Abstract
Tuberculosis has become a major health problem globally. This is worsened by the emergence of resistant strains of Mycobacterium tuberculosis showing ability to evade the effectiveness of the current antimycobacterial therapies. Therefore, the efforts carried out to explore new entities from many sources, including marine, are critical. This review summarizes several marine-derived macrolides that show promising activity against M. tuberculosis. We also provide information regarding the biosynthetic processes of marine macrolides, including the challenges that are usually experienced in this process. As most of the studies reporting the antimycobacterial activities of the listed marine macrolides are based on in vitro studies, the future direction should consider expanding the trials to in vivo and clinical trials. In addition, in silico studies should also be explored for a quick screening on marine macrolides with potent activities against mycobacterial infection. To sum up, macrolides derived from marine organisms might become therapeutical options for tackling antimycobacterial resistance of M. tuberculosis.
1. Introduction
Tuberculosis (TB) has become a global burden for years, especially in the developing areas of the world. A recent report by the World Health Organization showed that a total of 1.5 million TB-linked deaths was recorded in 2020 [1]. From the same report, it was also reported that the efforts for treating patients suffering from TB experienced an unprecedented obstacle because of COVID-19. This pandemic has limited access to provide the appropriate treatments for TB patients. For example, the number of patients treated with a drug regimen for drug-resistant TB dropped by 15%. Furthermore, the number of those receiving TB-preventive treatments also experienced a decrease of 21% [1].
Like other microbial infections, treatment of TB is always overshadowed by the case of resistance. It has been demonstrated that Mycobacterium tuberculosis, the causative agent of TB, has developed a number of modes used to evade the therapeutical actions of the current TB drugs [2,3,4]. Consequently, new strategies should be applied to counter this issue.
One of the reasonable strategies is to find and develop new TB drugs. It is noteworthy that the new TB drugs should try to explore drug candidates with a different core structure, potency, or mechanism than the currently used TB drugs. At this point, the efforts for discovering new TB drugs should examine the potencies of drug candidates originating from marine sources. However, this strategy is limited by the fear of excessive exploitation of natural marine resources. To counter this obstacle, chemical synthesis equipped with the knowledge of the structure–activity relationship could provide a breakthrough to avoid unexpected damages to the marine environment.
Macrolides are a type of polyketide antibiotic. They obstruct protein synthesis by binding to the bacterial 50S ribosomal subunit at the peptidyl transferase center formed by 23S rRNA [5]. Since the discovery of the first macrolide, pikromycin, in 1950 from an actinomycete, many macrolides have been discovered and classified into different groups [6]. Each macrolide molecule is distinguished by three structural elements: a macrocyclic lactone ring, multiple ketone and hydroxyl groups, and two deoxy sugars linked by a glycosidic bond [7]. The number of carbons in the lactone ring divides macrolides into several groups, including 12-membered rings (i.e., methymicin, neomethymycin, and litorin), 14-membered rings (i.e., erythromycin A and F, oleandomycin, anthracimycin, clarithromycin, roxithromycoin, and sporamicin), 15-membered rings (i.e., azithromycin), and 16-membered rings (i.e., tylosin, josamycin, kitasamycin, spiramycin, and midecamycin).
Although their use in the treatment of TB is not as frequent as the other TB drugs, e.g., isoniazid, ethambutol, and pyrazinamide, macrolides might act as a therapeutical option in a TB regimen. Several macrolides, e.g., erythromycin, azithromycin, and clarithromycin, have been used in treating a number of microbial infections, including TB [8]. However, it has been reported that the effectiveness of these macrolides in fighting the microbes decreases over time [9,10,11]. Therefore, the seeking of new macrolides from various sources, including from the ocean, with a promising antimicrobial action is pivotal.
As reviewed by Das et al., marine macrolides possess various potential biological activities, such as anti-inflammatory and anticancer activities, including their potent ability as antimicrobial agents [12]. Here, we review several macrolides obtained from marine organisms showing a promising potency as antimycobacterial agents. Although several studies have reported the presence and identification of marine-based macrolides, to the best of our knowledge, no studies have tried to collect valuable information regarding the specific uses of marine macrolides as antimycobacterial agents. At this point, this review aims to fill that gap and provides the lost brick needed to comprehensively understand the antimycobacterial potencies of marine-based macrolides.
2. Materials and Methods
Available articles from three bibliographical databases, e.g., Google Scholar, Scopus, and PubMed, were searched. The search criteria were represented by the keywords used, i.e., (“macrolide” OR “macrolides”) AND (“marine” OR “sea” OR “ocean” OR “coral” OR “algae” OR “sponge”) AND (“antimycobacterial” OR “mycobacterial” OR “Mycobacterium” OR “anti-Mycobacterium tuberculosis” OR “Mycobacterium tuberculosis” OR “M. tuberculosis” OR “tuberculosis” OR “mycobacterial infections”).
3. Structure–Activity Relationship of Macrolides
Structure–activity relationship (SAR) studies explore the relationships between the chemical structure and biological activity of a molecule [13]. Various SAR studies of macrolides have been reported to identify the correlation of functional elements essential for maintaining antibacterial activity [14,15]. Structurally, macrolides contain a macrocyclic lactone of different ring sizes and are decorated with one or more deoxy-sugar or amino sugar residues [16]. Macrolides display broad-spectrum activity against many Gram-positive bacteria by binding to bacterial 50S ribosomal subunits and interfering with the synthesis of protein [17]. The first-generation prototypical macrolide, erythromycin, is a naturally occurring antibiotic produced by Streptomyces erythreus, currently reclassified as Saccharopolyspora erythraea. Erythromycin has several co-metabolites (A, B, C, D), of which the derivative A (erythromycin A) is the most dominant and most used product in therapy [14,18].
Erythromycin possesses a 14-membered macrolactone ring called erythronolide and is attached with L-cladinose (a neutral sugar in C-3 position) and D-desosamine (an amino sugar in C-5 position). The integrity of erythronolide including 9-oxime is essential for antibacterial activity [14]. Moreover, the presence of the methyl groups in an alpha configuration in the C-4, C-6, C-8, and C-12 positions as well as the methyl groups in a beta configuration in the C-2 and C-10 positions is also essential for the appearance of activity. In addition, the hydroxyl groups in beta configuration in the C-6, C-11, and C-12 positions are essential for induction of antibacterial activity. Furthermore, the removal of D-desosamine in the C-5 position would cause a total loss of activity. The integrity of aglycone could be maintained by the presence of an ethyl group in an alpha configuration in the C-13 position, which would be essential for protection against the opening of the cyclic ester function (Figure 1A) [14,19].
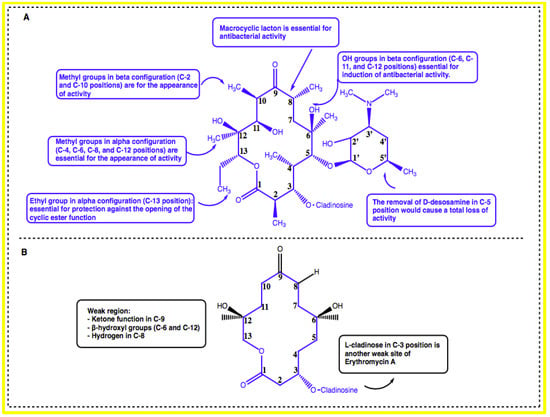
Figure 1.
(A) Structure–activity relationship study of erythromycin. (B) The weak regions of erythromycin.
The weak regions (inactivation sites) of erythromycin A have also been identified by SAR studies. The ketone function in the C-9 position, beta-hydroxyl groups in the C-6 and C-12 positions, and hydrogen in the C-8 position are responsible for the formation of inactive hemiketal and spiroketal derivatives, which are responsible for the digestive intolerance of erythromycin A. Additionally, the presence of L-cladinose in the C-3 position is another weak site of erythromycin A (Figure 1B). This entity could be hydrolyzed in gastric-acid medium to form an inactive 3-hydroxy erythromycin A derivative, which is responsible for the appearance of drug-resistant germs [14].
The second-generation macrolides, clarithromycin, roxithromycin, and dirithromycin, are semisynthetic derivatives of the 14-membered ring-macrolide erythromycin A and the 15-membered one (azithromycin). Clarithromycin has a methoxy group at the C-6 position of the lactone ring. Flurithromycin has a fluorine atom at the C-8 position (alpha to the ketone carbonyl group) and roxithromycin has an etheroxime chain at the C-9 position. Azithromycin has an azalide group at the C-9a position. These derivatives contain all modifications at the C-6 or C-9 positions of the macrocyclic lactone, consequently preventing the formation of hemiketal and spiroketal derivatives [20].
