Thallusin Quantification in Marine Bacteria and Algae Cultures
Abstract
1. Introduction
2. Results
2.1. Method Development
2.2. Quantification of Thallusin in Bacterial Cultures
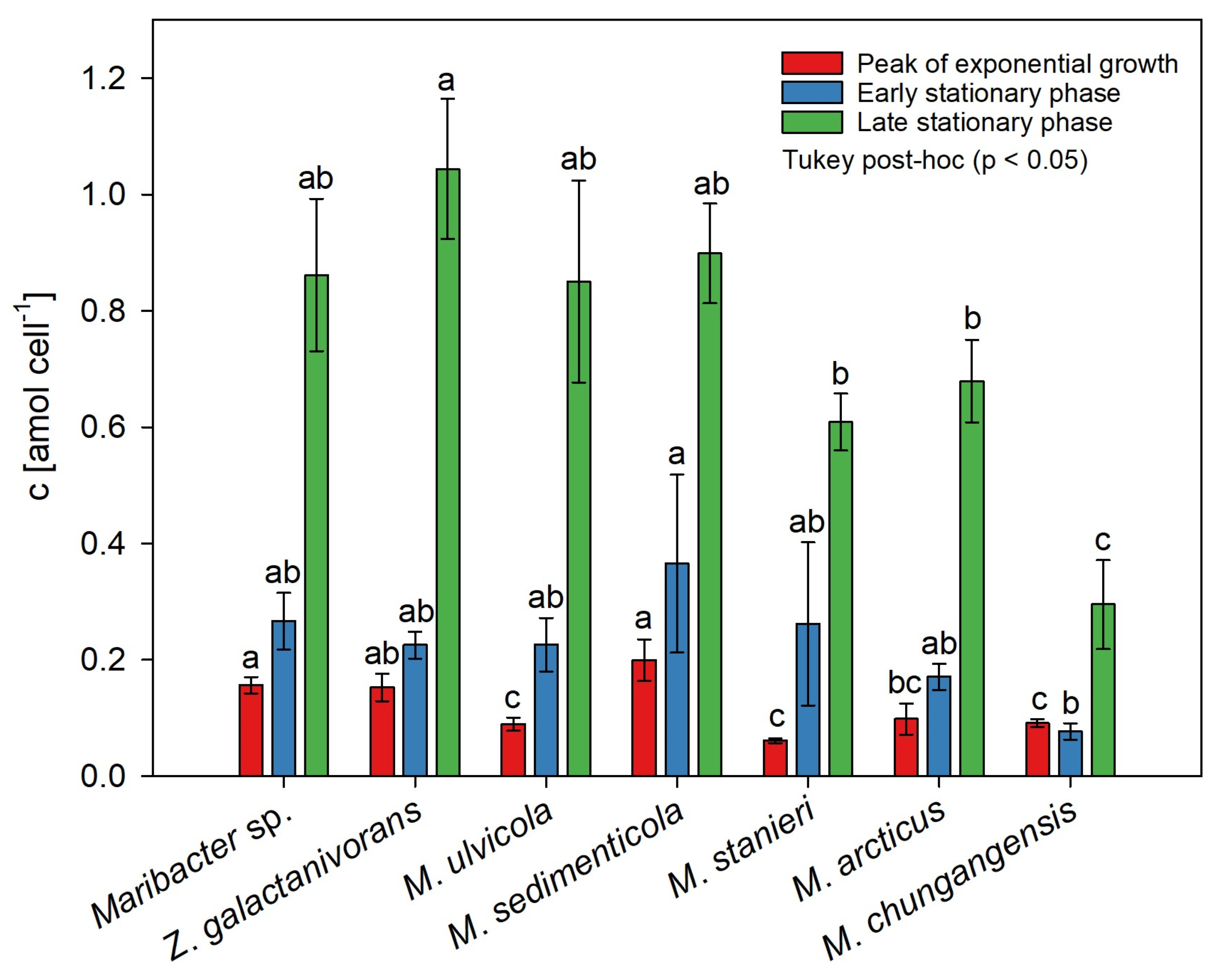
2.3. Species-Dependent Production of Thallusin
2.4. Quantification of Thallusin in Algal Cultures
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultivation Conditions
4.2. Chemicals, Standards, and Materials
4.3. Monitoring of Bacterial Growth
4.4. Sample Preparation
4.5. Solid Phase Extraction
4.6. Derivatization of Thallusin
4.7. Ultra-High-Performance Liquid Chromatography (UHPLC) Coupled with Electrospray Ionization (ESI) High-Resolution Mass Spectrometry (HRMS) Measurements
4.8. Calibration and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spoerner, M.; Wichard, T.; Bachhuber, T.; Stratmann, J.; Oertel, W. Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factors. J. Phycol. 2012, 48, 1433–1447. [Google Scholar] [CrossRef] [PubMed]
- Wichard, T. From model organism to application: Bacteria-induced growth and development of the green seaweed Ulva and the potential of microbe leveraging in algal aquaculture. Semin. Cell Dev. Biol. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, H.; Loret, E.P. Ulva lactuca, a source of troubles and potential riches. Mar. Drugs 2019, 17, 357. [Google Scholar] [CrossRef] [PubMed]
- Wichard, T. Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Front. Plant Sci. 2015, 6, 86. [Google Scholar] [CrossRef]
- Steinhagen, S.; Barco, A.; Wichard, T.; Weinberger, F. Conspecificity of the model organism Ulva mutabilis and Ulva compressa (Ulvophyceae, Chlorophyta). J. Phycol. 2019, 55, 25–36. [Google Scholar] [CrossRef]
- Alsufyani, T.; Califano, G.; Deicke, M.; Grueneberg, J.; Weiss, A.; Engelen, A.H.; Kwantes, M.; Mohr, J.F.; Ulrich, J.F.; Wichard, T. Macroalgal–bacterial interactions: Identification and role of thallusin in morphogenesis of the seaweed Ulva (Chlorophyta). J. Exp. Bot. 2020, 71, 3340–3349. [Google Scholar] [CrossRef]
- Akhtar, S.S.; Mekureyaw, M.F.; Pandey, C.; Roitsch, T. Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 2019, 10, 1777. [Google Scholar] [CrossRef]
- Su, Y.; Xia, S.; Wang, R.; Xiao, L. Phytohormonal quantification based on biological principles. Horm. Metab. Signal. Plants 2017, 13, 431–470. [Google Scholar]
- Dhiman, S.; Ulrich, J.F.; Wienecke, P.; Wichard, T.; Arndt, H.-D. Stereoselective total synthesis of (−)-thallusin for bioactivity profiling. Angew. Chem. Int. Ed. 2022, 61, e202206746. [Google Scholar] [CrossRef]
