Abstract
The marine environment is an important source of specialized metabolites with valuable biological activities. Xanthones are a relevant chemical class of specialized metabolites found in this environment due to their structural variety and their biological activities. In this work, a comprehensive literature review of marine xanthones reported up to now was performed. A large number of bioactive xanthone derivatives (169) were identified, and their structures, biological activities, and natural sources were described. To characterize the chemical space occupied by marine-derived xanthones, molecular descriptors were calculated. For the analysis of the molecular descriptors, the xanthone derivatives were grouped into five structural categories (simple, prenylated, O-heterocyclic, complex, and hydroxanthones) and six biological activities (antitumor, antibacterial, antidiabetic, antifungal, antiviral, and miscellaneous). Moreover, the natural product-likeness and the drug-likeness of marine xanthones were also assessed. Marine xanthone derivatives are rewarding bioactive compounds and constitute a promising starting point for the design of other novel bioactive molecules.
1. Introduction
The marine environment occupies more than half of the Earth’s surface and harbors the largest pool of biodiversity. Among others, marine microorganisms produce specialized metabolites used mostly in interspecies competition and defense from predators. The harsh conditions found in the sea spurs the development of specific biosynthetic pathways that produce metabolites bearing novel scaffolds, quite different from those found in terrestrial sources [1]. Specialized metabolites were optimized by evolution to establish flawless interactions with biological targets [2]. The sum of these factors makes the marine environment a prolific source of structurally diverse bioactive molecules that have a pharmacological interest [3].
Among the most relevant chemical classes of specialized metabolites isolated from the marine biodiversity, xanthones are a class of oxygen-heterocycles containing a heterocycle containing a dibenzo γ-pyrone moiety [4,5,6]. Depending on the nature and position of substituents, xanthone derivatives show a wide variety of biological activities making the xanthone scaffold a “privileged structure” in Medicinal Chemistry with a good potential for the discovery of novel hits, leads, and drugs [7].
The chemical space occupied by a collection of substances is usually mapped by molecular descriptors [8]. Molecular descriptors are generically defined as mathematical representations of molecular features and embrace a vast collection of molecular, physicochemical, and topological parameters. Each molecular feature is encoded by at least one molecular descriptor. Size is usually inferred by the molecular weight (MW); flexibility by the number of rotatable bonds; lipophilicity by partition coefficient between octanol and water (Log P); polarity by the topological polar surface area (TPSA); solubility by the logarithm of the solubility measured in mol L−1 (log S); and carbon saturation by the fraction of sp3 carbons (Fsp3). Besides providing a numerical expression for chemical features, molecular descriptors allow tracking the suitable pharmacodynamics and pharmacokinetics properties, i.e., allow pursuing drug-likeness. Sets of rules or filters have been proposed over time in order to predict pharmacokinetic behavior. The most common set is the Lipinski′s rule of five [9], but other approaches have been suggested by other authors, namely by Veber [10], Ghoose [11], Egan [12], and Gleeson [13]. More recently, Bickerton et al. proposed the quantitative estimate of drug-likeness (QED) based on the calculation of the desirability of eight molecular properties [14]. Due to its usefulness in mapping the drug-likeness territory, this model has also been expanded to natural products (NP), creating the concept of NP-likeness evaluated by a score that allows comparing the chemical space covered by NPs with the one covered by synthetic molecules (SM) [15]. The application of this score helps medicinal chemists design molecules that are inspired by nature and have a higher probability of having a suitable pharmacokinetic behavior [16].
In this work, we review 169 bioactive marine xanthone derivatives and present their structures, biological activities, and marine sources. The chemical space occupied by bioactive marine xanthones is mapped and framed according to the NP-likeness and drug-likeness concepts.
2. Bioactive Xanthones Isolated from the Marine Environment
The bibliographic research was conducted using Scopus®, Web of Science®, and Google Scholar® without any temporal restriction. The keywords used were “marine AND xanthone*”.
In total, 169 xanthones derivatives were identified, which were sorted into 5 different structural categories (Figure 1). Simple oxygenated xanthones, bearing substituents such as hydroxyl, carboxyl, and methoxy groups, were classified as “simple” xanthones. Xanthones bearing isoprenyl groups were classified as “prenylated” compounds. Xanthones bearing additional O-heterocyclic groups, such a pyran or furan ring, were classified as “O-heterocyclic” compounds. Xanthones bearing O-heterocyclic and isoprenyl groups were classified as “O-heterocyclic” because they have a higher similarity with this category. Dimeric, pseudo-dimeric (one xanthonic and a hydroxanthone nucleus connected by a C-C bond), and glycosylated xanthones were loosely classified as “complex”. Dihydro-, tetrahydro-, and hexahydroxanthones were included in the “hydroxanthone” category (Figure 1).

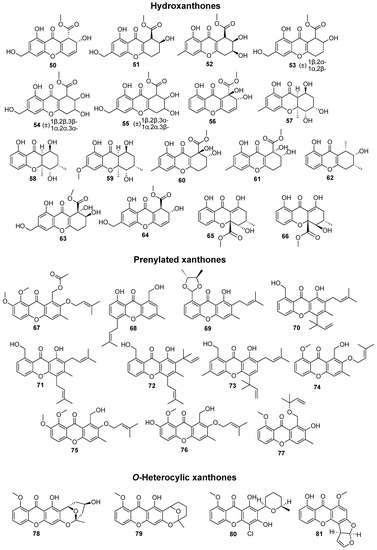
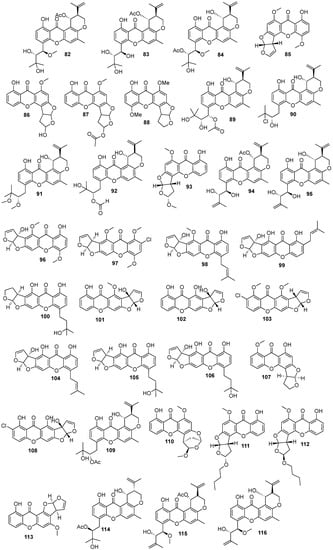
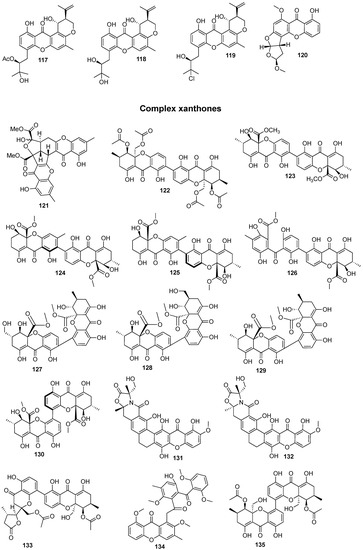
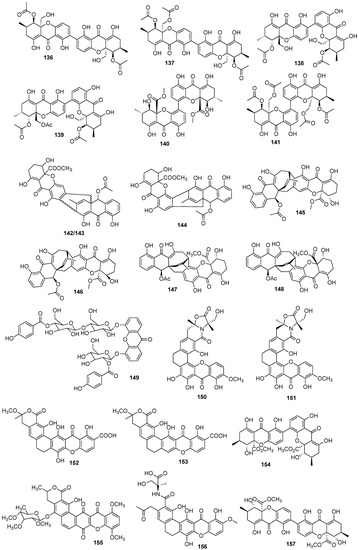
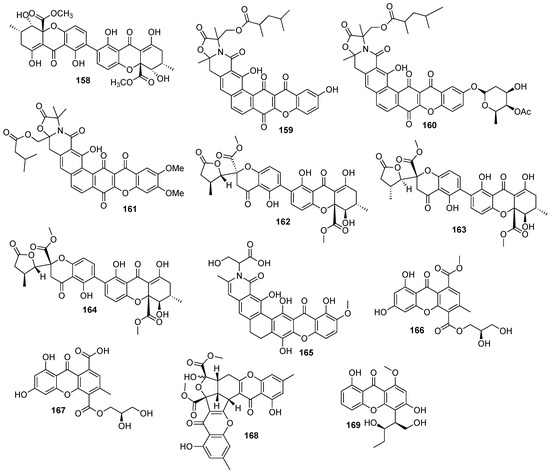
Figure 1.