Existing 14-, 15-, and 16-membered macrolides, although effective for other bacterial infections, did not display significant efficacy in treating tuberculosis [20]. Studies of the activity of macrolides against Mycobacterium tuberculosis are very restricted. Recently, Zhang et al. studied the antituberculosis potency of clarithromycin. They reported that a single methyl group in clarithromycin improves its potency against M. tuberculosis. Interestingly, they also found that the allosteric dynamics of A2062 by ribosome–clarithromycin complex may have great potential to increase the drug efficacy and may help to design the next generation of antituberculosis drugs to fight against multidrug-resistance tuberculosis [21].
Zhu et al. found that the appropriate substitutions on the C-9, C-11, C-12, or C6 positions in the macrolactone ring have good activity against M. tuberculosis. This group also showed that L-cladinosine located in the C-3 position is critical for macrolides in exerting their anti-mycobacterial activities (Figure 2) [20].
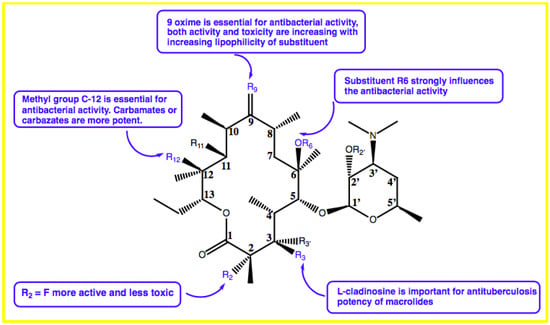
Figure 2.
Structure–activity relationship study of macrolides.
4. Biosynthesis of Marine Macrolides
4.1. Biosynthesis Challenges
Macrolides are extremely important as pharmaceutical leads, yet the significant slowing of innovative chemical development has become a major concern. Theoretically, macrocyclic systems can be created by cyclizing open, long-chain progenitors or cleaving internal polycyclic bonds. However, due to the structural complexity of macrocyclic natural compounds, difficulties in derivatizing them by chemical synthesis pose a barrier to pharmaceutical development. Moreover, several problems exist during the synthesis process to achieve the desired compounds. Thus, genetic modification can generate a huge number of congeners from known valuable natural substances.
Generally, the biosynthesis of marine macrolides, typified by erythromycin derivatives, is catalyzed by modular type I polyketide synthase (PKS) and modified by tailoring enzymes such as cytochrome P450 (CYP450), glycosyltransferase (GT), and oxidation enzymes such as monooxygenase (MO), methyltransferase (MT), and oxidoreductase (OR) [22,23,24]. The type I PKSs are multi-modular enzymes with non-iterative catalysis of one cycle of polyketide-chain elongation that are liable for consecutive condensation of activated coenzyme A (CoA) thioesters, including acetyl-CoA, propionyl-CoA, malonyl-CoA, or methylmalonyl-CoA. Each module minimally contains a set of functional domains, acyltransferase (AT), β-ketosynthase (KS), and acyl carrier protein (ACP) that are required for growing a polyketide chain and generating a β-ketoacyl-S-ACP intermediate [25,26]. In addition, the modules may also contain domains that consecutively modify the β-keto group to a hydroxyl group (ketoreductase, KR), a double bound (dehydrase, DH), and a single bond (enoyl reductase, ER) [26,27]. The ACP provides both intermediates of polyketide and building blocks to the catalytic domains for loading, extension, and processing of the keto-group utilizing thioester (TE) linkages and a tethered phosphopantetheine arm [28]. Mostly, a TE domain in the terminal module releases the intermediates, which are fully processed via cyclization or hydrolysis of macrolactone [26,29].
After PKS-mediated biosynthesis of aglycones, the post-PKS modification extensively occurs to generate the structure of the final macrolide. Figure 3 shows the precursor-directed biosynthesis of erythromycin, which is divided into two stages [26]. First, 6-deoxyerythronolide PKS (DEBS), the modular PKS complex, catalyzes the sequential condensation of six methylmalonyl-CoA precursors and one propionyl-CoA precursor to generate macrolide 6-deoxyerythronolide B (6-dEB). Second, 6-dEB is converted to erythronolide B by EryF hydroxylase. Then, EryBV glycosyltransferase transfers L-mycarose to erythronolide B, forming 3-O-mycarosylerythronolide B. EryCII activates EryCIII, completing the attachment of two deoxy sugars to the aglycone ring by transferring D-desosamine to C-5 hydroxyl, yielding erythromycin D as the first bioactive intermediate. Subsequently, methylation of 3′′-OH of L-mycarose by EryG methyltransferase and hydroxylation of C-12 by EryK hydroxylase generates the final product (erythromycin) [26].
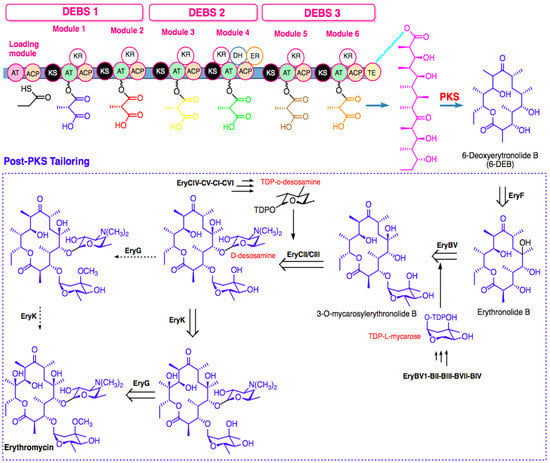
Figure 3.
Biosynthesis-scheme assembly of erythromycin.
Fundamentally, there are four steps throughout biosynthesis for diversification of a macrolide: (1) choice of the length of the chain and building block; (2) reduction and stereochemical arrangement of β-keto intermediates, including primary cyclization, branching, and alkylation; (3) rearrangement and secondary cyclization; and (4) tailoring of post-PKS [22,30]. Furthermore, it has been reported that the AT domain of modular PKSs controls the specific extender unit selected by each module, which naturally offer large portions of the polyketide structure, as these extender units are gathered into scaffolds of natural product. The ATs were able to discriminate between extender units to the PKS in the producing organism [31].
Biosynthesis of macrolides utilizing modular type I PKSs are usually bounded in scope and utility due to the limited substrate specificity of the polyketide biosynthetic machinery [26]. Recent information on the protein structure of enzymes and the advanced technique of genetic manipulation presents an immense opportunity for fine-tuning the step of post-PKS to yield structurally diversified or novel macrolides [32]. The diverse modularity in the genetic architecture of PKSs presents sufficient reason for expecting feasibility for engineering the enzymes to obtain new drug candidates by combinatorial biosynthesis [22,33].
Shinde et al. evaluated the combinatorial biosynthesis of glycosylated derivatives of a 12-membered macrolide by utilizing Streptomyces venezuelae YJ003 [34]. S. venezuelae has been developed as an important host for the combinatorial biosynthesis of new macrolides due to its amenability to genetic manipulation and faster growth rate than other streptomycetes. Their results revealed that combinatorial biosynthesis has promising potential to generate new glycosylated macrolides with improved antibacterial activities. L-rhamnosyl-10-deoxymethynolide exhibited outstanding activity against clinically isolated erythromycin-resistant pathogenic strains, as well as erythromycin-susceptible strains relative to YC-17 and its other analogs [34]. Similarly, by utilizing S. venezuelae-based combinatorial biosynthesis machinery, Jung et al. successfully revealed the bioconversion of 12-, 14-, and 16-membered ring macrolactones, including 10-deoxymethynolide, narbonolide, and tylactone, respectively, to glycosylated macrolides. The biosynthesis of TDP-3-dimethyl-D-chalcose or TDP-L-rhamnose together with DesVII/DesVIII, a novel narbomycin derivative (novel analog) decorated with L-rhamnose or 3-O-demethyl-D-chalcose, were obtained. These compounds showed greater antibacterial activity than narbomycin and the clinically relevant erythromycin [35].
Additionally, in a study by Ye et al., three 22-membered macrolides were discovered by deciphering the streamlined genome of mangrove-derived Streptomyces sp. HM190, which is a marine actinomycete. They found that PKS genes S1–S8 were proposed to be responsible for the production of three 22-membered macrolides. A total of 30 biosynthetic gene clusters (BGCs), especially gene cluster 5, were responsible for biosynthesis of the macrolide in a strain-specific 126,331bp genomic island belonging to the left-arm region [36].