- Matsuo, Y.; Imagawa, H.; Nishizawa, M.; Shizuri, Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science 2005, 307, 1598. [Google Scholar] [CrossRef]
- Yamamoto, H.; Takagi, Y.; Yamasaki, N.; Mitsuyama, T.; Kasai, Y.; Imagawa, H.; Kinoshita, Y.; Oka, N.; Hiraoka, M. Syntheses of thallusin analogues and their algal morphogenesis-inducing activities. Tetrahedron 2018, 74, 7173–7178. [Google Scholar] [CrossRef]
- Davies, P. Plant Hormones: Biosynthesis, Signal, Transduction, Action; Springer Science & Business Media Press: New York, NY, USA, 2004. [Google Scholar]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Bhalerao, R.P.; Bennett, M.J. The case for morphogens in plants. Nat. Cell Biol. 2003, 5, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Paque, S.; Weijers, D. Q&A: Auxin: The plant molecule that influences almost anything. BMC Biol. 2016, 14, 67. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Brütting, C.; Baldwin, I.T.; Kallenbach, M. High-throughput quantification of more than 100 primary- and secondary-metabolites, and phytohormones by a single solid-phase extraction based sample preparation with analysis by UHPLC–HESI–MS/MS. Plant Meth. 2016, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Düchting, P.; Weiler, E.W. A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 2002, 216, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Tarakhovskaya, E.R.; Maslov, Y.I.; Shishova, M.F. Phytohormones in algae. Russ. J. Plant Physiol. 2007, 54, 163–170. [Google Scholar] [CrossRef]
- Yalçın, S.; Şükran Okudan, E.; Karakaş, Ö.; Önem, A.N.; Sözgen Başkan, K. Identification and quantification of some phytohormones in seaweeds using UPLC-MS/MS. J. Liq. Chromatogr. Relat. Technol. 2019, 42, 475–484. [Google Scholar] [CrossRef]
- Mikami, K.; Mori, I.C.; Matsuura, T.; Ikeda, Y.; Kojima, M.; Sakakibara, H.; Hirayama, T. Comprehensive quantification and genome survey reveal the presence of novel phytohormone action modes in red seaweeds. J. Appl. Phycol. 2016, 28, 2539–2548. [Google Scholar] [CrossRef]
- Mori, I.C.; Ikeda, Y.; Matsuura, T.; Hirayama, T.; Mikami, K. Phytohormones in red seaweeds: A technical review of methods for analysis and a consideration of genomic data. Bot. Mar. 2017, 60, 153–170. [Google Scholar] [CrossRef]
- Almeida Trapp, M.; De Souza, G.D.; Rodrigues-Filho, E.; Boland, W.; Mithöfer, A. Validated method for phytohormone quantification in plants. Front. Plant Sci. 2014, 5, 417. [Google Scholar] [CrossRef] [PubMed]