Structures of bioactive marine xanthones.
From the considered 5 different structural types, “simple” (28.7%) and “complex” (28.7%) groups were the most prevalent, followed by “O-heterocyclic” (26.3%), “hydroxanthones” (9.9%), and “prenylated” (6.4%) (Figure 2a).
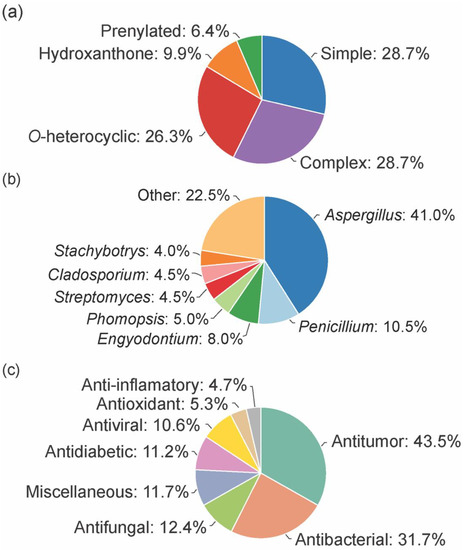
Figure 2.
Distribution of type (a) microorganism source (b) and activity (c) of the bioactive marine xanthones.
The bioactive marine xanthones were mostly isolated from marine fungi, namely from fungi belonging to the Aspergillus genus (41%, Figure 2b). Only a few examples were isolated from bacteria, and among them, the Streptomyces genus was the source that provided more bioactive xanthones (4.5%). Half of the reported bioactive marine xanthones (53%) were isolated from endophytic microorganisms associated with macroorganisms, like mangroves (39 marine xanthones), sponges (38 marine xanthones), algae (16 marine xanthones), corals (12 marine xanthones), jellyfish (1 marine xanthone), and seaweed (1 marine xanthone).
As specialized metabolites, xanthones are often used as chemical defense agents. The most prevalent described activities were antitumor (43.5%) and antimicrobial (antibacterial (31.7%), antifungal (12.4%), and antiviral (10.6%) which provides some sort of protection to other competitive or predator marine organisms (Figure 2c). Interestingly, 22% of the identified marine xanthones presented more than one biological activity (sum of all activities >100% in Figure 2c).
3. Chemical Space of Bioactive Marine Xanthones
Bioactive marine xanthones are produced by and act in living organisms, and to fulfill their specific biological task, they are structurally optimized by nature. Therefore, defining their chemical space is important for designing new molecules with desirable properties and pharmacological potential.
To describe the chemical space occupied by bioactive marine xanthones, several molecular descriptors, embracing different molecular, physico-chemical, and topological properties, were calculated using the RDkit (release 2021_03_5 Q1 2021) and SwissADME [17]. For each marine xanthone, the following molecular descriptors were calculated: molecular weight (MW), fraction of sp3 carbons (Fsp3), number of rotatable bonds (RB), lipophilicity (Log P), topological polar surface area (TPSA), and solubility (Log S) (Table S1). The molecular descriptors were analyzed accordingly to the structural type of xanthones (Figure 3).
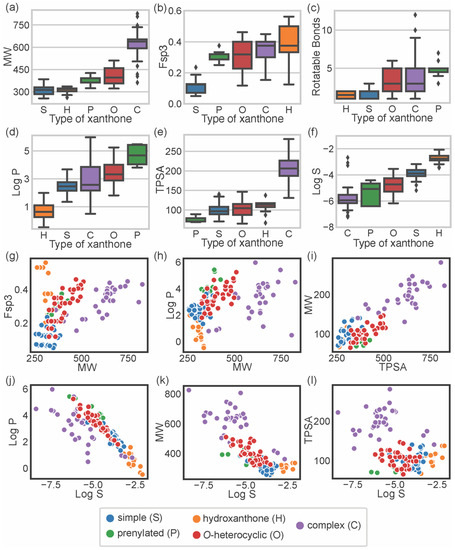
Figure 3.
Distribution of MW (a); Fsp3 (b); number of RBs (c); log P (d); TPSA (e); and log S (f) accordingly to the type of xanthone: “simple” (S, blue), “prenylated” (P, green), “hydroxanthone” (H, orange), O-heterocyclic (O, red), and complex (C, purple). Comparison between the values of Fsp3 carbons and MW (g); log P and MW (h); MW and TPSA (i); log P and log S (j); MW and log S (k); TPSA and log S (l).
Molecular size can be expressed in terms of MW, which is a predictor for pharmacokinetics behavior because bioavailability usually decreases as the molecular size increases [18]. In terms of size, the majority of the marine xanthones presented MWs within the range of 300 to 600 g·mol−1. “Complex” xanthones presented the highest mean value (621.7 g·mol−1) and the highest value dispersion (standard deviation of 85) due to the inclusion of several dimeric and glycosylated structures. On the other side, “simple” xanthones and “hydroxanthones” have the lowest MW mean values (306.9 and 311.2 g·mol−1, respectively) as these xanthones are composed by the xanthonic nucleus with simple substituents (such as hydroxyl or methyl or methoxy group).
Molecular flexibility depends primarily on carbon saturation (Fsp3) and on the number of rotatable bonds (RB). A high number of RB and/or Fsp3 means that the molecule has conformation flexibility which results in a less planar, less rigid, and more complex three-dimensional shape. Both RB and Fsp3 are important for determining oral bioavailability [10,19]. “Simple” xanthones have the lowest Fsp3 values (mean value of 0.11) as saturated bonds are only present in substituent groups (Figure 3b). “Prenylated” and “O-heterocyclic” xanthones have equal mean Fsp3 values (mean values of 0.31), but the latter showed higher value dispersion (interquartile range of 0.03 for “prenylated” and 0.17 for “O-heterocyclic” xanthones, Figure 3b), which is a consequence of higher 3D complexity rather than larger size (Figure 3g). “Complex” xanthones have a high value dispersion (standard deviation of 0.07, Figure 3b) due to their wide range of sizes (Figure 3g). Fsp3 values greater than 0.42 are considered to be suitable values for a drug [19], and half of the “hydroxanthones” obey this criterion. In agreement with Fsp3 analysis, “O-heterocyclic”, “complex”, and “prenylated” xanthones have a higher number of freely rotating bonds (median values of 3, 3, and 5 for “O-heterocyclic”, “complex”, and “prenylated”, respectively). “Hydroxanthones” have the highest Fsp3 values (median value of 0.38), but they have the lowest number of RB (median value of 1.50), meaning that saturated bonds belong to the cyclic system of the hydroxanthonic nucleus (Figure 3c). Good oral absorption is associated with a number of RBs < 10 [10], and the vast majority of the marine xanthones fulfill this criterion.
Lipophilicity, assessed by log P, is a key parameter that affects both pharmacodynamics and pharmacokinetics [20]. “Hydroxanthone” (mean value of 0.68) and “simple” xanthones (mean value of 2.48) have the lowest log P values as they are frequently substituted with hydrophilic groups (hydroxyl and carboxylic). “O-heterocyclic” xanthones presented log P values similar (median value of 3.32) to the xanthone itself (calculated log P of 2.95), meaning that the additional O-heterocyclic moiety does not contribute significantly to lipophilicity. “Prenylated” xanthones have the highest log P value (mean value of 4.68), significantly higher than the “O-heterocyclic”. “Complex” xanthones (median of 2.57) present the highest dispersion of log P values (standard deviation of 1.2), putting in evidence their structural diversity. The increase or decrease lipophilicity of marine xanthones is dependent on the substitution pattern, namely on the presence of hydrophilic or lipophilic substitutions. The size of the marine xanthone was not correlated with increasing lipophilicity as different sized molecules have quite similar log P values (Figure 3h), such as compound 10 (314 g·mol−1, log P 2.14) and compound 157 (638 g·mol−1, log P 2.16).
