Agarose-Degrading Characteristics of a Deep-Sea Bacterium Vibrio Natriegens WPAGA4 and Its Cold-Adapted GH50 Agarase Aga3420
Abstract
1. Introduction
2. Results
2.1. Identification and Taxonomy of Strain WPAGA4
2.2. The Determination and Optimization of the Agarose-Degrading Activity in the Broth
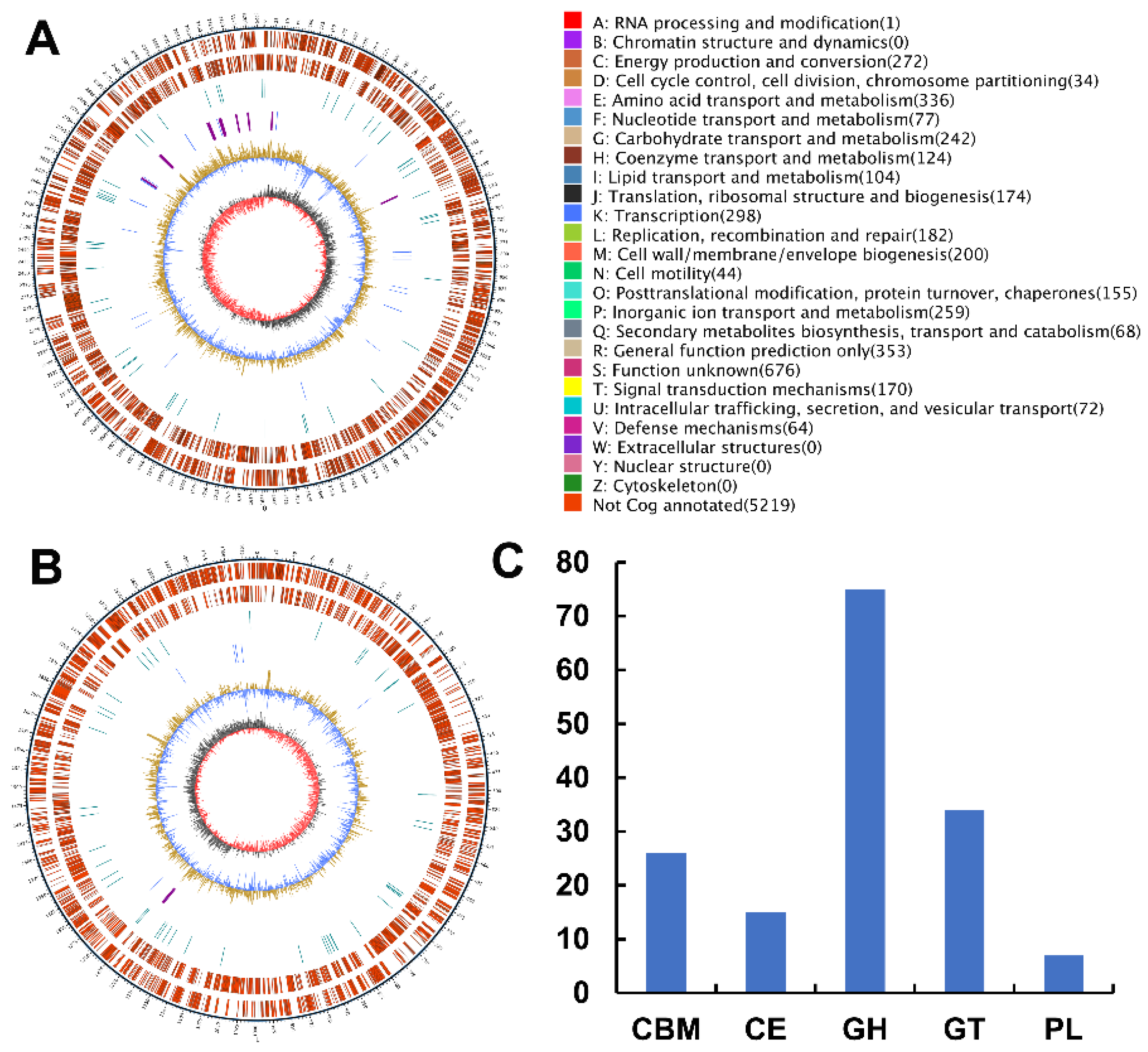
2.3. Analyses of the Genome Sequence and Putative Agarase Gene Aga3420 of Strain WPAGA4
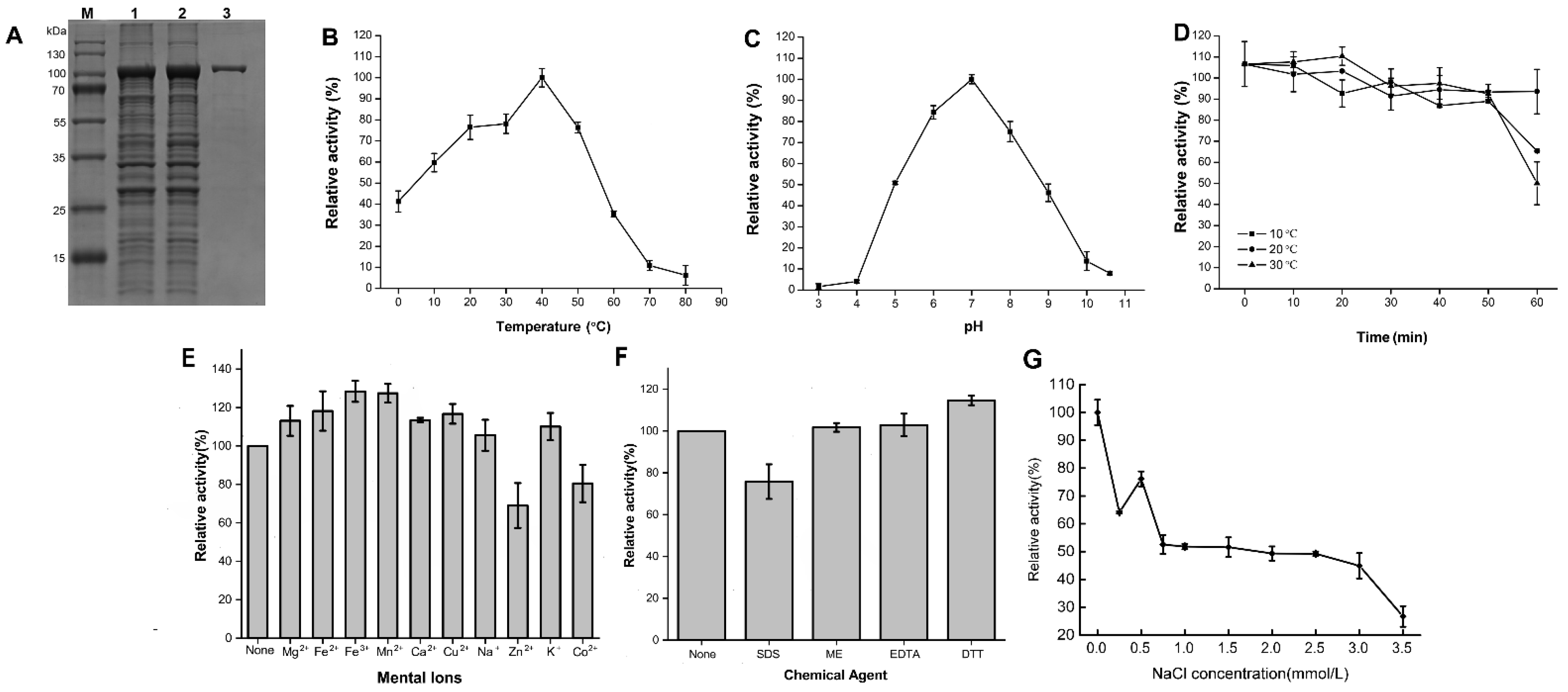
2.4. Expression and Characterization of the Recombinant Agarases
2.5. Agarose Degradation Products by rAga3420
3. Discussion
4. Materials and Methods
4.1. Isolation, Purification, and Identification
4.2. Genome Sequencing and Phylogenetic Analysis of Strain WPAGA4
4.3. The Determination and Optimization of the Agarase Activity in the Fermentation Broth
4.4. The Analysis of Agarase Gene Aga3420 in the Genome of WPAGA4
4.5. The Cloning and Expression of the Agarase Genes
4.6. Activity Determination and Characterization of rAga3420
4.7. The Determination of Degradation Products
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fu, X.T.; Kim, S.M. Agarase: Review of major sources, categories, purification method, enzyme characteristics and applications. Mar. Drugs 2010, 8, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-K.; Lim, Y.-Y.; Leow, A.T.-C.; Namasivayam, P.; Abdullah, J.O.; Ho, C.-L. Biosynthesis of agar in red seaweeds: A review. Carbohydr. Polym. 2017, 164, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Lee, C.-R.; Hong, S.-K. Implications of agar and agarase in industrial applications of sustainable marine biomass. Appl. Microbiol. Biotechnol. 2020, 104, 2815–2832. [Google Scholar] [CrossRef]
- Yun, E.J.; Yu, S.; Kim, K.H. Current knowledge on agarolytic enzymes and the industrial potential of agar-derived sugars. Appl. Microbiol. Biotechnol. 2017, 101, 5581–5589. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jin, X.; Wu, C.; Zhu, X.; Liu, M.; Call, D.R.; Zhao, Z. Genome-wide identification and functional characterization of β-agarases in Vibrio astriarenae strain HN897. Front. Microbiol. 2020, 11, 1404. [Google Scholar] [CrossRef]
- Xu, Z.-X.; Yu, P.; Liang, Q.-Y.; Mu, D.-S.; Du, Z.-J. Inducible expression of agar-degrading genes in a marine bacterium Catenovulum maritimus Q1T and characterization of a β-agarase. Appl. Microbiol. Biotechnol. 2020, 104, 10541–10553. [Google Scholar] [CrossRef]