- Alsufyani, T.; Weiss, A.; Wichard, T. Time course exo-metabolomic profiling in the green marine macroalga Ulva (Chlorophyta) for identification of growth phase-dependent biomarkers. Mar. Drugs 2017, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Avila-Zárraga, J.G.; Martínez, R. Efficient methylation of carboxylic acids with potassium hydroxide/methyl sulfoxide and iodomethane. Synth. Commun. 2001, 31, 2177–2183. [Google Scholar] [CrossRef]
- Ghaderiardakani, F.; Califano, G.; Mohr, J.F.; Abreu, M.H.; Coates, J.C.; Wichard, T. Analysis of algal growth- and morphogenesis-promoting factors in an integrated multi-trophic aquaculture system for farming Ulva spp. Aquac. Environ. Interact. 2019, 11, 375–391. [Google Scholar] [CrossRef]
- Abrahim, A.; Hartman, R.; Ge, Z.; Mao, B.; Marcoux, J. Development of a derivatization method, coupled with reverse phase HPLC, for monitoring the formation of an enolate intermediate. J. Liq. Chromatogr. Relat. Technol. 2002, 25, 1049–1062. [Google Scholar] [CrossRef]
- Lee, S.T.; Mitchell, R.B.; Wang, Z.; Heiss, C.; Gardner, D.R.; Azadi, P. Isolation, characterization, and quantification of steroidal saponins in switchgrass (Panicum virgatum L.). J. Agric. Food Chem. 2009, 57, 2599–2604. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Sato, Y.; Sakurai, T.; Yamasaki, T.; Yamamoto, H.; Hiraoka, M. Development of blade cells and rhizoid cells aseptically isolated from the multicellular leafy seaweed Gayralia oxysperma. Cytologia 2022, 87, 17–22. [Google Scholar] [CrossRef]
- Maruyama, A.; Maeda, M.; Simidu, U. Distribution and classification of marine bacteria with the ability of cytokinin and auxin production. Bull. Jap. Soc. Microb. Ecol. 1990, 5, 1–8. [Google Scholar] [CrossRef][Green Version]
- Ashen, J.B.; Cohen, J.D.; Goff, L.J. GC-SIM-MS detection and quantification of free indole-3-acetic acid in bacterial galls on the marine alga Prionitis lanceolata (Rhodophyta). J. Phycol. 1999, 35, 493–500. [Google Scholar] [CrossRef]
- De Clerck, O.; Kao, S.-M.; Bogaert, K.A.; Blomme, J.; Foflonker, F.; Kwantes, M.; Vancaester, E.; Vanderstraeten, L.; Aydogdu, E.; Boesger, J.; et al. Insights into the Evolution of Multicellularity from the Sea Lettuce Genome. Curr. Biol. 2018, 28, 2921–2933.e2925. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Kumar, M.; Brahmbhatt, H.; Reddy, C.R.K.; Seth, A.; Jha, B. Simultaneous determination of different endogenetic plant growth regulators in common green seaweeds using dispersive liquid–liquid microextraction method. Plant Physiol. Biochem. 