The construction of macrocyclic structures of marine macrolides remains a challenging problem in medicinal chemistry [37]. However, several new synthetic methods have been discovered to overcome this hurdle and mostly emphasize the key macrolide ring-forming reactions [37]. For example, the total synthesis of borrelidin has been achieved by utilizing a samarium-(II) iodide-mediated intramolecular Reformatsky-type reaction for macrocyclization at C11–C12 after esterification between two segments [38]. Additionally, Terwilliger et al. reported the first synthesis of divergolide I (a naphthoquinone ansamycin). They demonstrated that the biomimetic cyclization of a protodivergolide (a macrocyclic precursor) could be surprisingly enantioselective and relatively short (less than 20 linear steps) [39].
4.2. Metabolic Engineering
The main constraint for macrolide biosynthesis in the producer-host is affluent availability of cofactors and precursors, which generally derive from primary metabolism, including glycolysis, tricarboxylic acid cycle (TCA), pentose phosphate pathway, and amino-acid/nucleic-acid metabolism [22,40], as well as the lower expression level of biosynthetic genes and regulation of the biosynthetic gene/genes [41]. Basically, correlation and regulation of precursor supply for increasing the number of natural products are focused on metabolism of carbohydrates, intracellular cofactor supplies, and fatty-acid precursors [42]. Numerous studies have been reported to illustrate the genetic circuit-guided pathway engineering approaches for increasing the important secondary metabolites (Table 1), such as heterologous overexpression of the S-adenosyl-L-methionine (SAM) synthetase metK and enhanced production of pikromycin, avermectin, and actinorhodin [43,44].

Table 1.
Metabolic-engineering strategies for improving the production of marine macrolides.
One metabolic-engineering strategy for enhanced production of erythromycin involves blocking the flow of propionate into the TCA cycle through a mutB knockout in the industrial S. erythraea strain HL3168 E3 [45]. Similarly, the engineering of the methylmalonyl-CoA metabolite node of S. erythraea through duplication of the mmCoA mutase operon (meaA, mutB, meaB, mutR) led to an elevated level of erythromycin in the oil-based medium [46]. On the other hand, from comparative proteomic and transcriptomic approaches, a putative regulatory protein (SACE_5599) exhibited significantly higher expression in the industrial S. erythraea strain ABE1441 as compared to the wild-type strain and was also involved in erythromycin production [47].
Furthermore, metabolic engineering was employed to improve the formation of novel erythromycin analogues by altering the tailoring pathway modularity in the biosynthesis of erythromycin analogs heterologously engineered in E. coli [48].
Overall, three steps are critical, including (1) bioinformatics analysis of assembly-line enzymes. The first step toward successful assembly-line enzyme engineering is a thorough understanding of the enzyme's domains and modules. Because of their huge size, these genes are frequently misannotated in public genome databases, and their highly repetitive sequences tend to induce problems in open-reading frame annotation or in next-generation sequencing itself. (2) Heterologous expression vector construction: The expression vector is essential for heterologous expression to be successful. Because of their ease of use in gene manipulation, E. coli-Streptomyces shuttle vectors are the most commonly used. (3) Modification: Due to the lack of a robust DNA restriction system, the Streptomyces strain has been widely employed as a heterologous expression host [49,50].
5. Mechanism of Resistance of Mycobacterium tuberculosis
Tuberculosis is a world health problem that is exacerbated by the emergence of resistant strains of M. tuberculosis. The pattern of resistance found in TB cases can be classified based on the class of drugs involved. Multidrug-resistant TB (MDR-TB) is a case of TB caused by strains that are resistant to two first-line drugs, namely, isoniazid and rifampicin [57]. A more resistant strain was discovered in 2006, termed extensively drug-resistant TB (XDR-TB). In the case of XDR-TB, resistance increases to include fluoroquinolones and at least one of the second-line injectables, including amikacin, capreomycin, and kanamycin. Recently, a more worrying case occurred, namely, the emergence of TB strains that are not sensitive to all available treatments, also known as totally drug-resistant TB (TDR-TB) (Figure 4) [58,59].
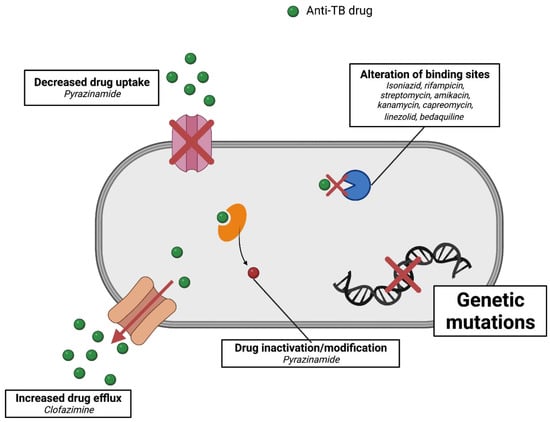
Figure 4.
Summary of the mechanisms of resistance of M. tuberculosis to particular anti-TB drugs.
A deeper understanding of the mechanism of resistance in M. tuberculosis strains is imperative to aid the development of new drugs and the detection of resistance levels in patients. Early detection is very important to ensure better disease management and spread prevention. Principally, M. tuberculosis resistance is acquired through mutations in genes that play a role in the expression of drug targets [4]. Patients can become infected with resistant TB through two scenarios. First, the host becomes infected with resistant strains, resulting in primary-drug resistance. Second, resistance develops during disease progression due to the emergence of new mutations, also known as secondary-drug resistance [2,3]. Furthermore, the mechanisms of resistance of M. tuberculosis to particular anti-TB drugs are discussed in the following sections and summarized in Figure 4.
5.1. Isoniazid
Isoniazid (INH) is one of the first-line drugs in the treatment of TB. This compound inhibits the synthesis of the cell wall of M. tuberculosis by preventing the formation of mycolic acid [60]. This process is related to the inhibitory activity of the inhA protein. INH is a prodrug that is activated by the enzyme KatG, which is produced intracellularly by M. tuberculosis. This enzymatic process pairs INH with NADH and renders it active. Therefore, mutations in the inhA, katG, and ndh genes are considered to be the main mechanisms of resistance to INH [61,62]. In particular, mutations in katG, which are more common in MDR cases, are associated with high levels of resistance [63]. Meanwhile, mutations that occur in inhA will change the structure of the INH-binding target site [64]. Mutations in inhA also affect the performance of other drugs that have a similar therapeutic action, such as ethionamide [65]. Mutations in the ahpC promoter gene that plays a role in the synthesis of alkyl hydroperoxidase reductase enzymes were also considered as markers of resistance to INH. However, it was later found that this gene modification compensates for changes in catalase–peroxidase activity and is not directly related to INH resistance [66]. Mutations in kasA have also been reported in INH-resistant strains, although their exact role remains unclear [67].
5.2. Rifampicin
Rifampicin plays a key role in the treatment of drug-sensitive TB (DS-TB) because of its effectiveness against both active and slow-metabolizing bacilli [68]. Rifampicin acts by binding to the beta subunit of RNA polymerase, inhibiting mRNA elongation [69]. Mutations in the gene encoding the protein, rpoB, have been reported as the main mechanism of resistance to rifampicin. The mutation is particularly clustered at codon 507–533, also known as the rifampicin-resistance-determining region. Approximately 96% of cases of rifampicin resistance are associated with this mutation [11,70]. Interestingly, single resistance to rifampicin is rare. Cross-resistance to other drugs, particularly INH, has been reported frequently [71]. Therefore, rpoB mutations are often used as a surrogate marker for the determination of MDR strains. This mutation also directly reduces the effectiveness of other rifamycin-derived drugs [9].
5.3. Ethambutol
Ethambutol inhibits arabinogalactan synthesis in the cell walls of actively multiplying bacilli [72]. Resistance to ethambutol is related to disruption of the embB gene encoding the enzyme arabynosyl transferase, specifically at codon 306 [73]. However, about one-third of ethambutol-resistant isolates were found to have no alterations to embB306, suggesting that another possible mechanism is involved [2,4]. Previous reports have shown that simultaneous mutations between embB/embC and a gene involved in the biosynthesis of decaprenylphosphoryl-β-D-arabinose lead to variable changes in ethambutol MIC. It is important to note, however, that embB mutations alone elicit variable, but not high, resistance levels [10]. Another gene reported to play a role in ethambutol resistance is ubiA, which encodes decaprenyl-phosphate 5-phosphoribosyltransferase synthase. This enzyme is also involved in the synthesis of bacterial-cell walls. Mutations in ubiA, when co-occurring with embB mutations, can lead to high levels of ethambutol resistance [74].
