and Removal
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blobel, G. Protein targeting (Nobel lecture). ChemBioChem 2000, 1, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Jonet, M.A.; Mahadi, N.M.; Murad, A.M.; Rabu, A.; Bakar, F.D.; Rahim, R.A.; Low, K.O.; Illias, R.M. Optimization of a heterologous signal peptide by site-directed mutagenesis for improved secretion of recombinant proteins in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2012, 22, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Rusch, S.L.; Kendall, D.A. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 2007, 46, 9665–9673. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, C.J. Signal peptides: New markers in cardiovascular disease? Biomark Med. 2014, 8, 1013–1019. [Google Scholar] [CrossRef]
- Cheng, K.W.; Wang, F.; Lopez, G.A.; Singamsetty, S.; Wood, J.; Dickson, P.I.; Chou, T.F. Evaluation of artificial signal peptides for secretion of two lysosomal enzymes in CHO cells. Biochem. J. 2021, 478, 2309–2319. [Google Scholar] [CrossRef]
- Sun, X.; Shen, W.; Gao, Y.; Cai, M.; Zhou, M.; Zhang, Y. Heterologous expression and purification of a marine alginate lyase in Escherichia coli. Protein Expr. Purif. 2019, 153, 97–104. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Qin, Y.; Shen, P.; Peng, Q. Structures, properties and application of alginic acid: A review. Int. J. Biol. Macromol. 2020, 162, 618–628. [Google Scholar] [CrossRef]
- Suda, K.; Tanji, Y.; Hori, K.; Unno, H. Evidence for a novel Chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999, 180, 45–53. [Google Scholar] [CrossRef]
- Rahman, M.M.; Inoue, A.; Tanaka, H.; Ojima, T. Isolation and characterization of two alginate lyase isozymes, AkAly28 and AkAly33, from the common sea hare Aplysia kurodai. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 157, 317–325. [Google Scholar] [CrossRef]
- Singh, R.P.; Gupta, V.; Kumari, P.; Kumar, M.; Reddy, C.R.K.; Prasad, K.; Jha, B. Purification and partial characterization of an extracellular alginate lyase from Aspergillus oryzae isolated from brown seaweed. J. Appl. Phycol. 2011, 23, 755–762. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, E.W.; Yu, W.G.; Han, F. Purification and characterization of a new alginate lyase from a marine bacterium Vibrio sp. Biotechnol. Lett. 2013, 35, 703–708. [Google Scholar] [CrossRef]
- Madgwick, J.; Haug, A.; Larsen, B. Alginate lyase in the brown alga Laminaria digitata (Huds.) Lamour. Acta. Chem. Scand. 1973, 27, 711–712. [Google Scholar] [CrossRef]
- Wang, D.M.; Kim, H.T.; Yun, E.J.; Kim, D.H.; Park, Y.C.; Woo, H.C.; Kim, K.H. Optimal production of 4-deoxy-L-erythro-5-hexoseulose uronic acid from alginate for brown macro algae saccharification by combining endo- and exo-type alginate lyases. Bioprocess Biosyst. Eng. 2014, 37, 2105–2111. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Rollet, C.; Gal, L.; Guzzo, J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: A comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 290, 135–142. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Ma, L.; Lu, H.; Sprinkle, A.; Parsek, M.R.; Wozniak, D.J. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 2007, 189, 8353–8356. [Google Scholar] [CrossRef]
- Alkawash, M.A.; Soothill, J.S.; Schiller, N.L. Alginate lyase enhances antibiotic killing ofmucoid Pseudomonas aeruginosa in biofilms. APMIS 2006, 114, 131–138. [Google Scholar] [CrossRef]