2011, 49, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, T.; Mori, I.C.; Ikeda, Y.; Hirayama, T.; Mikami, K. Comprehensive phytohormone quantification in the red alga Pyropia yezoensis by liquid chromatography–mass spectrometry. In Protocols for Macroalgae Research; Charrier, B., Wichard, T., Reddy, C., Eds.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Yalçın, S.; Okudan, E.Ş.; Karakaş, Ö.; Önem, A.N. Determination of major phytohormones in fourteen different seaweeds utilizing SPE–LC–MS/MS. J. Chromatogr. Sci. 2019, 58, 98–108. [Google Scholar] [CrossRef]
- Kojima, M.; Kamada-Nobusada, T.; Komatsu, H.; Takei, K.; Kuroha, T.; Mizutani, M.; Ashikari, M.; Ueguchi-Tanaka, M.; Matsuoka, M.; Suzuki, K.; et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009, 50, 1201–1214. [Google Scholar] [CrossRef]
- Wolter, L.A.; Mitulla, M.; Kalem, J.; Daniel, R.; Simon, M.; Wietz, M. CAZymes in Maribacter dokdonensis 62–1 from the patagonian shelf: Genomics and physiology compared to related Flavobacteria and a co-occurring Alteromonas strain. Front. Microbiol. 2021, 12, 628055. [Google Scholar] [CrossRef]
- Matsuda, R.; Handayani, M.L.; Sasaki, H.; Takechi, K.; Takano, H.; Takio, S. Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol. 2018, 200, 255–265. [Google Scholar] [CrossRef]
- Zluhan-Martínez, E.; López-Ruíz, B.A.; García-Gómez, M.L.; García-Ponce, B.; de la Paz Sánchez, M.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Integrative roles of phytohormones on cell proliferation, elongation and differentiation in the Arabidopsis thaliana primary root. Front. Plant Sci. 2021, 12, 659155. [Google Scholar] [CrossRef]
- Ahmed, A.; Hasnain, S. Auxin-producing Bacillus sp.: Auxin quantification and effect on the growth of Solanum tuberosum. Pure Appl. Chem. 2010, 82, 313–319. [Google Scholar] [CrossRef]
- Ali, B.; Sabri, A.N.; Ljung, K.; Hasnain, S. Auxin production by plant associated bacteria: Impact on endogenous IAA content and growth of Triticum aestivum L. Lett. Appl. Microbiol. 2009, 48, 542–547. [Google Scholar] [CrossRef]
- Benítez García, I.; Dueñas Ledezma, A.K.; Martínez Montaño, E.; Salazar Leyva, J.A.; Carrera, E.; Osuna Ruiz, I. Identification and quantification of plant growth regulators and antioxidant compounds in aqueous extracts of Padina durvillaei and Ulva lactuca. Agronomy 2020, 10, 866. [Google Scholar] [CrossRef]
- Paliwal, C.; Jutur, P.P. Dynamic allocation of carbon flux triggered by task-specific chemicals is an effective non-gene disruptive strategy for sustainable and cost-effective algal biorefineries. Chem. Eng. J. 2021, 418, 129413. [Google Scholar] [CrossRef]