5.4. Pyrazinamide
Pyrazinamide is part of the first-line regimen in the treatment of TB. An interesting feature of this drug is its ability to target semi-dormant bacilli in TB lesions [75]. The use of this drug allows a reduction in the duration of TB treatment to 6 months. Pyrazinamide penetrates bacterial cells and is activated by pyrazinamide/nicotinamidase to the active form of pyrazinoic acid [76]. Normally, pyrazinoic acid would be subject to the efflux mechanism. However, under acidic conditions, as in TB lesions, protonated pyrazinoic acid allows re-entry into cells [77]. Resistance to pyrazinamide is generally associated with mutations in the pncA gene encoding the enzyme pyrazinamide/nicotinamidease [78]. However, a small percentage of resistant isolates showed no mutations in the pncA gene, suggesting that other types of mutations may be involved [79].
5.5. Streptomycin
Streptomycin inhibits bacterial protein synthesis by binding irreversibly to the 30s ribosomal subunit and is active in slow-growing bacilli [80]. This antibiotic was the first drug introduced in the treatment of TB and resistance to it is growing rapidly [81]. Streptomycin-resistant strains have been reported to have mutations in the rpsL and rrs genes, which are associated with ribosomal protein rRNA. Nearly three-quarters of the resistant isolates were found to have this mutation [82].
5.6. Fluoroquinolones
Fluoroquinolones are second line in the treatment of TB. This class of antibiotics prevents bacterial-cell replication by inhibiting DNA gyrase. Ciprofloxacin and ofloxacin are examples of previous-generation drugs that have been used for TB [83]. Two fourth-generation fluoroquinolones, moxifloxacin and gatifloxacin, are new therapeutic candidates for DR-TB [84]. gyrA and gyrB are the genes that code for DNA gyrase; thus, mutations in them could result in resistance to quinolones. Single mutations in gyrA or gyrB lead to significant resistance, and multiple mutations lead to a higher increase in MIC [85,86].
5.7. Second-Line Injectables
Amikacin and kanamycin (aminoglycosides) and capreomycin (cyclic polypeptides) belong to the second-line injectable group for the treatment of TB. These drugs have the same target of action, despite the different antibacterial classes [2,3]. Therefore, the mechanism of resistance to these compounds is also interrelated. These drugs act by inhibiting protein synthesis through the modification 16s rRNA on the bacterial ribosome. Mutations in the rrs gene are associated with high levels of resistance. The most common molecular mechanism is the A-G polymorphism at position 1401 of the rrs gene [87]. For capreomycin, mutations in the tylA gene involved in ribose methylation in rRNA were also found to trigger additional resistance [88]. Cross-resistance among these three drugs has also been reported [89,90].
5.8. Para-Amino Aalicylic Acid (PAS)
Previously one of the first choices in the treatment of TB, this para-amino benzoic acid analogue is now part of second-line drugs. The bactericidal activity of the drug is obtained through inhibition of folate synthesis [3]. Approximately 40% of PAS-resistant strains exhibit mutations in the thyA gene [91]. Also recently, mutations in the folC gene encoding dihydrofolate synthase were found to correlate with resistance to PAS in laboratory isolates of M. tuberculosis [92].
5.9. Novel and Repurposed Drugs
Several new drugs have been applied in the treatment of TB. However, although relatively new, resistance has been reported with some of these drugs. Linezolid, an oxazolidinone, is an early-stage inhibitor in protein synthesis. The drug binds to the V domain of the 50s subunit of the bacterial ribosome [93]. Two genes found to be associated with linezolid resistance were rrl and rplC [94]. Another drug that has also been used as a new agent for TB is bedaquiline from the diarylquinolines class. Bedaquiline acts by inhibiting bacterial-cell respiration by targeting ATP synthase in M. tuberculosis [95]. Mutations in the atpE gene related to this process have been associated with high levels of resistance to bedaquiline [96,97].
One example of a repurposed drug in TB therapy is clofazimine, which was previously used in the treatment of leprosy. This drug has now become part of a short-course treatment regimen based on WHO recommendations. The exact mechanism of this drug has not been established. However, previous studies have suggested that the formation of reactive oxygen species after clofazimine is reduced by NADH dehydrogenase [98]. One mechanism associated with resistance to clofazimine is an off-target mutation in rvo678 that causes increased efflux of the drug pumped out of mycobacterial cells [99].
6. Marine Macrolides to Counter Mycobacterium tuberculosis Resistance
In this section, we have listed several compounds classified as macrolides isolated from marine organisms (Figure 5 and Table 2). These macrolides, either in a single administration or in combination with other compounds, have a promising activity for tackling mycobacterial infection so that they can hopefully provide a breakthrough for countering M. tuberculosis-resistance cases. The structures of the listed marine macrolides are presented in Figure 6.
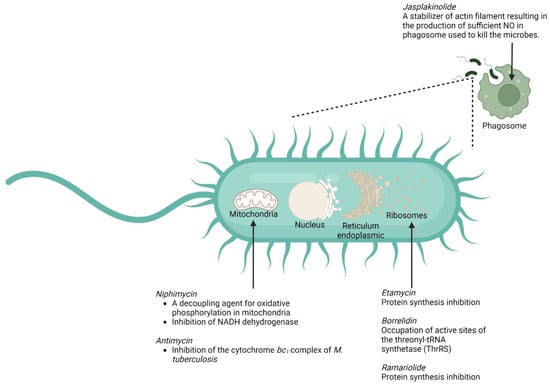
Figure 5.
Summary of the mechanisms of action of several marine macrolides against M. tuberculosis.

Table 2.
Several marine macrolides exhibiting anti-mycobacterial activities.
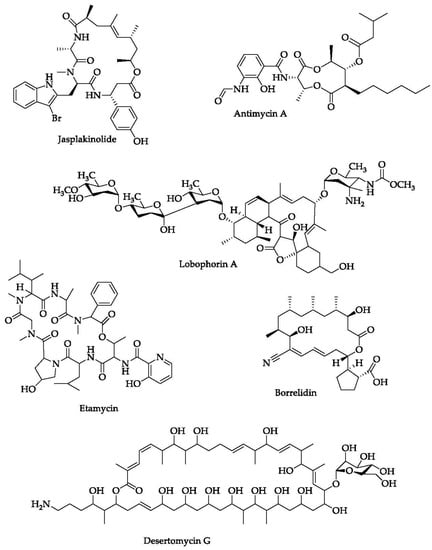
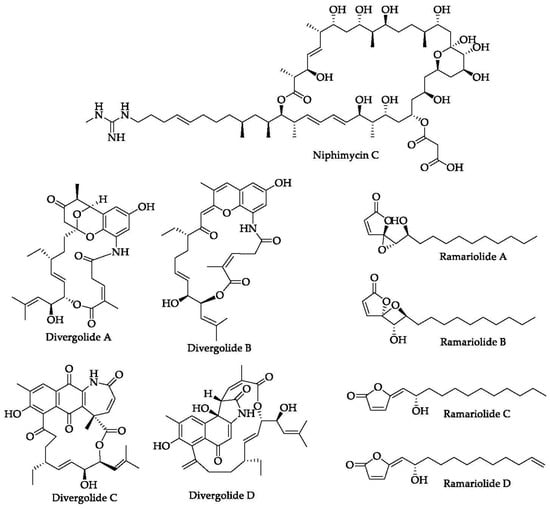
Figure 6.
Chemical structures of several macrolides isolated from marine organisms.
Although disturbance of bacterial-protein synthesis in ribosomes is still the main mechanism by which the macrolides (e.g., etamycin, ramariolide, and borrelidin) exert their antimycobacterial action [118,119,120], several marine macrolides also offer a number of relatively different ways to act as an antimicrobe. One of those mechanisms is associated with the ability of the macrolide (jasplakinolide) to disturb the regulation of actin filament, leading to the success of the macrophages in killing the bacterium [101]. Another mechanism that might be exploited by the marine macrolide (antimycin and niphimycin) is linked to their ability to perturb the activity of the mitochondria in M. tuberculosis [106,107,121]. The summary of the putative mechanisms by which several marine macrolides exert their antimycobacterial actions is provided in Figure 5.