- Hunt, D.E.; Gevers, D.; Vahora, N.M.; Polz, M.F. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 2008, 74, 44–51. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, H.; Wang, X.; Austin, B. Significance of Vibrio species in the marine organic carbon cycle—A review. Sci. China Earth Sci. 2018, 61, 1357–1368. [Google Scholar] [CrossRef]
- Chi, W.-J.; Chang, Y.-K.; Hong, S.-K. Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 2012, 94, 917–930. [Google Scholar] [CrossRef]
- Wu, Y.-R.; Zhou, Z.-R.; Zhao, M.; Lin, B.; Zhong, M.; Hu, Z. Molecular characterization of the thermostability and carbohydrate-binding module from a newly identified GH118 family agarase, AgaXa. Process Biochem. 2017, 52, 192–199. [Google Scholar] [CrossRef]
- Choi, U.; Jung, S.; Hong, S.-K.; Lee, C.-R. Characterization of a novel neoagarobiose-producing GH42 β-agarase, AgaJ10, from Gayadomonas joobiniege G7. Appl. Biochem. Biotechnol. 2019, 189, 1–12. [Google Scholar] [CrossRef]
- Jung, S.; Lee, C.-R.; Chi, W.-J.; Bae, C.-H.; Hong, S.-K. Biochemical characterization of a novel cold-adapted GH39 β-agarase, AgaJ9, from an agar-degrading marine bacterium Gayadomonas joobiniege G7. Appl. Microbiol. Biotechnol. 2017, 101, 1965–1974. [Google Scholar] [CrossRef]
- Chen, X.-L.; Hou, Y.-P.; Jin, M.; Zeng, R.-Y.; Lin, H.-T. Expression and characterization of a novel thermostable and pH-stable β-agarase from deep-sea bacterium Flammeovirga sp. OC4. J. Agric. Food Chem. 2016, 64, 7251–7258. [Google Scholar] [CrossRef]
- Sun, H.; Gao, L.; Xue, C.; Mao, X. Marine-polysaccharide degrading enzymes: Status and prospects. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2767–2796. [Google Scholar] [CrossRef]
- Dong, C.; Lin, B.; Song, Y.; Peng, T.; Zhong, M.; Li, J.; Hu, Z. Characterization and activity enhancement of a novel exo-type agarase Aga575 from Aquimarina agarilytica ZC1. Appl. Microbiol. Biotechnol. 2021, 105, 8287–8296. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Yan, R.; Zeng, R.; Zuo, Y.; Wang, D.; Qu, W. Expression and Characterization of a Novel Cold-Adapted and Stable β-Agarase Gene agaW1540 from the Deep-Sea Bacterium Shewanella sp. WPAGA9. Mar. Drugs 2021, 19, 431. [Google Scholar] [CrossRef]