- Yoon, H.J.; Hashimoto, W.; Miyake, O.; Okamoto, M.; Mikami, B.; Murata, K. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr. Purif. 2000, 19, 84–90. [Google Scholar] [CrossRef]
- Zhu, B.W.; Ni, F.; Sun, Y.; Ning, L.M.; Yao, Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol. Biofuels 2019, 12, 13. [Google Scholar] [CrossRef]
- Ogura, K.; Yamasaki, M.; Mikami, B.; Hashimoto, W.; Murata, K. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J. Mol. Biol. 2008, 380, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, Q.; Lu, D.; Han, W.; Li, F. A Novel Bifunctional Endolytic Alginate Lyase with Variable Alginate-Degrading Modes and Versatile Monosaccharide-Producing Properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Bi, X.; Ren, Y.; Han, Q.; Zhou, Y.; Han, Y.; Yao, R.; Li, S. Characterization of an Alkaline Alginate Lyase with pH-Stable and Thermo-Tolerance Property. Mar. Drugs 2019, 17, 308. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, Z.; Li, S.; Su, H.; Tang, L.; Tan, Y.; Yu, W.; Han, F. Characterization of a new endo-type polysaccharide lyase (PL) family 6 alginate lyase with cold-adapted and metal ions-resisted property. Int. J. Biol. Macromol. 2018, 120 Pt A, 729–735. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, J.; Li, X.; Peng, Q.; Lu, H.; Du, Y. Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. J. Ind. Microbiol. Biotechnol. 2013, 40, 113–122. [Google Scholar] [CrossRef]
- Li, H.F.; Wang, S.L.; Zhang, Y.Y.; Chen, L.H. High-Level Expression of a Thermally Stable Alginate Lyase Using Pichia pastoris, Characterization and Application in Producing Brown Alginate Oligosaccharide. Mar. Drugs 2018, 16, 158. [Google Scholar] [CrossRef]
- Zhou, H.X.; Xu, S.S.; Yin, X.J.; Wang, F.L.; Li, Y. Characterization of a New Bifunctional and Cold-Adapted Polysaccharide Lyase (PL) Family 7 Alginate Lyase from Flavobacterium sp. Mar. Drugs 2020, 18, 388. [Google Scholar] [CrossRef]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an alginate lyase, FlAlyA, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef]
- Zhu, B.; Tan, H.; Qin, Y.; Xu, Q.; Du, Y.; Yin, H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, M.; Yin, H.; Du, Y.; Ning, L. Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase. Mar. Drugs 2016, 14, 108. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uchimura, K.; Miyazaki, M.; Nogi, Y.; Horikoshi, K. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 2009, 13, 121–129. [Google Scholar] [CrossRef]
- Uchimura, K.; Miyazaki, M.; Nogi, Y.; Kobayashi, T.; Horikoshi, K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 2010, 12, 526–533. [Google Scholar] [CrossRef]
- Huang, G.; Wang, Q.; Lu, M.; Xu, C.; Li, F.; Zhang, R.; Liao, W.; Huang, S. AlgM4: A New Salt-Activated Alginate Lyase of the PL7 Family with Endolytic Activity. Mar. Drugs 2018, 16, 120. [Google Scholar] [CrossRef]
- Chen, X.L.; Dong, S.; Xu, F.; Dong, F.; Li, P.Y.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z.; Xie, B.B. Characterization of a New Cold-Adapted and Salt-Activated Polysaccharide Lyase Family 7 Alginate Lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, 1120. [Google Scholar] [CrossRef]
- Meng, Q.; Tian, X.; Jiang, B.; Zhou, L.; Chen, J.; Zhang, T. Characterization and enhanced extracellular overexpression of a new salt-activated alginate lyase. J. Sci. Food Agric. 2021, 101, 5154–5162. [Google Scholar] [CrossRef]
- Mahajan, S.; Sunsunwal, S.; Gautam, V.; Singh, M.; Ramya, T. Biofilm inhibitory effect of alginate lyases on mucoid P. aeruginosa from a cystic fibrosis patient. Biochem. Biophys. Rep. 2021, 26, 101028. [Google Scholar] [CrossRef]