Most of the compounds listed above show their antimycobacterial activities. However, those activities have been confirmed through in vitro studies using various M. tuberculosis variants. Based on the MIC values, niphimycin (4–16 μg/mL) and etamycin (0.097–25 μg/mL) showed more potency in inhibiting the growth of several tested mycobacteria relative to the other compounds.
Moreover, both entities demonstrated fewer cytotoxic effects compared to the related control groups, indicating their safety in the tested models. For example, the cytotoxic activity of niphimycin C and niphimycin Iα on K562, HepG2, MCF-7, and HeLa cells was 8.5–10.2 μM and 6.8–23.9 μM, respectively, whereas the standard drug (doxorubicin) yielded 1.1–3.5 μM.
As these results were collected through various in vitro models, further in vivo and clinical studies must be conducted to confirm their efficacy and safety. To provide a quick screening on macrolides extracted from marine sources, in silico studies should also be considered. Although the others have less potency in inhibiting the growth of the mycobacteria, these compounds have the potential to be modified structurally to form the related compounds with better efficacy and safety properties.
From the list, marine Streptomyces sp. becomes the major source for marine macrolides. In addition, marine microorganisms residing in corals, sponges, or other marine plants also have the potential to be the sources for isolating antibiotics. Figure 7 shows several marine organisms that have become the main sources for isolating marine microorganisms, including Streptomyces sp., that are associated with the production of macrolides.
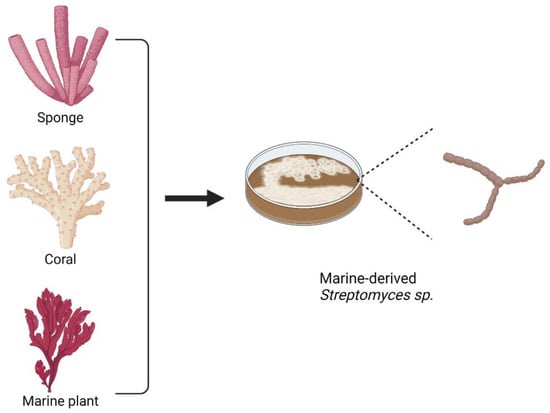
Figure 7.
Examples of organisms as the main sources for obtaining macrolide-producing marine microorganisms, including Streptomyces sp.
7. Concluding Remarks
TB is still a burden globally not only for its health-related impacts but also for its influence in other aspects, such as social and economic aspects. This condition is exacerbated by the ability of M. tuberculosis to evade the action of the current TB drugs. Efforts to discover novel drugs used to counter the resistant strains of TB should be carried out at an accelerated pace. At this point, the exploration of marine-derived compounds for their antimycobacterial activity should be taken into consideration.
The potencies of macrolides isolated from marine organisms in treating TB have attracted interests in the recent times given that several antibiotics for TB, e.g., isoniazid, ethambutol, and the currently used macrolides, show increased rates of resistance against the microbe. It becomes clear that the ocean is storing an enormous number of compounds that are unique not only in terms of their structure but also their biological activities, compared to their counterparts originating from terrestrial organisms.
To date, the number of marine-derived compounds that have been proven to possess antimycobacterial activities is minimal. However, these activities are mostly investigated through various in vitro assays, whereas in vivo, in silico, and clinical studies of marine macrolides for countering TB are very limited. Obviously, this condition should direct the research on this topic to be more expanded, with further studies carried out to decipher the antimycobacterial potencies of the compounds in in vivo and clinical settings.
Studies aiming to investigate the molecular mechanism of the compounds should also be considered. It is also obvious that studies focusing on synthesizing marine natural marine products possessing activity against mycobacterial infections have to be carried out to avoid excessive marine exploitation.
Author Contributions
Conceptualization, F.N., S.S.M., T.B.E., and J.S.-G.; methodology, F.N., S.S.M., A.M., and M.S.; writing—original draft preparation, F.N., S.S.M., A.M., A.F., R.N.U., and M.S.; writing—review and editing, F.N., S.S.M., A.M., A.F., M.S., C.M.G.L., H.C., T.B.E., and J.S.-G.; supervision, F.N. and J.S.-G.; project administration, F.N., T.B.E., and J.S.-G.; funding acquisition, F.N. and J.S.-G. All authors revised the manuscript into its final form and gave the approval for submission. All authors have read and agreed to the published version of the manuscript.
Funding
Research carried out in F.N.’s lab is supported by PDUPT Research Grant 2022 (No. 020/E5/PG.02.00PT/2022) from the Directorate General of Higher Education, Ministry of Education, Culture, Research, and Technology, Indonesia.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Global Tuberculosis Report 2020; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Almeida Da Silva, P.E.; Palomino, J.C. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: Classical and new drugs. J. Antimicrob. Chemother. 2011, 66, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Dookie, N.; Rambaran, S.; Padayatchi, N.; Mahomed, S.; Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis: A review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 2018, 73, 1138–1151. [Google Scholar] [CrossRef] [PubMed]
- Palomino, J.C.; Martin, A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 2014, 3, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, S.M.; Karlsson, M.; Johansson, L.B.; Vester, B. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol. Microbiol. 2001, 41, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Muxfeldt, H.; Shrader, S.; Hansen, P.; Brockmann, H. The structure of pikromycin. J. Am. Chem. Soc. 1968, 90, 4748–4749. [Google Scholar] [CrossRef]
- Berry, M. The macrolide antibiotics. Q. Rev. Chem. Soc. 1963, 17, 343–361. [Google Scholar] [CrossRef]
- van der Paardt, A.-F.; Wilffert, B.; Akkerman, O.W.; de Lange, W.C.M.; van Soolingen, D.; Sinha, B.; van der Werf, T.S.; Kosterink, J.G.W.; Alffenaar, J.-W.C. Evaluation of macrolides for possible use against multidrug-resistant Mycobacterium tuberculosis. Eur. Respir. J. 2015, 46, 444. [Google Scholar] [CrossRef]
- Fonseca, J.; Knight, G.; McHugh, T. The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int. J. Infect. Dis. 2015, 32, 94–100. [Google Scholar] [CrossRef]
- Safi, H.; Lingaraju, S.; Amin, A.; Kim, S.; Jones, M.; Holmes, M.; McNeil, M.; Peterson, S.N.; Chatterjee, D.; Fleischmann, R. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013, 45, 1190–1197. [Google Scholar] [CrossRef]
- Somoskovi, A.; Parsons, L.M.; Salfinger, M. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2001, 2, 1–5. [Google Scholar] [CrossRef]
- Das, R.; Rauf, A.; Mitra, S.; Emran, T.B.; Hossain, M.J.; Khan, Z.; Naz, S.; Ahmad, B.; Meyyazhagan, A.; Pushparaj, K.; et al. Therapeutic potential of marine macrolides: An overview from 1990 to 2022. Chem. Biol. Interact. 2022, 365, 110072. [Google Scholar] [CrossRef]
- Gianti, E.; Zauhar, R.J. Chapter 7—Structure–activity relationships and drug design. In Remington, 23rd ed.; Adejare, A., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 129–153. [Google Scholar]
- Ouattara, M.; Songuigama, C.; N’Guessan, D.J.-P. Pharmacochemical Aspects of the Evolution from Erythromycin to Neomacrolides, Ketolides and Neoketolides. Open J. Med. Chem. 2020, 10, 57–112. [Google Scholar] [CrossRef]
- Pavlović, D.; Mutak, S.; Andreotti, D.; Biondi, S.; Cardullo, F.; Paio, A.; Piga, E.; Donati, D.; Lociuro, S. Synthesis and Structure-Activity Relationships of α-Amino-γ-lactone Ketolides: A Novel Class of Macrolide Antibiotics. ACS Med. Chem. Lett. 2014, 5, 1133–1137. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Zou, J.; Yan, X.; Chen, J.; Cao, X.; Wu, J.; Liu, Y.; Wang, T. Marine-Derived Macrolides 1990–2020: An Overview of Chemical and Biological Diversity. Mar Drugs 2021, 19, 180. [Google Scholar] [CrossRef]