- Franz, A.K.; Danielewicz, M.A.; Wong, D.M.; Anderson, L.A.; Boothe, J.R. Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity. ACS Chem. Biol. 2013, 8, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Polikovsky, M.; Califano, G.; Dunger, N.; Wichard, T.; Golberg, A. Engineering bacteria -seaweed symbioses for modulating the photosynthate content of Ulva (Chlorophyta): Significant for the feedstock of bioethanol production. Algal Res. 2020, 49, 101945. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Kim, S.B.; Han, S.K.; Lysenko, A.M.; Rohde, M.; Rhee, M.-S.; Frolova, G.M.; Falsen, E.; Mikhailov, V.V.; Bae, K.S. Maribacter gen. nov., a new member of the family Flavobacteriaceae, isolated from marine habitats, containing the species Maribacter sedimenticola sp. nov., Maribacter aquivivus sp. nov., Maribacter orientalis sp. nov. and Maribacter ulvicola sp. nov. Int. J. Syst. Evol. 2004, 54, 1017–1023. [Google Scholar] [CrossRef]
- Barbeyron, T.; L’Haridon, S.; Corre, E.; Kloareg, B.; Potin, P. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int. J. Syst. Evol. 2001, 51, 985–997. [Google Scholar] [CrossRef]
- Weerawongwiwat, V.; Kang, H.; Jung, M.Y.; Kim, W. Maribacter chungangensis sp. nov., isolated from a green seaweed, and emended descriptions of the genus Maribacter and Maribacter arcticus. Int. J. Syst. Evol. Microbiol. 2013, 63, 2553–2558. [Google Scholar] [CrossRef]
- Hahnke, R.L.; Harder, J. Phylogenetic diversity of Flavobacteria isolated from the North Sea on solid media. Syst. Appl. Microbiol. 2013, 36, 497–504. [Google Scholar] [CrossRef]
- Weiss, A.; Costa, R.; Wichard, T. Morphogenesis of Ulva mutabilis (Chlorophyta) induced by Maribacter species (Bacteroidetes, Flavobacteriaceae). Bot. Mar. 2017, 60, 197–206. [Google Scholar] [CrossRef]
- Hahnke, R.L.; Bennke, C.M.; Fuchs, B.M.; Mann, A.J.; Rhiel, E.; Teeling, H.; Amann, R.; Harder, J. Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea. Environ. Microbiol. 2015, 17, 3515–3526. [Google Scholar] [CrossRef]
- Nahor, O.; Morales-Reyes, C.F.; Califano, G.; Wichard, T.; Golberg, A.; Israel, Á. Flow cytometric measurements as a proxy for sporulation intensity in the cultured macroalga Ulva (Chlorophyta). Bot. Mar. 2021, 64, 83–92. [Google Scholar] [CrossRef]