- Daboor, S.M.; Raudonis, R.; Cohen, A.; Rohde, J.R.; Cheng, Z. Marine Bacteria, A Source for Alginolytic Enzyme to Disrupt Pseudomonas aeruginosa Biofilms. Mar. Drugs 2019, 17, 307. [Google Scholar] [CrossRef]
- Ghadam, P.; Akhlaghi, F.; Ali, A.A. One-step purification and characterization of alginate lyase from a clinical Pseudomonas aeruginosa with destructive activity on bacterial biofilm. Iran J. Basic Med. Sci. 2017, 20, 467–473. [Google Scholar] [CrossRef]
- Blanco-Cabra, N.; Paetzold, B.; Ferrar, T.; Mazzolini, R.; Torrents, E.; Serrano, L.; LLuch-Senar, M. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020, 10, 9390. [Google Scholar] [CrossRef]
- Yin, R.; Yi, Y.J.; Chen, Z.; Wang, B.X.; Li, X.H.; Zhou, Y.X. Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features. Mar. Drugs 2021, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.S.; Salunke, B.K.; Kim, B.S. A rapid, sensitive, simple plate assay for detection of microbial alginate lyase activity. Enzym. Microb. Technol. 2015, 77, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta. Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Swift, S.M.; Hudgens, J.W.; Heselpoth, R.D.; Bales, P.M.; Nelson, D.C. Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 2014, 9, e112939. [Google Scholar] [CrossRef]
- Thorn, C.R.; Raju, D.; Lacdao, I.; Gilbert, S.; Sivarajah, P.; Howell, P.L.; Prestidge, C.A.; Thomas, N. Protective Liquid Crystal Nanoparticles for Targeted Delivery of PslG: A Biofilm Dispersing Enzyme. ACS Infect. Dis. 2021, 7, 2102–2115. [Google Scholar] [CrossRef]
- Grela, E.; Kozlowska, J.; Grabowiecka, A. Current methodology of MTT assay in bacteria—A review. Acta. Histochem. 2018, 120, 303–311. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.T.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef]



| Assignment | The Peak Area Ratio of ALYI1 (%) | The Peak Area Ratio of ALYI1-1 (%) |
|---|---|---|
| β-sheet | 36.8 | 35.8 |
| β-turn | 11.6 | 25.4 |
| 𝛼-Helix | 23.6 | 19.0 |
| Random | 28.0 | 19.8 |
| Kinetic Parameter | rALYI1 | rALYI1-1 |
|---|---|---|
| Vmax (μmol min−1 mg−1) | 16.9 ± 0.1 | 4.1 ± 0.2 |
| Km (mol/L) | 0.2 ± 0.1 | 1.2 ± 0.1 |
| Kcat (s−1) | 130.1 ± 3.8 | 5.3 ± 0.3 |
| Kcat/Km(s−1 mol−1) | 541.9 ± 20.2 | 4.3 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.-J.; Yun, S.-T.; Wang, X.-C.; Peng, L.-Y.; Dou, C.; Zhou, Y.-X. Insights into the Influence of Signal Peptide on the Enzymatic Properties of Alginate Lyase AlyI1 with Removal Effect on Pseudomonas aeruginosa Biofilm. Mar. Drugs 2022, 20, 753. https://doi.org/10.3390/md20120753
Zhang M-J, Yun S-T, Wang X-C, Peng L-Y, Dou C, Zhou Y-X. Insights into the Influence of Signal Peptide on the Enzymatic Properties of Alginate Lyase AlyI1 with Removal Effect on Pseudomonas aeruginosa Biofilm. Marine Drugs. 2022; 20(12):753. https://doi.org/10.3390/md20120753
Chicago/Turabian StyleZhang, Ming-Jing, Shuai-Ting Yun, Xiao-Chen Wang, Li-Yang Peng, Chuan Dou, and Yan-Xia Zhou. 2022. "Insights into the Influence of Signal Peptide on the Enzymatic Properties of Alginate Lyase AlyI1 with Removal Effect on Pseudomonas aeruginosa Biofilm" Marine Drugs 20, no. 12: 753. https://doi.org/10.3390/md20120753
APA StyleZhang, M.-J., Yun, S.-T., Wang, X.-C., Peng, L.-Y., Dou, C., & Zhou, Y.-X. (2022). Insights into the Influence of Signal Peptide on the Enzymatic Properties of Alginate Lyase AlyI1 with Removal Effect on Pseudomonas aeruginosa Biofilm. Marine Drugs, 20(12), 753. https://doi.org/10.3390/md20120753