- Lee, Y.R.; Jung, S.; Chi, W.J.; Bae, C.H.; Jeong, B.C.; Hong, S.K.; Lee, C.R. Biochemical Characterization of a Novel GH86 β-Agarase Producing Neoagarohexaose from Gayadomonas joobiniege G7. J. Microbiol. Biotechnol. 2018, 28, 284–292. [Google Scholar] [CrossRef]
- Chi, W.-J.; Seo, J.W.; Hong, S.-K. Characterization of two thermostable β-agarases from a newly isolated marine agarolytic bacterium, Vibrio sp. S1. Biotechnol. Bioprocess Eng. 2019, 24, 799–809. [Google Scholar] [CrossRef]
- Li, C.; Li, C.; Li, L.; Yang, X.; Chen, S.; Qi, B.; Zhao, Y. Comparative Genomic and Secretomic Analysis Provide Insights Into Unique Agar Degradation Function of Marine Bacterium Vibrio fluvialis A8 Through Horizontal Gene Transfer. Front. Microbiol. 2020, 11, 1934. [Google Scholar] [CrossRef]
- Liao, L.; Xu, X.W.; Jiang, X.W.; Cao, Y.; Yi, N.; Huo, Y.Y.; Wu, Y.H.; Zhu, X.F.; Zhang, X.Q.; Wu, M. Cloning, expression, and characterization of a new beta-agarase from Vibrio sp. strain CN41. Appl. Environ. Microbiol. 2011, 77, 7077–7079. [Google Scholar] [CrossRef]
- Shi, Y.-L.; Lu, X.-Z.; Yu, W.-G. A new β-agarase from marine bacterium Janthinobacterium sp. SY12. World J. Microbiol. Biotechnol. 2008, 24, 2659–2664. [Google Scholar] [CrossRef]
- Zhang, W.-w.; Sun, L. Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134. Appl. Environ. Microbiol. 2007, 73, 2825–2831. [Google Scholar] [CrossRef] [PubMed]
- Arnosti, C.; Wietz, M.; Brinkhoff, T.; Hehemann, J.H.; Probandt, D.; Zeugner, L.; Amann, R. The biogeochemistry of marine polysaccharides: Sources, inventories, and bacterial drivers of the carbohydrate cycle. Annu. Rev. Mar. Sci. 2021, 13, 81–108. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.G.; Carpenter, S.D.; Deming, J.W. Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can. J. Microbiol. 2009, 55, 63–72. [Google Scholar] [CrossRef]
- Raguénès, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 1997, 47, 989–995. [Google Scholar] [CrossRef]
- Zhang, N.X.; Zhang, D.C.; Qiao, N.H. Vibrio profundi sp. nov., isolated from a deep-sea seamount. Antonie Van Leeuwenhoek 2019, 112, 1603–1610. [Google Scholar] [CrossRef]
- Lasa, A.; Auguste, M.; Lema, A.; Oliveri, C.; Borello, A.; Taviani, E.; Bonello, G.; Doni, L.; Millard, A.D.; Bruto, M.; et al. A deep-sea bacterium related to coastal marine pathogens. Environ. Microbiol. 2021, 23, 5349–5363. [Google Scholar] [CrossRef]