- Deicke, M.; Bellenger, J.P.; Wichard, T. Direct quantification of bacterial molybdenum and iron metallophores with ultra-high-performance liquid chromatography coupled to time-of-flight mass spectrometry. J. Chromatogr. A 2013, 1298, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Reichenbächer, M.; Einax, J.W. The lack-of-fit test by ANOVA. In Challenges in Analytical Quality Assurance; Springer: Berlin/Heidelberg, Germany, 2011; pp. 136–138. [Google Scholar]
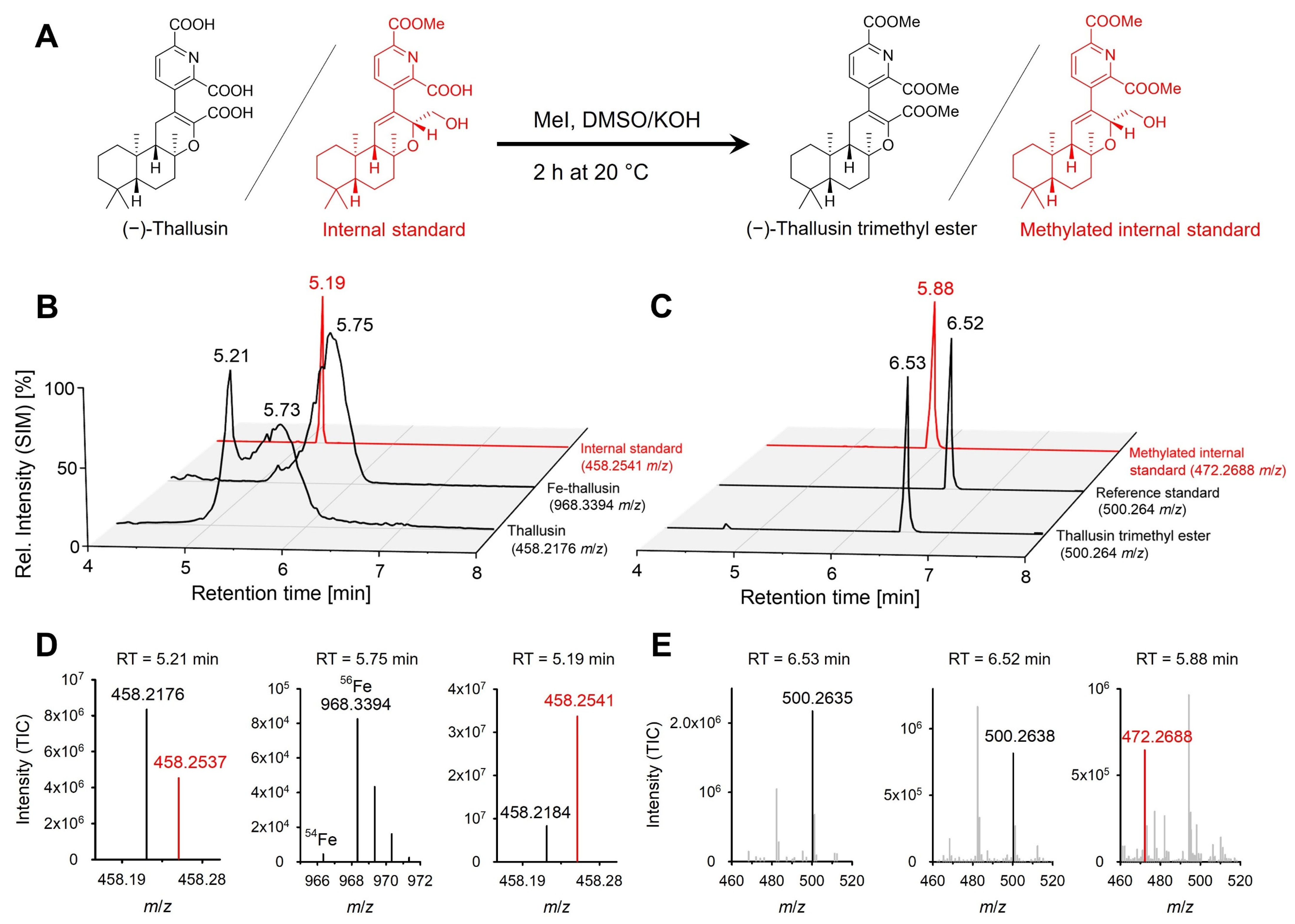




| Parameter | Value |
|---|---|
| Intercept, a0 | 0 |
| Slope, a1 | 0.177 |
| Calibration error, sy,x | 0.008 |
| Analytical standard deviation, sx,0 | 0.047 µmol L−1 |
| Critical measured value, yc | 0.013 |
| Relative standard deviation, sr | 0.758% |
| Number of calibration standards, nc | 7 |
| Limit of detection (LOD) 1 | 0.0756 µmol L−1 |
| Limit of quantification (LOQ) 1 | 0.2217 µmol L−1 |
| Thallusin Concentration [nmol L−1] | |||||
|---|---|---|---|---|---|
| Name | Strain | NCBI/GenBank Accession Number | OD620 = 0.15 | OD620 = 0.25 | Late Stationary Phase |
| Maribacter sp. | MS6 | EU359911 | 1.46 ± 0.14 | 4.29 ± 0.83 | 11.21 ± 1.68 |
| Maribacter arcticus | DSMZ 23546T | AY771762 | 0.89 ± 0.31 | 2.82 ± 0.37 | 9.08 ± 0.95 |
| Maribacter chungangensis | CCUG 61948T | JN036550 | 0.82 ± 0.08 | 1.05 ± 0.19 | 3.64 ± 0.96 |
| Maribacter sedimenticola | DSMZ 19840T | AY271623 | 1.71 ± 0.34 | 3.87 ± 1.60 | 9.18 ± 0.90 |
| Maribacter stanieri | DSMZ 19891T | EU246691 | 0.67 ± 0.04 | 3.59 ± 1.98 | 6.79 ± 0.45 |
| Maribacter ulvicola | DSMZ 15366T | AY271626 | 1.41 ± 0.15 | 3.96 ± 0.87 | 12.67 ± 2.58 |
| Zobellia galactanivorans | DsijT | GCA_ 000973105 | 1.38 ± 0.22 | 3.57 ± 0.50 | 14.81 ± 1.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulrich, J.F.; Gräfe, M.S.; Dhiman, S.; Wienecke, P.; Arndt, H.-D.; Wichard, T. Thallusin Quantification in Marine Bacteria and Algae Cultures. Mar. Drugs 2022, 20, 690. https://doi.org/10.3390/md20110690
Ulrich JF, Gräfe MS, Dhiman S, Wienecke P, Arndt H-D, Wichard T. Thallusin Quantification in Marine Bacteria and Algae Cultures. Marine Drugs. 2022; 20(11):690. https://doi.org/10.3390/md20110690
Chicago/Turabian StyleUlrich, Johann F., Melina S. Gräfe, Seema Dhiman, Paul Wienecke, Hans-Dieter Arndt, and Thomas Wichard. 2022. "Thallusin Quantification in Marine Bacteria and Algae Cultures" Marine Drugs 20, no. 11: 690. https://doi.org/10.3390/md20110690
APA StyleUlrich, J. F., Gräfe, M. S., Dhiman, S., Wienecke, P., Arndt, H.-D., & Wichard, T. (2022). Thallusin Quantification in Marine Bacteria and Algae Cultures. Marine Drugs, 20(11), 690. https://doi.org/10.3390/md20110690