- Karpiński, T.M. Marine Macrolides with Antibacterial and/or Antifungal Activity. Mar Drugs 2019, 17, 241. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Wu, J.; Skalina, K.; Pfeifer, B.A. Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 2010, 17, 1232–1240. [Google Scholar] [CrossRef]
- Mabe, S.; Eller, J.; Champney, W.S. Structure-activity relationships for three macrolide antibiotics in Haemophilus influenzae. Curr. Microbiol. 2004, 49, 248–254. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Krasnykh, O.; Pan, D.; Petukhova, V.; Yu, G.; Liu, Y.; Liu, H.; Hong, S.; Wang, Y.; Wan, B.; et al. Structure-activity relationships of macrolides against Mycobacterium tuberculosis. Tuberculosis 2008, 88, S49–S63. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Z.; Sun, Y.; Cui, P.; Liang, J.; Xing, Q.; Wu, J.; Xu, Y.; Zhang, W.; Zhang, Y.; et al. Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides. Emerg Microbes Infect 2022, 11, 293–305. [Google Scholar] [CrossRef]
- Dhakal, D.; Sohng, J.K.; Pandey, R.P. Engineering actinomycetes for biosynthesis of macrolactone polyketides. Microb. Cell Factories 2019, 18, 137. [Google Scholar] [CrossRef]
- Dutta, S.; Whicher, J.R.; Hansen, D.A.; Hale, W.A.; Chemler, J.A.; Congdon, G.R.; Narayan, A.R.; Håkansson, K.; Sherman, D.H.; Smith, J.L.; et al. Structure of a modular polyketide synthase. Nature 2014, 510, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Whicher, J.R.; Dutta, S.; Hansen, D.A.; Hale, W.A.; Chemler, J.A.; Dosey, A.M.; Narayan, A.R.H.; Håkansson, K.; Sherman, D.H.; Smith, J.L.; et al. Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature 2014, 510, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Keatinge-Clay, A.T. Polyketide Synthase Modules Redefined. Angew. Chem. Int. Ed. Engl. 2017, 56, 4658–4660. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Yoon, Y.J. Recent advances in the discovery and combinatorial biosynthesis of microbial 14-membered macrolides and macrolactones. J. Ind. Microbiol. Biotechnol. 2019, 46, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006, 106, 3468–3496. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Schnarr, N.A.; Kim, C.Y.; Cane, D.E.; Khosla, C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc. 2006, 128, 3067–3074. [Google Scholar] [CrossRef]
- Xu, W.; Qiao, K.; Tang, Y. Structural analysis of protein-protein interactions in type I polyketide synthases. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 98–122. [Google Scholar] [CrossRef]
- Cummings, M.; Breitling, R.; Takano, E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol. Lett. 2014, 351, 116–125. [Google Scholar] [CrossRef]
- Koryakina, I.; Kasey, C.; McArthur, J.B.; Lowell, A.N.; Chemler, J.A.; Li, S.; Hansen, D.A.; Sherman, D.H.; Williams, G.J. Inversion of Extender Unit Selectivity in the Erythromycin Polyketide Synthase by Acyltransferase Domain Engineering. ACS Chem. Biol. 2017, 12, 114–123. [Google Scholar] [CrossRef]
- Nepal, K.K.; Wang, G. Streptomycetes: Surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products. Biotechnol. Adv. 2019, 37, 1–20. [Google Scholar] [CrossRef]
- Weissman, K.J.; Leadlay, P.F. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 2005, 3, 925–936. [Google Scholar] [CrossRef]
- Shinde, P.B.; Han, A.R.; Cho, J.; Lee, S.R.; Ban, Y.H.; Yoo, Y.J.; Kim, E.J.; Kim, E.; Song, M.C.; Park, J.W.; et al. Combinatorial biosynthesis and antibacterial evaluation of glycosylated derivatives of 12-membered macrolide antibiotic YC-17. J. Biotechnol. 2013, 168, 142–148. [Google Scholar] [CrossRef]
- Jung, W.S.; Han, A.R.; Hong, J.S.; Park, S.R.; Choi, C.Y.; Park, J.W.; Yoon, Y.J. Bioconversion of 12-, 14-, and 16-membered ring aglycones to glycosylated macrolides in an engineered strain of Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 2007, 76, 1373–1381. [Google Scholar] [CrossRef]
- Ye, Y.; Anwar, N.; Mao, X.; Wu, S.; Yan, C.; Zhao, Z.; Zhang, R.; Nie, Y.; Zhang, J.; Wang, J.; et al. Discovery of Three 22-Membered Macrolides by Deciphering the Streamlined Genome of Mangrove-Derived Streptomyces sp. HM190. Front. Microbiol. 2020, 11, 1464. [Google Scholar] [CrossRef]
- Kumar, C.N.S.S.P. Total Synthesis of Macrolides. In Organic Synthesis; Belakatte Parameshwarappa, N., Ed.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Nagamitsu, T.; Takano, D.; Marumoto, K.; Fukuda, T.; Furuya, K.; Otoguro, K.; Takeda, K.; Kuwajima, I.; Harigaya, Y.; Ōmura, S. Total Synthesis of Borrelidin. J. Org. Chem. 2007, 72, 2744–2756. [Google Scholar] [CrossRef]
- Terwilliger, D.W.; Trauner, D. Selective Synthesis of Divergolide I. J. Am. Chem. Soc. 2018, 140, 2748–2751. [Google Scholar] [CrossRef]
- Zhang, M.M.; Wang, Y.; Ang, E.L.; Zhao, H. Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 2016, 33, 963–987. [Google Scholar] [CrossRef]
- Luo, Y.; Li, B.Z.; Liu, D.; Zhang, L.; Chen, Y.; Jia, B.; Zeng, B.X.; Zhao, H.; Yuan, Y.J. Engineered biosynthesis of natural products in heterologous hosts. Chem. Soc. Rev. 2015, 44, 5265–5290. [Google Scholar] [CrossRef]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Dhakal, D.; Sohng, J.K. An insight into the “-omics” based engineering of streptomycetes for secondary metabolite overproduction. Biomed. Res. Int. 2013, 2013, 968518. [Google Scholar] [CrossRef]
- Lechner, A.; Brunk, E.; Keasling, J.D. The Need for Integrated Approaches in Metabolic Engineering. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hong, M.; Chu, J.; Huang, M.; Ouyang, L.; Tian, X.; Zhuang, Y. Blocking the flow of propionate into TCA cycle through a mutB knockout leads to a significant increase of erythromycin production by an industrial strain of Saccharopolyspora erythraea. Bioprocess Biosyst. Eng. 2017, 40, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Reeves, A.R.; Brikun, I.A.; Cernota, W.H.; Leach, B.I.; Gonzalez, M.C.; Weber, J.M. Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab. Eng. 2007, 9, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Kirm, B.; Magdevska, V.; Tome, M.; Horvat, M.; Karničar, K.; Petek, M.; Vidmar, R.; Baebler, S.; Jamnik, P.; Fujs, Š.; et al. SACE_5599, a putative regulatory protein, is involved in morphological differentiation and erythromycin production in Saccharopolyspora erythraea. Microb. Cell. Fact. 2013, 12, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, Y.; Fang, L.; Pfeifer, B.A. Tailoring pathway modularity in the biosynthesis of erythromycin analogs heterologously engineered in E. coli. Sci. Adv. 2015, 1, e1500077. [Google Scholar] [CrossRef]
- Wilkinson, B.; Micklefield, J. Mining and engineering natural-product biosynthetic pathways. Nat. Chem. Biol. 2007, 3, 379–386. [Google Scholar] [CrossRef]
- Zhang, L.; Awakawa, T.; Abe, I. Understanding and Manipulating Assembly Line Biosynthesis by Heterologous Expression in Streptomyces. In Engineering Natural Product Biosynthesis; Springer: Berlin/Heidelberg, Germany, 2022; pp. 223–238. [Google Scholar]
- Wu, H.; Chen, M.; Mao, Y.; Li, W.; Liu, J.; Huang, X.; Zhou, Y.; Ye, B.C.; Zhang, L.; Weaver, D.T.; et al. Dissecting and engineering of the TetR family regulator SACE_7301 for enhanced erythromycin production in Saccharopolyspora erythraea. Microb. Cell Fact 2014, 13, 158. [Google Scholar] [CrossRef]