- Labare, M.P.; Bays, J.T.; Butkus, M.A.; Snyder-Leiby, T.; Smith, A.; Goldstein, A.; Schwartz, J.D.; Wilson, K.C.; Ginter, M.R.; Bare, E.A.; et al. The effects of elevated carbon dioxide levels on a Vibrio sp. isolated from the deep-sea. Environ. Sci. Pollut. Res. Int. 2010, 17, 1009–1015. [Google Scholar] [CrossRef]
- Uchimura, K.; Miyazaki, M.; Nogi, Y.; Kobayashi, T.; Horikoshi, K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 2010, 12, 526–533. [Google Scholar] [CrossRef]
- Hamamoto, T.; Horikoshi, K. Characterisation of an amylase from a psychrotrophic Vibrio isolated from a deep-sea mud sample. FEMS Microbiol. Lett. 1991, 84, 79–84. [Google Scholar] [CrossRef]
- Ohta, Y.; Hatada, Y.; Ito, S.; Horikoshi, K. High-level expression of a neoagarobiose-producing beta-agarase gene from Agarivorans sp. JAMB-A11 in Bacillus subtilis and enzymic properties of the recombinant enzyme. Biotechnol. Appl. Biochem. 2005, 41 Pt 2, 183–191. [Google Scholar]
- Dong, J.; Tamaru, Y.; Araki, T. Molecular cloning, expression, and characterization of a beta-agarase gene, agaD, from a marine bacterium, Vibrio sp. strain PO-303. Biosci. Biotechnol. Biochem. 2007, 71, 38–46. [Google Scholar] [CrossRef]
- Kang, N.Y.; Choi, Y.L.; Cho, Y.S.; Kim, B.K.; Jeon, B.S.; Cha, J.Y.; Kim, C.H.; Lee, Y.C. Cloning, expression and characterization of a beta-agarase gene from a marine bacterium, Pseudomonas sp. SK38. Biotechnol. Lett. 2003, 25, 1165–1170. [Google Scholar] [CrossRef]
- Gao, B.; Jin, M.; Li, L.; Qu, W.; Zeng, R. Genome Sequencing Reveals the Complex Polysaccharide-Degrading Ability of Novel Deep-Sea Bacterium Flammeovirga pacifica WPAGA1. Front. Microbiol. 2017, 8, 600. [Google Scholar] [CrossRef]
- Miyazaki, M.; Nogi, Y.; Ohta, Y.; Hatada, Y.; Fujiwara, Y.; Ito, S.; Horikoshi, K. Microbulbifer agarilyticus sp. nov. and Microbulbifer thermotolerans sp. nov., agar-degrading bacteria isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 5, 1128–1133. [Google Scholar] [CrossRef]
- Xiao, Q.; Liu, C.; Ni, H.; Zhu, Y.; Jiang, Z.; Xiao, A. β-Agarase immobilized on tannic acid-modified Fe3O4 nanoparticles for efficient preparation of bioactive neoagaro-oligosaccharide. Food Chem. 2019, 272, 586–595. [Google Scholar] [CrossRef]
- Xiao, A.; Xiao, Q.; Lin, Y.; Ni, H.; Zhu, Y.; Cai, H. Efficient immobilization of agarase using carboxyl-functionalized magnetic nanoparticles as support. Electron. J. Biotechnol. 2017, 25, 13–20. [Google Scholar] [CrossRef]