Molecular polarity, evaluated as the sum of surfaces of polar atoms in a molecule (TPSA), has been used to predict the permeability of drugs [21]. Different xanthone types have quite similar mean TPSA values (ranged from 74.8 Å2 for “prenylated” up to 110.7 Å2 for “hydroxanthones”), with the exception of complex xanthones that have significantly higher values (mean value of 203.9 Å2) (Figure 3e). “Complex” xanthones are the only type that violates the preconized 140 Å2 limit value [10]. In marine xanthones, the polarity is mostly related to MW, as TPSA values increase almost linearly with the MW (Figure 3i). In the case of marine xanthones, this is attributed to the increased number of polar atoms, such as oxygen or nitrogen, with increasing MW.
The water solubility, expressed as log S, is an important parameter for drug bioavailability. Compounds with poor water solubility have poor absorption and oral bioavailability, the evaluation of their bioactivity might be erratic, and the formulation development will be challenging [22]. The solubility trend observed with marine xanthones was: “hydroxanthones” > “simple” > “O-heterocyclic” > “prenylated” > “complex”. “Hydroxanthones” were the most soluble group (mean value of −2.74), while “complex” xanthones were the most poorly soluble group (mean value of −5.84) (Figure 3f). “Simple” and “hydroxanthones” are above the log S value of −4, which is considered an acceptable value for a drug [23]. This trend is related to lipophilicity (Figure 3j) and size (Figure 3k). The poor solubility of complex xanthones is ascribed to their high molecular weight and high lipophilicity, while the smaller and/or more hydrophilic hydroxanthones have good water solubility (Figure 3f). Solubility of marine xanthones seems not to be affected by polarity as log S and TPSA were not correlated (Figure 3l).
NP-likeness allows measuring the similarity of a molecule to natural products [15]. The NP-likeness score quantifies this similarity; the higher the score, the higher the resemblance of that molecule to an NP [15]. NP-likeness scores of marine xanthones were calculated using the web service NaPles [24]. Figure 4a depicts the probability density function, based on kernel density estimation (KDE), of NP-likeness score for all NPs and synthetic molecules (SMs), as well as the score of marine xanthones. As expected, the NP-likeness score of marine xanthones falls within the range of NP. NP-likeness score of marine xanthones was analyzed considering the different types of xanthones (Figure 4b). Xanthone itself is similar to SMs (NP-likeness score of 0.19), while hexahydroxanthone presents a high score (NP-likeness score of 1.57). Within marine xanthones, “simple” xanthones have the lowest similarity with NP molecules (mean score of 1.09). The extension of the degree of substitution, from the simple hydroxyl groups in “simple” xanthones, up to an additional xanthonic nucleus present in “complex” xanthones, leads to the increase in the similarity to NP. This is in agreement with the fact that usually, NP are structurally more complex than synthetic molecules [25].
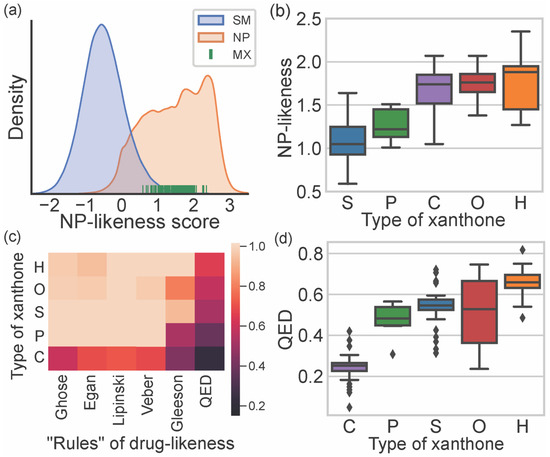
Figure 4.
(a) KDE distribution plot NP-likeness score of synthetic molecules (SM), natural products (NP), and NP-likeness score of marine xanthones. (b) Distribution of NP-likeness score accordingly to the type of xanthone: “simple” (S, blue), “prenylated” (P, green), “hydroxanthone” (H, orange), “O-heterocyclic” (O, red), and “complex” (C, purple). (c) Heatmap of the compliance with rules of drug-likeness for the xanthone types. (d) Distribution of QED index accordingly to the type of xanthone.
Drug-likeness allows estimating the probability of a molecule to become a drug administered orally [17]. The classical approach to drug-likeness is normally based on a set of criteria to which the compounds under study should obey. This approach provides a binary “yes or no” assessment, depending on if the compound obeys or not the preconized limit values. Drug-likeness of marine xanthones were evaluated considering the classical Lipinski [9], Veber [10], Ghoose [11], Egan [12], and Gleeson rules [13]. In this study, a compliance value, defined as 0 when a compound does not obey any of the preconized criteria of that rule and 1 when a compound fulfills all criteria, was calculated for each marine xanthones (Table S2). Figure 4c displays the obtained mean compliance values of each type of marine xanthones for each rule. A lighter color in the heatmap plotted in Figure 4c means higher compliance, while a darker color means less compliance. “Hydroxanthone”, “O-heterocyclic”, “simple”, and “prenylated” xanthones meet most of the criteria defined by classical rules, except for the Gleeson rules. On the contrary, “complex” xanthones violate at least one criterion in all the considered rules (Figure 4c). Among the classical rules, the Gleeson rules [13] were the best to discriminate the different types of xanthones. “Simple” and “hydroxanthones” obey most of the criteria, “prenylated” obey just some, and “complex” xanthones do not obey the generality of Gleeson’s proposed criteria.
Classical rules have many exceptions, and there are many examples, namely among NP or NP-inspired, of successful drugs that violate them [26]. Quantitative estimate of drug-likeness (QED) is an alternative way for assessing drug-likeness. QED index is generated considering eight properties, namely MW, log P, TPSA, RB, number of hydrogen donors and acceptors, number of aromatic rings, and number of alerts for undesirable substructures [14]. Compared with classical drug-likeness rules, the QED method is more flexible because it does not use cutoffs but a continuous score index of drug-likeness. When all properties are unfavorable, the QED index is 0, and when all properties are favorable, the score is 1 [14]. The obtained QED indexes for marine xanthones clearly differentiate the distinct types (Table S2, Figure 4c,d), enabling sorting the marine xanthones in the following ascending order of drug-likeness: “complex”, “prenylated”, “simple”, “O-heterocyclic”, and “hydroxanthones”. The low drug-likeness of “complex” marine xanthones is related to their high MW (Figure 3a) and low solubility (Figure 3f), while the drug-likeness of “hydroxanthones” is ascribed to their low lipophilicity (Figure 3d) and good water solubility (Figure 3f).
Considering the reported biological activities of marine xanthones, the relationship between these and the molecular descriptors was established. Figure 5a–d displays the relationship of the most relevant molecular descriptors (MW, log S, log P, and Fsp3) with the most representative biological activities (antitumor, antibacterial, antifungal, antiviral, and antidiabetic). As the number of xanthones reported for each biological activity is different, the results should only be compared when the number of reported molecules is similar (antitumor vs. antibacterial and antifungal vs. antiviral vs. antidiabetic).
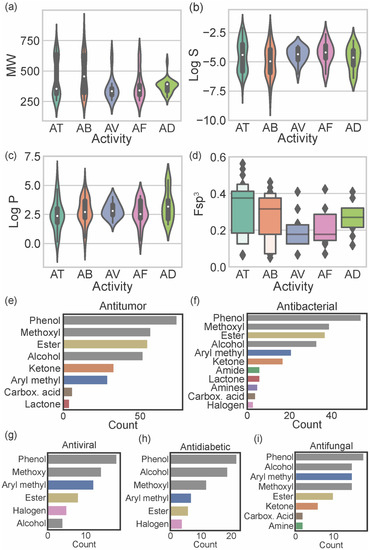
Figure 5.