- Yi, J.S.; Kim, M.; Kim, E.J.; Kim, B.G. Production of pikromycin using branched chain amino acid catabolism in Streptomyces venezuelae ATCC 15439. J. Ind. Microbiol. Biotechnol. 2018, 45, 293–303. [Google Scholar] [CrossRef]
- Dhakal, D.; Le, T.T.; Pandey, R.P.; Jha, A.K.; Gurung, R.; Parajuli, P.; Pokhrel, A.R.; Yoo, J.C.; Sohng, J.K. Enhanced production of nargenicin A(1) and generation of novel glycosylated derivatives. Appl. Biochem. Biotechnol. 2015, 175, 2934–2949. [Google Scholar] [CrossRef]
- Bian, X.; Tang, B.; Yu, Y.; Tu, Q.; Gross, F.; Wang, H.; Li, A.; Fu, J.; Shen, Y.; Li, Y.Z.; et al. Heterologous Production and Yield Improvement of Epothilones in Burkholderiales Strain DSM 7029. ACS Chem. Biol. 2017, 12, 1805–1812. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, J.; Lu, C.; Shen, Y. Heterologous expression of galbonolide biosynthetic genes in Streptomyces coelicolor. Antonie Leeuwenhoek 2015, 107, 1359–1366. [Google Scholar] [CrossRef]
- Hashimoto, T.; Hashimoto, J.; Kozone, I.; Amagai, K.; Kawahara, T.; Takahashi, S.; Ikeda, H.; Shin-Ya, K. Biosynthesis of Quinolidomicin, the Largest Known Macrolide of Terrestrial Origin: Identification and Heterologous Expression of a Biosynthetic Gene Cluster over 200 kb. Org. Lett. 2018, 20, 7996–7999. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention, C. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs--worldwide, 2000–2004. MMWR Morb. Mortal. Wkly. Rep. 2006, 55, 301–305. [Google Scholar]
- Gandhi, N.R.; Moll, A.; Sturm, A.W.; Pawinski, R.; Govender, T.; Lalloo, U.; Zeller, K.; Andrews, J.; Friedland, G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 2006, 368, 1575–1580. [Google Scholar] [CrossRef]
- Udwadia, Z.F.; Amale, R.A.; Ajbani, K.K.; Rodrigues, C. Totally drug-resistant tuberculosis in India. Clin. Infect. Dis. 2012, 54, 579–581. [Google Scholar] [CrossRef]
- Hong, B.-L.; D’Cunha, R.; Li, P.; Al-Shaer, M.H.; Alghamdi, W.A.; An, G.; Peloquin, C. A systematic review and meta-analysis of isoniazid pharmacokinetics in healthy volunteers and patients with tuberculosis. Clin. Ther. 2020, 42, e220–e241. [Google Scholar] [CrossRef]
- Barry III, C.E.; Slayden, R.A.; Mdluli, K. Mechanisms of isoniazid resistance in Mycobacterium tuberculosis. Drug Resist. Updates 1998, 1, 128–134. [Google Scholar] [CrossRef]
- Zhang, Y.; Heym, B.; Allen, B.; Young, D.; Cole, S. The catalase—Peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 1992, 358, 591–593. [Google Scholar] [CrossRef]
- Hazbón, M.H.; Brimacombe, M.; Bobadilla del Valle, M.; Cavatore, M.; Guerrero, M.I.; Varma-Basil, M.; Billman-Jacobe, H.; Lavender, C.; Fyfe, J.; García-García, L. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2006, 50, 2640–2649. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.V.; Reich, R.; Dou, S.-J.; Jasperse, L.; Pan, X.; Wanger, A.; Quitugua, T.; Graviss, E.A. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 1241–1250. [Google Scholar] [CrossRef]
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; De Lisle, G.; Jacobs Jr, W.R. hA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994, 263, 227–230. [Google Scholar] [CrossRef]
- Sherman, D.R.; Mdluli, K.; Hickey, M.J.; Arain, T.M.; Morris, S.L.; Barry III, C.E.; Stover, C.K. Compensatory ahp C gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 1996, 272, 1641–1643. [Google Scholar] [CrossRef] [PubMed]
- Slayden, R.; Barry 3rd, C. The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberculosis 2002, 82, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Fox, W. The chemotherapy of pulmonary tuberculosis: A review. Chest 1979, 76, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Telenti, A.; Imboden, P.; Marchesi, F.; Schmidheini, T.; Bodmer, T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob. Agents Chemother. 1993, 37, 2054–2058. [Google Scholar] [CrossRef]
- Imperiale, B.R.; Di Giulio, Á.B.; Adrián Cataldi, Á.; Morcillo, N.S. Evaluation of Mycobacterium tuberculosis cross-resistance to isoniazid, rifampicin and levofloxacin with their respective structural analogs. J. Antibiot. 2014, 67, 749–754. [Google Scholar] [CrossRef]
- Takayama, K.; Kilburn, J.O. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 1989, 33, 1493–1499. [Google Scholar] [CrossRef]
- Sreevatsan, S.; Stockbauer, K.E.; Pan, X.; Kreiswirth, B.N.; Moghazeh, S.L.; Jacobs Jr, W.R.; Telenti, A.; Musser, J.M. Ethambutol resistance in Mycobacterium tuberculosis: Critical role of embB mutations. Antimicrob. Agents Chemother. 1997, 41, 1677–1681. [Google Scholar] [CrossRef]
- Tulyaprawat, O.; Chaiprasert, A.; Chongtrakool, P.; Suwannakarn, K.; Ngamskulrungroj, P. Association of ubiA mutations and high-level of ethambutol resistance among Mycobacterium tuberculosis Thai clinical isolates. Tuberculosis 2019, 114, 42–46. [Google Scholar] [CrossRef]
- Mitchison, D. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 1985, 66, 219–225. [Google Scholar] [CrossRef]
- Konno, K.; Feldmann, F.M.; McDermott, W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 1967, 95, 461–469. [Google Scholar]
- Zhang, Y.; Mitchison, D. The curious characteristics of pyrazinamide: A review. Int. J. Tuberc. Lung Dis. 2003, 7, 6–21. [Google Scholar]
- Juréen, P.; Werngren, J.; Toro, J.-C.; Hoffner, S. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2008, 52, 1852–1854. [Google Scholar] [CrossRef]
- Cheng, S.-J.; Thibert, L.; Sanchez, T.; Heifets, L.; Zhang, Y. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: Spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 2000, 44, 528–532. [Google Scholar] [CrossRef]
- Moazed, D.; Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 1987, 327, 389–394. [Google Scholar] [CrossRef]
- Crofton, J.; Mitchison, D. Streptomycin resistance in pulmonary tuberculosis. Br. Med. J. 1948, 2, 1009. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, S.H. Evolution of drug resistance in Mycobacterium tuberculosis: Clinical and molecular perspective. Antimicrob. Agents Chemother. 2002, 46, 267–274. [Google Scholar] [CrossRef]
- Berning, S.E. The role of fluoroquinolones in tuberculosis today. Drugs 2001, 61, 9–18. [Google Scholar] [CrossRef]
- Ruan, Q.; Liu, Q.; Sun, F.; Shao, L.; Jin, J.; Yu, S.; Ai, J.; Zhang, B.; Zhang, W. Moxifloxacin and gatifloxacin for initial therapy of tuberculosis: A meta-analysis of randomized clinical trials. Emerg. Microbes Infect. 2016, 5, e12. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef] [PubMed]
- Takiff, H.E.; Salazar, L.; Guerrero, C.; Philipp, W.; Huang, W.M.; Kreiswirth, B.; Cole, S.T.; Jacobs, W.R., Jr.; Telenti, A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 1994, 38, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Georghiou, S.B.; Magana, M.; Garfein, R.S.; Catanzaro, D.G.; Catanzaro, A.; Rodwell, T.C. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: A systematic review. PLoS ONE 2012, 7, e33275. [Google Scholar] [CrossRef] [PubMed]
- Johansen, S.K.; Maus, C.E.; Plikaytis, B.B.; Douthwaite, S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2’-O-methylations in 16S and 23S rRNAs. Mol Cell 2006, 23, 173–182. [Google Scholar] [CrossRef]
- Jugheli, L.; Bzekalava, N.; de Rijk, P.; Fissette, K.; Portaels, F.; Rigouts, L. High level of cross-resistance between kanamycin, amikacin, and capreomycin among Mycobacterium tuberculosis isolates from Georgia and a close relation with mutations in the rrs gene. Antimicrob. Agents Chemother. 2009, 53, 5064–5068. [Google Scholar] [CrossRef]