- Liu, N.; Mao, X.; Yang, M.; Mu, B.; Wei, D. Gene cloning, expression and characterisation of a new β-agarase, AgWH50C, producing neoagarobiose from Agarivorans gilvus WH0801. World J. Microbiol. Biotechnol. 2014, 30, 1691–1698. [Google Scholar] [CrossRef]
- Gao, B.; Li, L.; Wu, H.; Zhu, D.; Jin, M.; Qu, W.; Zeng, R. A Novel Strategy for Efficient Agaro-Oligosaccharide Production Based on the Enzymatic Degradation of Crude Agarose in Flammeovirga pacifica WPAGA1. Front. Microbiol. 2019, 10, 1231. [Google Scholar] [CrossRef]
- Anggraeni, S.R.; Ansorge-Schumacher, M.B. Characterization and Modeling of Thermostable GH50 Agarases from Microbulbifer elongatus PORT2. Mar. Biotechnol. 2021, 23, 809–820. [Google Scholar] [CrossRef]
- Horikoshi, K. Barophiles: Deep-sea microorganisms adapted to an extreme environment. Curr. Opin. Microbiol. 1998, 1, 291–295. [Google Scholar] [CrossRef]
- Bhatia, R.K.; Ullah, S.; Hoque, M.Z.; Ahmad, I.; Yang, Y.-H.; Bhatt, A.K.; Bhatia, S.K. Psychrophiles: A source of cold-adapted enzymes for energy efficient biotechnological industrial processes. J. Environ. Chem. Eng. 2021, 9, 104607. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.-P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Ríos-Ríos, K.L.; Montilla, A.; Olano, A.; Villamiel, M. Physicochemical changes and sensorial properties during black garlic elaboration: A review. Trends Food Sci. Technol. 2019, 88, 459–467. [Google Scholar] [CrossRef]
- Chen, X.; Fu, X.; Huang, L.; Xu, J.; Gao, X. Agar oligosaccharides: A review of preparation, structures, bioactivities and application. Carbohydr. Polym. 2021, 265, 118076. [Google Scholar] [CrossRef]
- Hong, S.J.; Lee, J.-H.; Kim, E.J.; Yang, H.J.; Chang, Y.-K.; Park, J.-S.; Hong, S.-K. In vitro and in vivo investigation for biological activities of neoagarooligosaccharides prepared by hydrolyzing agar with β-agarase. Biotechnol. Bioprocess Eng. 2017, 22, 489–496. [Google Scholar] [CrossRef]
- Di, W.; Qu, W.; Zeng, R. Cloning, expression, and characterization of thermal-stable and pH-stable agarase from mangrove sediments. J. Basic Microbiol. 2018, 58, 302–309. [Google Scholar] [CrossRef]
- Kim, J.-D.; Lee, D.-G.; Lee, S.-H. Cloning, expression, and characterization of a thermotolerant β-agarase from Simiduia sp. SH-4. Biotechnol. Bioprocess Eng. 2018, 23, 525–531. [Google Scholar] [CrossRef]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.-W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef]