Distribution of MW (a); log S (b); log P (c); and fraction of sp3 carbons (d) accordingly to the biological activity reported for marine xanthones: antitumor (AT), antibacterial (AB), antiviral (AV), antifungal (AF), and antidiabetic (AD). Analysis of the functional group frequently found on antitumor (e); antibacterial (f); antiviral (g); antifungal (h), and antidiabetic (i) marine xanthones.
Antitumor marine xanthones have a bimodal distribution of MW with 2 subsets of different sized compounds (one mode of 314.3 g mol−1 and another mode of 636.6 g mol−1) (Figure 5a). The presence of a bimodal distribution is also observed for the solubility of antitumor marine xanthones (modes of −3.45 and −5.66) (Figure 5b), which is not surprising considering that log S and MW are strictly correlated (Figure 3k). However, log P have a unimodal distribution with a mode of 2.33 (Figure 5c). Similarly, antibacterial marine xanthones also showed a bimodal distribution of MW (modes of 336.3 and 628.6 314.3 g mol−1) and log S values (modes of −4.23 and −5.84) (Figure 5a,b) and a unimodal distribution of log P values (mode of 2.37) (Figure 5c). The features of the large-sized subset of antitumor/antibacterial marine xanthone, i.e., “obese” molecules that apparently violate drug-likeness but with a suitable log P value, is a trait of NPs molecules. Despite being often cited as exceptions to classical drug-likeness rules, NP molecules largely comply in terms of log P [27]. This is attributed to the way in which natural evolution took place, producing bioactive compounds that retain low hydrophobicity, even for molecules with high MW [27]. The major difference between the physicochemical properties of antitumor and antibacterial xanthones was in terms of carbon saturation. Antibacterial marine xanthones have lower and more dispersed Fsp3 values than antitumor xanthones (median value of 0.38 and 0.32 for antibacterial and antitumor, respectively), raising the hypothesis that more rigidity might be an important aspect for the antibacterial activity (Figure 5d).
Antiviral, antifungal, and antidiabetic marine xanthones presented a unimodal distribution of MW values representing only one set of similar-sized compounds (mode values ranged from 334.7 to 394.5 g mol−1) (Figure 5a). Antidiabetic xanthones have a narrower probability distribution of the MW values (Figure 5a), and antiviral xanthones have a narrower dispersion of log S probability distribution (Figure 5b) and of log P probability distribution (Figure 5c). Antiviral and antifungal marine xanthones tend to be quite rigid molecules as they show the lowest Fsp3 values (median values of 0.18, Figure 5d). Antiviral, antifungal, and antidiabetic marine fulfill the limited preconized by the drug-likeness filters independently of the descriptor.
The most representative heteroatom in marine xanthones is the oxygen atom, distributed by phenols (present in 96.5% of the reported marine xanthones), methoxy groups (67.8%), alcohols (63.7%), esters (58.5%), ketones (26.9%), and carboxylic acids (7.0%) (Table S3). Other heteroatoms, different from oxygen, present in marine xanthones are halogens (9.4%), mainly the chloride atom, followed by the nitrogen atom, distributed by amines (10 marine xanthones) and amides (8 marine xanthones). The distribution of chemical functional groups by biological activities of marine xanthones was analyzed (Figure 5e–i). The most prevalent functional groups in antitumor, antibacterial, and antiviral xanthones are phenolic and methoxy groups (grey bars on Figure 5e–g). The most prevalent functional groups in antidiabetic and antifungal are phenolic and alcohol groups (grey bars on Figure 5h,i). Esters are very common in xanthones independent of biological activity. Ketones and aryl methyl groups are common in antitumor, antibacterial, and antifungal marine xanthones (Figure 5e,f,i). Halogens are present in some antiviral and antidiabetic xanthones. Nitrogen-containing groups, like amide and amines, are present in antibacterial and antifungal xanthones. Amine groups, which are protonated at physiological pH, could be important for the anti-infective activity of the marine xanthones bearing this group.
4. Biological Activities of Marine Xanthones
A total of 74 marine xanthones described in literature were evaluated for antitumor activity, measuring their growth inhibitory activity in different tumor cell lines (Table 1). The cervical carcinoma cell line (HeLA), human lung carcinoma (A549), human breast adenocarcinoma cell line (MCF-7), and human leukemia cell line (HL-60) were the most used tumor cell lines in the biological assays (Figure 6). The number of xanthones with a half maximum inhibitory concentration (IC50) lower than 10 µM varied depending on the tested cell lines. For instance, the number of xanthones screened against HL-60, MCF-7, and Huh7 cells was almost the same, but the number of most potent xanthones was higher identified against HL-60 cells. None of the marine xanthones assayed against C4-2B, 22RV1, and RWPE-1 presented an IC50 lower than 10 µM, while for MGC-803, the most part presented an IC50 lower than 10 µM.

Table 1.
Antitumor marine xanthones.
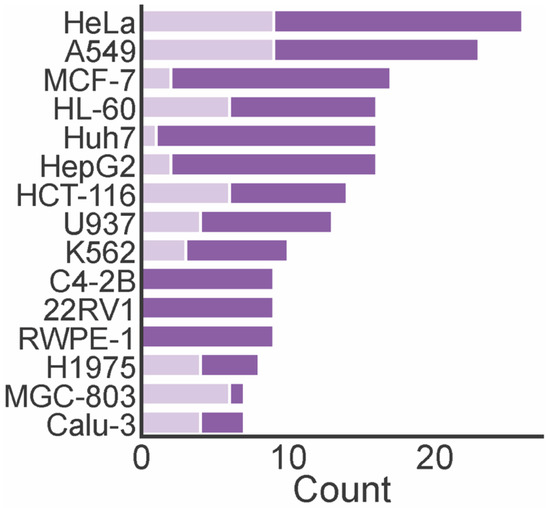
Figure 6.
The number of antitumor marine xanthone and the most frequently assayed tumor cell lines. Dark purple bar represents the total count of the assayed xanthones. Light purple bar represents the count of xanthones with IC50 lower than 10 µM.
A total of 54 marine xanthones described in literature were evaluated for antibacterial activity, measuring the growth inhibitory activity of different bacteria (Table 2). Gram-positive bacteria were more exploited (144 assessments) than Gram-negative (78 assessments) (Figure 7a). The number of xanthones with a minimum inhibitory concentration (MIC) lower than 4 µg mL−1 varied depending on the tested bacteria. Marine xanthones revealed a great selectivity for growth inhibition of Gram-positive bacteria as the percentage of marine xanthones with MIC lower than 4 is significantly higher in these types of bacteria (28% Gram-positive vs. 8% Gram-negative). This could be ascribed to a target potentially involving the peptidoglycan layer that is present in Gram-positive bacteria and absent in Gram-negative bacteria. The majority of marine xanthones were evaluated against S. aureus and E. coli (Figure 7b). Marine xanthones were particularly active against S. aureus, B. subtilis, E. faecalis, which are all Gram-positive bacteria.

Table 2.
Antibacterial marine xanthone.
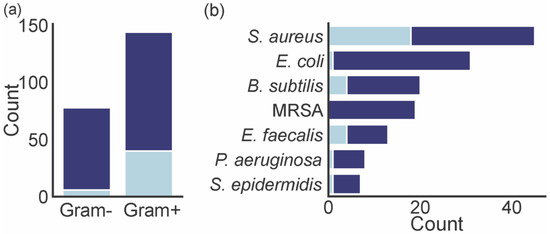
Figure 7.
(a) The number of antibacterial marine xanthones evaluated against Gram-positive and Gram-negative bacteria. Dark blue bar represents the total count of the assayed xanthones. Light blue bar represents the count of xanthones with MIC lower than 4 µg mL−1. (b) The bacteria that were assayed for antibacterial activity. Dark blue bar represents the total count of the assayed xanthones. Light blue bar represents the count of xanthones with MIC lower than 4 µg mL−1.