- Krüüner, A.; Jureen, P.; Levina, K.; Ghebremichael, S.; Hoffner, S. Discordant resistance to kanamycin and amikacin in drug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 2971–2973. [Google Scholar] [CrossRef]
- Mathys, V.; Wintjens, R.; Lefevre, P.; Bertout, J.; Singhal, A.; Kiass, M.; Kurepina, N.; Wang, X.-M.; Mathema, B.; Baulard, A.; et al. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2009, 53, 2100–2109. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, X.D.; Erber, L.N.; Luo, M.; Guo, A.Z.; Yang, S.S.; Gu, J.; Turman, B.J.; Gao, Y.R.; Li, D.F.; et al. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 1479–1487. [Google Scholar] [CrossRef]
- Singh, B.; Cocker, D.; Ryan, H.; Sloan, D.J. Linezolid for drug-resistant pulmonary tuberculosis. Cochrane Database Syst. Rev. 2019, 3, Cd012836. [Google Scholar] [CrossRef]
- Richter, E.; Rüsch-Gerdes, S.; Hillemann, D. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 1534–1536. [Google Scholar] [CrossRef]
- Huitric, E.; Verhasselt, P.; Andries, K.; Hoffner, S.E. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother 2007, 51, 4202–4204. [Google Scholar] [CrossRef]
- Huitric, E.; Verhasselt, P.; Koul, A.; Andries, K.; Hoffner, S.; Andersson, D.I. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 2010, 54, 1022–1028. [Google Scholar] [CrossRef]
- Petrella, S.; Cambau, E.; Chauffour, A.; Andries, K.; Jarlier, V.; Sougakoff, W. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob. Agents Chemother. 2006, 50, 2853–2856. [Google Scholar] [CrossRef]
- Lechartier, B.; Cole, S.T. Mode of Action of Clofazimine and Combination Therapy with Benzothiazinones against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 4457–4463. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Uplekar, S.; Cole, S.T. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 2979–2981. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Fratti, R.A.; Poschet, J.F.; Timmins, G.S.; Master, S.S.; Burgos, M.; Marletta, M.A.; Deretic, V. Mycobacteria inhibit nitric oxide synthase recruitment to phagosomes during macrophage infection. Infect. Immun. 2004, 72, 2872–2878. [Google Scholar] [CrossRef]
- Holzinger, A.; Blaas, K. Actin-Dynamics in Plant Cells: The Function of Actin-Perturbing Substances: Jasplakinolide, Chondramides, Phalloidin, Cytochalasins, and Latrunculins. Methods Mol. Biol. 2016, 1365, 243–261. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Guo, H.; Hou, W.; Yang, N.; Ren, B.; Liu, M.; Dai, H.; Liu, X.; Song, F.; et al. Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 2013, 97, 3885–3892. [Google Scholar] [CrossRef]
- Lin, Z.; Koch, M.; Pond, C.D.; Mabeza, G.; Seronay, R.A.; Concepcion, G.P.; Barrows, L.R.; Olivera, B.M.; Schmidt, E.W. Structure and activity of lobophorins from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121–126. [Google Scholar] [CrossRef]
- Chandra, P.; He, L.; Zimmerman, M.; Yang, G.; Köster, S.; Ouimet, M.; Wang, H.; Moore, K.J.; Dartois, V.; Schilling, J.D.; et al. Inhibition of Fatty Acid Oxidation Promotes Macrophage Control of Mycobacterium tuberculosis. mBio 2020, 11. [Google Scholar] [CrossRef]
- Cumming, B.M.; Addicott, K.W.; Adamson, J.H.; Steyn, A.J. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. Elife 2018, 7, e39169. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.T.; Uddin, M.S.; Jeandet, P.; Emran, T.B.; Mitra, S.; Albadrani, G.M.; Sayed, A.A.; Abdel-Daim, M.M.; Simal-Gandara, J. Anti-Alzheimer’s molecules derived from marine life: Understanding molecular mechanisms and therapeutic potential. Mar. Drugs 2021, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Rotsaert, F.A.J.; Ding, M.G.; Trumpower, B.L. Differential efficacy of inhibition of mitochondrial and bacterial cytochrome bc1 complexes by center N inhibitors antimycin, ilicicolin H and funiculosin. Biochim. Biophys. Acta (BBA) Bioenerg. 2008, 1777, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Koyama, N.; Kanamoto, A.; Tomoda, H. Discovery of nosiheptide, griseoviridin, and etamycin as potent anti-mycobacterial agents against Mycobacterium avium complex. Molecules 2019, 24, 1495. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Maloney, K.N.; Nam, S.J.; Haste, N.M.; Raju, R.; Aalbersberg, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E.; Fenical, W. Fijimycins A-C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 6557–6562. [Google Scholar] [CrossRef]
- Stoye, A.; Nagalingam, G.; Britton, W.J.; Payne, R.J. Synthesis of Norfijimycin A with Activity against Mycobacterium tuberculosis. Aust. J. Chem. 2017, 70, 229–232. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Martín, J.; Otero, L.; Palacios-Gutiérrez, J.J.; Fernández, J.; Mohamedi, Y.; Fontanil, T.; Salmón, M.; et al. Desertomycin G, a New Antibiotic with Activity against Mycobacterium tuberculosis and Human Breast Tumor Cell Lines Produced by Streptomyces althioticus MSM3, Isolated from the Cantabrian Sea Intertidal Macroalgae Ulva sp. Mar. Drugs 2019, 17, 114. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, M.; Wu, C.; Tan, Y.; Li, J.; Hao, X.; Duan, Y.; Guan, Y.; Shang, X.; Wang, Y.; et al. Identification and Proposed Relative and Absolute Configurations of Niphimycins C-E from the Marine-Derived Streptomyces sp. IMB7-145 by Genomic Analysis. J. Nat. Prod. 2018, 81, 178–187. [Google Scholar] [CrossRef]
- Gutierrez-Lugo, M.T.; Bewley, C.A. Susceptibility and mode of binding of the Mycobacterium tuberculosis cysteinyl transferase mycothiol ligase to tRNA synthetase inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 2480–2483. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Shao, J.; Sun, C.; Song, Y.; Li, Q.; Lu, L.; Hu, Y.; Gui, C.; Zhang, H.; Ju, J. Borrelidins F-I, cytotoxic and cell migration inhibiting agents from mangrove-derived Streptomyces rochei SCSIO ZJ89. Bioorg. Med. Chem. 2018, 26, 1488–1494. [Google Scholar] [CrossRef]
- Yu, M.; Li, Y.; Banakar, S.P.; Liu, L.; Shao, C.; Li, Z.; Wang, C. New Metabolites from the Co-culture of Marine-Derived Actinomycete Streptomyces rochei MB037 and Fungus Rhinocladiella similis 35. Front. Microbiol. 2019, 10, 915. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Maier, A.; Fiebig, H.H.; Görls, H.; Lin, W.H.; Peschel, G.; Hertweck, C. Divergolides A–D from a Mangrove Endophyte Reveal an Unparalleled Plasticity in ansa-Macrolide Biosynthesis. Angew. Chem. 2011, 123, 1668–1672. [Google Scholar] [CrossRef]
- Centko, R.M.; Ramón-García, S.; Taylor, T.; Patrick, B.O.; Thompson, C.J.; Miao, V.P.; Andersen, R.J. Ramariolides A–D, Antimycobacterial Butenolides Isolated from the Mushroom Ramaria cystidiophora. J. Nat. Prod. 2012, 75, 2178–2182. [Google Scholar] [CrossRef] [PubMed]
- Bahbah, E.I.; Ghozy, S.; Attia, M.S.; Negida, A.; Emran, T.B.; Mitra, S.; Albadrani, G.M.; Abdel-Daim, M.M.; Uddin, M.S.; Simal-Gandara, J. Molecular Mechanisms of Astaxanthin as a Potential Neurotherapeutic Agent. Mar. Drugs 2021, 19, 201. [Google Scholar] [CrossRef]
- Hanh, B.T.B.; Kim, T.H.; Park, J.W.; Lee, D.G.; Kim, J.S.; Du, Y.E.; Yang, C.S.; Oh, D.C.; Jang, J. Etamycin as a Novel Mycobacterium abscessus Inhibitor. Int. J. Mol. Sci. 2020, 21, 6908. [Google Scholar] [CrossRef]
- Lukarska, M.; Palencia, A. Chapter Eleven—Aminoacyl-tRNA synthetases as drug targets. In The Enzymes; Ribas de Pouplana, L., Kaguni, L.S., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 48, pp. 321–350. [Google Scholar]
- Song, X.; Yuan, G.; Li, P.; Cao, S. Guanidine-Containing Polyhydroxyl Macrolides: Chemistry, Biology, and Structure-Activity Relationship. Molecules 2019, 24, 3913. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).