- Simpson, S.; Batley, G. Sediment Quality Assessment: A Practical Guide; CSIRO Publishing: Clayton, Australia, 2016. [Google Scholar]
- Suzuki, M.T.; Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996, 62, 625–630. [Google Scholar] [CrossRef]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Chai, C.; Chen, C.; Zhu, L.; Liu, J.; Hui, F. Genome Sequence Resource of Albifimbria verrucaria Causing the Leaf Spot Disease of the Spinach Plant Spinacia oleracea. Plant Dis. 2022, 106, 2511–2513. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinform. 2018, 62, e51. [Google Scholar] [CrossRef]
- Berns, R.S. Methods for characterizing CRT displays. Displays 1996, 16, 173–182. [Google Scholar] [CrossRef]
- Bertelli, C.; Brinkman, F.S.L. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. In The Protein Protocols Handbook; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; pp. 17–24. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Kim, J.H.; Yun, E.J.; Yu, S.; Kim, K.H.; Kang, N.J. Different Levels of Skin Whitening Activity among 3,6-Anhydro-l-galactose, Agarooligosaccharides, and Neoagarooligosaccharides. Mar. Drugs 2017, 15, 321. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, J.; Zeng, R.; Wang, D.; Wang, W.; Tong, X.; Qu, W. Agarose-Degrading Characteristics of a Deep-Sea Bacterium Vibrio Natriegens WPAGA4 and Its Cold-Adapted GH50 Agarase Aga3420. Mar. Drugs 2022, 20, 692. https://doi.org/10.3390/md20110692
Zhang M, Wang J, Zeng R, Wang D, Wang W, Tong X, Qu W. Agarose-Degrading Characteristics of a Deep-Sea Bacterium Vibrio Natriegens WPAGA4 and Its Cold-Adapted GH50 Agarase Aga3420. Marine Drugs. 2022; 20(11):692. https://doi.org/10.3390/md20110692
Chicago/Turabian StyleZhang, Mengyuan, Jianxin Wang, Runying Zeng, Dingquan Wang, Wenxin Wang, Xiufang Tong, and Wu Qu. 2022. "Agarose-Degrading Characteristics of a Deep-Sea Bacterium Vibrio Natriegens WPAGA4 and Its Cold-Adapted GH50 Agarase Aga3420" Marine Drugs 20, no. 11: 692. https://doi.org/10.3390/md20110692
APA StyleZhang, M., Wang, J., Zeng, R., Wang, D., Wang, W., Tong, X., & Qu, W. (2022). Agarose-Degrading Characteristics of a Deep-Sea Bacterium Vibrio Natriegens WPAGA4 and Its Cold-Adapted GH50 Agarase Aga3420. Marine Drugs, 20(11), 692. https://doi.org/10.3390/md20110692