A total of 21 marine xanthones were evaluated for the antifungal activity against 17 different fungi, measuring the growth inhibitory activity of different fungi (Table 3). Among the evaluated fungi, Fusarium (12 xanthones), Colletotrichum (8 xanthones), Candida (5 xanthones), and Microbotryum (4 xanthones) were the most frequent genus.

Table 3.
Antifungal marine xanthone.
A total of 18 marine xanthones were evaluated against different viral targets of H1N1 (10 xanthones), HSV-2 (10 xanthones), HSV-1 (7 xanthones), HIV-1 (3 xanthones), EV71 (2 xanthones), H3N2 (2 xanthones), TMV (1 xanthone) (Table 4).

Table 4.
Antiviral marine xanthone.
A total of 19 marine xanthones were evaluated for antidiabetic activity using two different approaches: the assessment of α-glucosidase or protein tyrosine phosphatases inhibition activity and the assessment of the induction of the pancreatic β-cells proliferation in a zeafish model (Table 5).

Table 5.
Antidiabetic marine xanthone.
A total of 9 marine xanthones were evaluated for anti-oxidant activity through the DPPH assay and ABTS or trolox equivalent antioxidant capacity (TEAC) assay (Table 6).

Table 6.
Antioxidant marine xanthone.
A total of 8 marine xanthones were evaluated for anti-inflammatory activity by measuring the inhibitory activity against cyclooxygenase (COX), by measuring the inhibition of inflammatory response induced by nitric oxide (NO) and NF-κΒ (factor nuclear kappa B), and by measuring the decrease in IL-6 cytokine production on LPS-stimulated macrophages (Table 7).

Table 7.
Anti-inflammatory marine xanthone.
The remaining biological activities were classified as miscellaneous (Table 8). Seven marine xanthones were evaluated for their immunosuppressive activity through the assessment of the inhibition of proliferation of mouse splenic lymphocytes stimulated with Con-A and LPS. One xanthone was evaluated for its anti-Alzheimer activity through the assessment of acetylcholinesterase inhibition. Three marine xanthones were evaluated for their antiprotozoal activity against Trypanosoma brucei, Trypanosoma cruzi, Leshnmania donovani, and Plamodium falciparum. Nine marine xanthones were evaluated for their aquatic pathogens biocide activity against Vibrio sp.

Table 8.
Marine xanthone with miscellaneous biological activities.
5. Conclusions
As far as we know, 169 bioactive marine xanthone derivatives were reported in the literature up to 2021. They were isolated from microorganisms, mainly from Aspergillus sp., which normally live in an endophytic relationship with microorganisms (e.g., algae, sponge, mangrove, among others).
The chemical space occupied by bioactive marine xanthones was described through molecular descriptors. For each structural category, the distribution of the MW, Fsp3, number of RB, Log P, TPSA, and Log S values were described and analyzed. The descriptors were framed accordingly to the NP and drug-likeness concepts. Among the different structural categories of xanthones, “hydroxanthones” and “O-heterocyclic” xanthones are those that better resemble NPs and the ones that better fulfill the drug-likeness criteria. Therefore, hydroxanthones” and “O-heterocyclic” xanthones represent the most promising starting point for a hit-to-lead expansion.
In terms of biological activities, a total of 13 different activities were reported for marine xanthones. The antitumor and antibacterial activities were the most predominant. Potent antitumor marine xanthones (IC50 < 10 µM) were identified mostly against HeLa (cervical carcinoma), A549 (non-small cell lung carcinoma), HL-60 (acute myeloid leukemia), HCT-116 (colon carcinoma), and MCG-803 (Gastric mucinous adenocarcinoma) cells lines. The most potent antibacterial marine xanthones (MIC < 4 µg mL−1) were identified predominantly against Gram-positive bacteria, namely against S. aureus, B. subtilis, and E. faecalis.
Xanthones isolated from marine and terrestrial organisms share a similar biosynthetic pathway. However, marine-derived xanthones have not been as exploited as terrestrial-derived xanthones in traditional drug discovery campaigns. The numerous and relevant bioactive xanthones isolated from the marine environment could inspire the development of new drugs. The data concerning this review allow us to go deeper in understanding molecular properties, at different levels, of such an important family of marine NP.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/md20010058/s1, Table S1: Molecular descriptors of the bioactive marine xanthones, Table S2: NP-likeness and drug-likeness scores of the bioactive marine xanthones, Table S3: Chemical functional groups present in bioactive marine xanthones.
Author Contributions
Conceptualization, J.X.S. and C.M.M.A.; writing—original draft preparation, J.X.S., D.R.P.L., A.L.D.; writing—review and editing, C.M.M.A., S.R., M.M.M.P.; project administration, S.R., M.M.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by FCT/MCTES—Foundation for Science and Technology from the Minister of Science, Technology and Higher Education and European Regional Development Fund (ERDF) under the projects, co-financed by COMPETE 2020, Portugal 2020, PTDC/SAU-PUB/28736/2017 (POCI-01-0145-FEDER-028736), PTDC/CTA-AMB/0853/2021, and within the scope of UIDB/04423/2020, UID/QUI/5000612019, and UIDP/04423/2020 (Group of Natural Products and Medicinal Chemistry). J.X.S. thanks for the FCT Ph.D. Programs, specifically by the BiotechHealth Program (PD/00016/2012), and for the grants (SFRH/BD/98105/2013 and SFRH/BD/116167/2016). DRPL thanks FCT for her Ph.D. grant (SFRH/BD/140844/2018). Gisela Adriano, Liliana Teixeira, and Sara Cravo for the technical support. Maria Sorokina for kindly providing the NP-likeness scores of NP and SM.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shang, J.; Hu, B.; Wang, J.; Zhu, F.; Kang, Y.; Li, D.; Sun, H.; Kong, D.-X.; Hou, T. Cheminformatic Insight into the Differences between Terrestrial and Marine Originated Natural Products. J. Chem. Inf. Model. 2018, 58, 1182–1193. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, D.R.P.; Magalhães, Á.F.; Soares, J.X.; Pinto, J.; Azevedo, C.M.G.; Vieira, S.; Henriques, A.; Ferreira, H.; Neves, N.; Bousbaa, H.; et al. Yicathins B and C and Analogues: Total Synthesis, Lipophilicity and Biological Activities. ChemMedChem 2020, 15, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, D.R.P.; Soares, J.X.; Costa, J.C.; Magalhães, Á.F.; Azevedo, C.M.G.; Pinto, M.M.M.; Afonso, C.M.M. Structures, Activities and Drug-Likeness of Anti-Infective Xanthone Derivatives Isolated from the Marine Environment: A Review. Molecules 2019, 24, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, M.M.M.; Castanheiro, R.A.P.; Kijjoa, A. Xanthones from marine-derived microorganisms: Isolation, structure elucidation and biological activities. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 1–21. ISBN 978-0-470-02731-8. [Google Scholar]
- Pinto, M.M.M.; Palmeira, A.; Fernandes, C.; Resende, D.I.S.P.; Sousa, E.; Cidade, H.; Tiritan, M.E.; Correia-da-Silva, M.; Cravo, S. From Natural Products to New Synthetic Small Molecules: A Journey through the World of Xanthones. Molecules 2021, 26, 431. [Google Scholar] [CrossRef]
- Oprea, T.I.; Gottfries, J. Chemography: The Art of Navigating in Chemical Space. J. Comb. Chem. 2001, 3, 157–166. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.P. Generation of a Set of Simple, Interpretable ADMET Rules of Thumb. J. Med. Chem. 2008, 51, 817–834. [Google Scholar] [CrossRef] [PubMed]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the Chemical Beauty of Drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ertl, P.; Roggo, S.; Schuffenhauer, A. Natural Product-Likeness Score and Its Application for Prioritization of Compound Libraries. J. Chem. Inf. Model. 2008, 48, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Jayaseelan, K.V.; Moreno, P.; Truszkowski, A.; Ertl, P.; Steinbeck, C. Natural Product-Likeness Score Revisited: An Open-Source, Open-Data Implementation. BMC Bioinform. 2012, 13, 106. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsson, P.; Kihlberg, J. How Big Is Too Big for Cell Permeability? J. Med. Chem. 2017, 60, 1662–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Cherukupalli, S.; Jing, L.; Liu, X.; Zhan, P. Fsp3: A New Parameter for Drug-Likeness. Drug Discov. 2020, 25, 1839–1845. [Google Scholar] [CrossRef]
- Hann, M.M.; Keserü, G.M. Finding the Sweet Spot: The Role of Nature and Nurture in Medicinal Chemistry. Nat. Rev. Drug Discov. 2012, 11, 355–365. [Google Scholar] [CrossRef]
- Caron, G.; Ermondi, G. Molecular Descriptors for Polarity: The Need for Going beyond Polar Surface Area. Future Med. Chem. 2016, 8, 2013–2016. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Edward, H.K.; Guy, T.C. Drug-Like Property Concepts in Pharmaceutical Design. Curr. Pharm. Des. 2009, 15, 2184–2194. [Google Scholar] [CrossRef]
- Meanwell, N.A. Improving Drug Candidates by Design: A Focus on Physicochemical Properties As a Means of Improving Compound Disposition and Safety. Chem. Res. Toxicol. 2011, 24, 1420–1456. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Steinbeck, C. NaPLeS: A Natural Products Likeness Scorer—Web Application and Database. J. Cheminformatics 2019, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Stratton, C.F.; Newman, D.J.; Tan, D.S. Cheminformatic Comparison of Approved Drugs from Natural Product versus Synthetic Origins. Bioorg. Med. Chem. Lett. 2015, 25, 4802–4807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doak, B.C.; Over, B.; Giordanetto, F.; Kihlberg, J. Oral Druggable Space beyond the Rule of 5: Insights from Drugs and Clinical Candidates. Chem. Biol. 2014, 21, 1115–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesan, A. The Impact of Natural Products upon Modern Drug Discovery. Curr. Opin. Chem. Biol. 2008, 12, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.F.; Wang, J.E.; Zhang, X.Y.; Nong, X.H.; Xu, X.Y.; Qi, S.H. Cytotoxic Polyketides from the Deep-Sea-Derived Fungus Engyodontium Album DFFSCS021. Mar. Drugs 2014, 12, 5902–5915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.X.; Qiu, S.; She, Z.; Lin, Y. A New Xanthone Derivative from the Marine Fungus Phomopsis sp. (No. SK7RN3G1). Chem. Nat. Compd. 2013, 49, 31–33. [Google Scholar] [CrossRef]
- Krick, A.; Kehraus, S.; Gerhäuser, C.; Klimo, K.; Nieger, M.; Maier, A.; Fiebig, H.H.; Atodiresei, I.; Raabe, G.; Fleischhauer, J.; et al. Potential Cancer Chemopreventive in Vitro Activities of Monomeric Xanthone Derivatives from the Marine Algicolous Fungus Monodictys Putredinis. J. Nat. Prod. 2007, 70, 353–360. [Google Scholar] [CrossRef]
- Wang, C.N.; Lu, H.M.; Gao, C.H.; Guo, L.; Zhan, Z.Y.; Wang, J.J.; Liu, Y.H.; Xiang, S.T.; Wang, J.; Luo, X.W. Cytotoxic Benzopyranone and Xanthone Derivatives from a Coral Symbiotic Fungus Cladosporium Halotolerans GXIMD 02502. Nat. Prod. Res. 2020, 27, 1–8. [Google Scholar] [CrossRef]
- Wang, W.Y.; Gao, M.L.; Luo, Z.H.; Liao, Y.Y.; Zhang, B.B.; Ke, W.Q.; Shao, Z.Z.; Li, F.; Chen, J.M. Secondary Metabolites Isolated from the Deep Sea-Derived Fungus Aspergillus Sydowii C1-S01-A7. Nat. Prod. Res. 2019, 33, 3077–3082. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.Q.; Li, D.; Peng, J.X.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Penicitols A-C and Penixanacid A from the Mangrove-Derived Penicillium Chrysogenum HDN11-24. J. Nat. Prod. 2015, 78, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Lin, X.P.; Lu, X.; Wan, J.T.; Zhou, X.F.; Liao, S.R.; Tu, Z.C.; Xu, S.H.; Liu, Y.H. Sesquiterpenoids and Xanthones Derivatives Produced by Sponge-Derived Fungus Stachybotry sp. HH1 ZSDS1F1-2. J. Antibiot. 2015, 68, 121–125. [Google Scholar] [CrossRef]
- Yamazaki, H.; Rotinsulu, H.; Kaneko, T.; Murakami, K.; Fujiwara, H.; Ukai, K.; Namikoshi, M. A New Dibenz[b,e]Oxepine Derivative, 1-Hydroxy-10-Methoxy-Dibenz[b,e]Oxepin-6,11-Dione, from a Marine-Derived Fungus, Beauveria Bassiana TPU942. Mar. Drugs 2012, 10, 2691–2697. [Google Scholar] [CrossRef] [Green Version]
- Tao, H.M.; Wei, X.Y.; Lin, X.P.; Zhou, X.F.; Dong, J.D.; Yang, B. Penixanthones A and B, Two New Xanthone Derivatives from Fungus Penicillium sp. SYFz-1 Derived of Mangrove Soil Sample. Nat. Prod. Res. 2017, 31, 2218–2222. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.Q.; Qin, X.C.; Lin, X.P.; Kaliyaperumal, K.; Zhou, X.F.; Liu, J.; Ju, Z.R.; Tu, Z.C.; Liu, Y.H. Sydoxanthone C and Acremolin B Produced by Deep-Sea-Derived Fungus Aspergillus sp. SCSIO Ind09F01. J. Antibiot. 2015, 68, 703–706. [Google Scholar] [CrossRef]
- Elnaggar, M.S.; Ebada, S.S.; Ashour, M.L.; Ebrahim, W.; Muller, W.E.G.; Mandi, A.; Kurtan, T.; Singab, A.; Lin, W.H.; Liu, Z.; et al. Xanthones and Sesquiterpene Derivatives from a Marine-Derived Fungus Scopulariopsis sp. Tetrahedron 2016, 72, 2411–2419. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Guo, W.Q.; Che, Q.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Versicones E-H and Arugosin K Produced by the Mangrove-Derived Fungus Aspergillus Versicolor HDN11-84. J. Antibiot. 2017, 70, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Lin, Y.C.; She, Z.G.; Du, D.S.; Chan, W.L.; Zheng, Z.H. Paeciloxanthone, a New Cytotoxic Xanthone from the Marine Mangrove Fungus Paecilomyces sp. (Tree1-7). J. Asian Nat. Prod. Res. 2008, 10, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Pontius, A.; Krick, A.; Kehraus, S.; Brun, R.; König, G.M. Antiprotozoal Activities of Heterocyclic-Substituted Xanthones from the Marine-Derived Fungus Chaetomium sp. J. Nat. Prod. 2008, 71, 1579–1584. [Google Scholar] [CrossRef]
- Zhu, F.; Lin, Y.C. Three Xanthones from a Marine-Derived Mangrove Endophytic Fungus. Chem. Nat. Compd. 2007, 43, 132–135. [Google Scholar] [CrossRef]
- Lee, Y.M.; Li, H.; Hong, J.; Cho, H.Y.; Bae, K.S.; Kim, M.A.; Kim, D.K.; Jung, J.H. Bioactive Metabolites from the Sponge-Derived Fungus Aspergillus Versicolor. Arch. Pharm. Res. 2010, 33, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Jiang, X.; Liu, X.P.; He, C.W.; Di, Y.X.; Lu, S.J.; Huang, H.L.; Lin, B.; Wang, D.; Fan, B.Y. Antibacterial Anthraquinone Dimers from Marine Derived Fungus Aspergillus sp. Fitoterapia 2019, 133, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Yang, M.Y.; Zhang, Y.H.; Shao, C.L.; Wang, C.Y.; Hu, L.D.; Cao, F.; Zhu, H.J. Absolute Configurations of 14,15-Hydroxylated Prenylxanthones from a Marine-Derived Aspergillus sp. Fungus by Chiroptical Methods. Sci. Rep. 2018, 8, 10621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.X.; Zhu, T.J.; Du, L.; Zhao, B.Y.; Li, D.H.; Gu, Q.Q. Sterigmatocystins from the Deep-Sea-Derived Fungus Aspergillus Versicolor. J. Antibiot. 2011, 64, 193–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.A.; Lin, X.; Zhou, X.; Chen, M.; Huang, X.; Yang, B.; Tao, H. Xanthones and Quinolones Derivatives Produced by the Deep-Sea-Derived Fungus Penicillium sp. SCSIO Ind16F01. Molecules 2017, 22, 1999. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Yuan, J.; Huang, X.S.; Wen, W.T.; Zhu, X.; Liu, Y.Y.; Li, H.X.; Lu, Y.J.; He, L.; Tan, H.M.; et al. New Dimeric Members of the Phomoxanthone Family: Phomolactonexanthones A, B and Deacetylphomoxanthone C Isolated from the Fungus Phomopsis sp. Mar. Drugs 2013, 11, 4961–4972. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.; Kimishima, A.; Setiawan, A.; Arai, M. Secalonic Acid D as a Selective Cytotoxic Substance on the Cancer Cells Adapted to Nutrient Starvation. J. Nat. Med. 2020, 74, 495–500. [Google Scholar] [CrossRef]
- Zhen, X.; Gong, T.; Wen, Y.H.; Yan, D.J.; Chen, J.J.; Zhu, P. A Chrysoxanthones A-C, Three New Xanthone-Chromanone Heterdimers from Sponge-Associated Penicillium Chrysogenum HLS111 Treated with Histone Deacetylase Inhibitor. Mar. Drugs 2018, 16, 357. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, Y.P.; Li, X.X.; Lu, Z.H.; Zheng, Q.H.; Liu, Q.Y. Isolation of 4,4′-Bond Secalonic Acid D from the Marine-Derived Fungus Penicillium Oxalicum with Inhibitory Property against Hepatocellular Carcinoma. J. Antibiot. 2019, 72, 34–44. [Google Scholar] [CrossRef]
- Wu, G.W.; Qi, X.; Mo, X.M.; Yu, G.H.; Wang, Q.; Zhu, T.J.; Gu, Q.Q.; Liu, M.; Li, J.; Li, D.H. Structure-Based Discovery of Cytotoxic Dimeric Tetrahydroxanthones as Potential Topoisomerase I Inhibitors from a Marine-Derived Fungus. Eur. J. Med. Chem. 2018, 148, 268–278. [Google Scholar] [CrossRef]
- Liu, L.L.; He, L.S.; Xu, Y.; Han, Z.; Li, Y.X.; Zhong, J.L.; Guo, X.R.; Zhang, X.X.; Ko, K.M.; Qian, P.Y. Caspase-3-Dependent Apoptosis of Citreamicin ε-Induced HeLa Cells Is Associated with Reactive Oxygen Species Generation. Chem. Res. Toxicol. 2013, 26, 1055–1063. [Google Scholar] [CrossRef]
- Yeon, J.T.; Kim, H.; Kim, K.J.; Lee, J.; Won, D.H.; Nam, S.J.; Kim, S.H.; Kang, H.; Son, Y.J. Acredinone C and the Effect of Acredinones on Osteoclastogenic and Osteoblastogenic Activity. J. Nat. Prod. 2016, 79, 1730–1736. [Google Scholar] [CrossRef] [PubMed]
- Pavão, G.B.; Venâncio, V.P.; de Oliveira, A.L.L.; Hernandes, L.C.; Almeida, M.R.; Antunes, L.M.G.; Debonsi, H.M. Differential Genotoxicity and Cytotoxicity of Phomoxanthone A Isolated from the Fungus Phomopsis Longicolla in HL60 Cells and Peripheral Blood Lymphocytes. Toxicol. Vitro 2016, 37, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.Y.; Takagi, M.; Shin-Ya, K. New Xanthoquinodin-like Compounds, JBIR-97,-98 and-99, Obtained from Marine Sponge-Derived Fungus Tritirachium sp. SpB081112MEf2. J. Antibiot. 2010, 63, 615–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, K.; Chung, B.; Shin, Y.; Rheingold, A.L.; Moore, C.E.; Park, S.J.; Park, S.; Lee, S.K.; Oh, K.B.; Shin, J.; et al. Pentacyclic Antibiotics from a Tidal Mud Flat-Derived Actinomycete. J. Nat. Prod. 2015, 78, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, Y.; Kawahara, T.; Hitora, Y.; Tsukamoto, S. Ukixanthomycin A: A Hexacyclic Xanthone from the Mudflat-Derived Actinomycete Streptomyces sp. Heterocycles 2020, 100, 1686–1693. [Google Scholar] [CrossRef]
- Li, H.L.; Li, X.M.; Liu, H.; Meng, L.H.; Wang, B.G. Two New Diphenylketones and a New Xanthone from Talaromyces Islandicus EN-501, an Endophytic Fungus Derived from the Marine Red Alga Laurencia Okamurai. Mar. Drugs 2016, 14, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Cai, X.L.; Yang, H.; Xia, X.K.; Guo, Z.Y.; Yuan, J.; Li, M.F.; She, Z.G.; Lin, Y.C. The Bioactive Metabolites of the Mangrove Endophytic Fungus Talaromyces sp. ZH-154 Isolated from Kandelia Candel (L.) Druce. Planta Med. 2010, 76, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.R.; Miao, F.P.; Zhang, J.; Wang, G.; Yin, X.L.; Ji, N.Y. Three New Xanthone Derivatives from an Algicolous Isolate of Aspergillus Wentii. Magn. Reson. Chem. 2013, 51, 65–68. [Google Scholar] [CrossRef]
- Wang, J.H.; Ding, W.J.; Wang, R.M.; Du, Y.P.; Liu, H.L.; Kong, X.H.; Li, C.Y. Identification and Bioactivity of Compounds from the Mangrove Endophytic Fungus Alternaria sp. Mar. Drugs 2015, 13, 4492–4504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.B.; Chen, W.J.; Shan, T.Z.; Sun, B.Y.; Yan, P.C.; Jiang, W. Antibacterial Diphenyl Ether, Benzophenone and Xanthone Derivatives from Aspergillus Flavipes. Chem. Biodivers. 2020, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Gao, J.; Hu, J.; He, H.; Huang, P.; Zhang, L.; Song, F. One New Xanthenone from the Marine-Derived Fungus Aspergillus Versicolor MF160003. Nat. Prod. Res. 2020, 34, 2907–2912. [Google Scholar] [CrossRef]
- Khoshbakht, M.; Thanaussavadate, B.; Zhu, C.X.; Cao, Y.; Zakharov, L.N.; Loesgen, S.; Blakemore, P.R. Total Synthesis of Chalaniline B: An Antibiotic Aminoxanthone from Vorinostat-Treated Fungus Chalara sp. 6661. J. Org. Chem. 2021, 86, 7773–7780. [Google Scholar] [CrossRef]
- Zhang, W.; Krohn, K.; Zia-Ullah, F.U.; Pescitelli, G.; Di Bari, L.; Antus, S.; Kurtán, T.; Rheinheimer, J.; Draeger, S.; Schulz, B. New Mono- and Dimeric Members of the Secalonic Acid Family: Blennolides A–G Isolated from the Fungus Blennoria sp. Chem. Eur. J. 2008, 14, 4913–4923. [Google Scholar] [CrossRef]
- Song, F.H.; Ren, B.; Chen, C.X.; Yu, K.; Liu, X.R.; Zhang, Y.H.; Yang, N.; He, H.T.; Liu, X.T.; Dai, H.Q.; et al. Three New Sterigmatocystin Analogues from Marine-Derived Fungus Aspergillus Versicolor MF359. Appl. Microbiol. Biotechnol. 2014, 98, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- Fredimoses, M.; Zhou, X.F.; Ai, W.; Tian, X.P.; Yang, B.; Lin, X.P.; Liu, J.; Liu, Y.H. Emerixanthone E, a New Xanthone Derivative from Deep Sea Fungus Emericella sp. SCSIO 05240. Nat. Prod. Res. 2019, 33, 2088–2094. [Google Scholar] [CrossRef] [PubMed]
- Fredimoses, M.; Zhou, X.F.; Lin, X.P.; Tian, X.P.; Ai, W.; Wang, J.F.; Liao, S.R.; Liu, J.; Yang, B.; Yang, X.W.; et al. New Prenylxanthones from the Deep-Sea Derived Fungus Emericella sp. SCSIO 05240. Mar. Drugs 2014, 12, 3190–3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmstrøm, J.; Christophersen, C.; Barrero, A.F.; Enrique Oltra, J.; Justicia, J.; Rosales, A. Bioactive Metabolites from a Marine-Derived Strain of the Fungus Emericella Variecolor. J. Nat. Prod. 2002, 65, 364–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.Q.; Lin, S.T.; Kumaravel, K.; Zhou, H.; Wang, S.Y.; Liu, Y.H. Polyketide-Derived Metabolites from the Sponge-Derived Fungus Aspergillus sp. F40. Phytochem. Lett. 2018, 27, 74–77. [Google Scholar] [CrossRef]
- Erbert, C.; Lopes, A.A.; Yokoya, N.S.; Furtado, N.; Conti, R.; Pupo, M.T.; Lopes, J.L.C.; Debonsi, H.M. Antibacterial Compound from the Endophytic Fungus Phomopsis Longicolla Isolated from the Tropical Red Seaweed Bostrychia Radicans. Bot. Mar. 2012, 55, 435–440. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, X.P.; Ju, Z.R.; Liao, X.J.; Huang, X.J.; Zhang, C.; Zhao, B.X.; Xu, S.H. Aspergchromones A and B, Two New Polyketides from the Marine Sponge-Associated Fungus Aspergillus sp. SCSIO XWS03F03. J. Asian Nat. Prod. Res. 2017, 19, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Sun, Y.L.; Zhang, X.Y.; Han, Z.; Gao, H.C.; He, F.; Qian, P.Y.; Qi, S.H. Antifouling and Antibacterial Polyketides from Marine Gorgonian Coral-Associated Fungus Penicillium sp. SCSGAF 0023. J. Antibiot. 2013, 66, 219–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Wiese, J.; Wenzel-Storjohann, A.; Malien, S.; Schmaljohann, R.; Imhoff, J.F. Engyodontochones, Antibiotic Polyketides from the Marine Fungus Engyodontium Album Strain LF069. Chem. Eur. J. 2016, 22, 7452–7462. [Google Scholar] [CrossRef] [PubMed]
- Eltamany, E.E.; Abdelmohsen, U.R.; Ibrahim, A.K.; Hassanean, H.A.; Hentschel, U.; Ahmed, S.A. New Antibacterial Xanthone from the Marine Sponge-Derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 2014, 24, 4939–4942. [Google Scholar] [CrossRef]
- Liu, L.L.; Xu, Y.; Han, Z.; Li, Y.X.; Lu, L.; Lai, P.Y.; Zhong, J.L.; Guo, X.R.; Zhang, X.X.; Qian, P.Y. Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces Caelestis. Mar. Drugs 2012, 10, 2571–2583. [Google Scholar] [CrossRef] [Green Version]
- Malet-Cascón, L.; Romero, F.; Espliego-Vázquez, F.; Grávalos, D.; Fernández-Puentes, J.L. IB-00208, a New Cytotoxic Polycyclic Xanthone Produced by a Marine-Derived Actinomadura. I. Isolation of the Strain, Taxonomy and Biological Activities. J. Antibiot. 2003, 56, 219–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peoples, A.J.; Zhang, Q.B.; Millett, W.P.; Rothfeder, M.T.; Peseatore, B.C.; Madden, A.A.; Ling, L.L.; Moore, C.M. Neocitreamicins I and II, Novel Antibiotics with Activity against Methicillin-Resistant Staphylococcus Aureus and Vancomycin-Resistant Enterococci. J. Antibiot. 2008, 61, 457–463. [Google Scholar] [CrossRef]
- Maiese, W.M.; Lechevalier, M.P.; Lechevalier, H.A.; Korshalla, J.; Goodman, J.; Wildey, M.J.; Kuck, N.; Greenstein, M. LL-E19085 Alpha, a Novel Antibiotic from Micromonospora Citrea: Taxonomy, Fermentation and Biological Activity. J. Antibiot. 1989, 42, 846–851. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Lateff, A.; Klemke, C.; König, G.M.; Wright, A.D. Two New Xanthone Derivatives from the Algicolous Marine Fungus Wardomyces Anomalus. J. Nat. Prod. 2003, 66, 706–708. [Google Scholar] [CrossRef]
- Shao, C.; Wang, C.; Wei, M.; Gu, Y.; Xia, X.; She, Z.; Lin, Y. Structure Elucidation of Two New Xanthone Derivatives from the Marine Fungus Penicillium sp. (ZZF 32#) from the South China Sea. Magn. Reson. Chem. 2008, 46, 1066–1069. [Google Scholar] [CrossRef]
- Wang, X.; Mao, Z.G.; Song, B.B.; Chen, C.H.; Xiao, W.W.; Hu, B.; Wang, J.W.; Jiang, X.B.; Zhu, Y.H.; Wang, H.J. Advances in the Study of the Structures and Bioactivities of Metabolites Isolated from Mangrove-Derived Fungi in the South China Sea. Mar. Drugs 2013, 11, 3601–3616. [Google Scholar] [CrossRef] [Green Version]
- Höller, U.; König, G.M.; Wright, A.D. A New Tyrosine Kinase Inhibitor from a Marine Isolate of Ulocladium Botrytis and New Metabolites from the Marine Fungi Asteromyces Cruciatus and Varicosporina Ramulosa. Eur. J. Org. Chem. 1999, 2949–2955. [Google Scholar] [CrossRef]
- Li, C.Y.; Zhang, J.; Shao, C.L.; Ding, W.J.; She, Z.G.; Lin, Y.C. A New Xanthone Derivative from the Co-Culture Broth of Two Marine Fungi (Strain No. E33 and K38). Chem. Nat. Compd. 2011, 47, 382–384. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.H.; Lin, W.H.; Wang, B.G.; Proksch, P. Co-Cultivation-A Powerful Emerging Tool for Enhancing the Chemical Diversity of Microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.F.; He, W.J.; Huang, X.L.; Tian, X.P.; Liao, S.R.; Yang, B.; Wang, F.Z.; Zhou, X.J.; Liu, Y.H. Antifungal New Oxepine-Containing Alkaloids and Xanthones from the Deep-Sea-Derived Fungus Aspergillus Versicolor SCSIO 05879. J. Agric. Food Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.W.; Yang, J.; Chen, F.M.; Lin, X.P.; Chen, C.M.; Zhou, X.F.; Liu, S.W.; Liu, Y.H. Structurally Diverse Polyketides From the Mangrove-Derived Fungus Diaporthe Sp SCSIO 41011 With Their Anti-Influenza A Virus Activities. Front. Chem. 2018, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.H.; Zhang, H.B.; Zhong, M.J.; Ma, L.Y.; Liu, D.S.; Liu, W.Z.; Ren, H. Potential Antiviral Xanthones from a Coastal Saline Soil Fungus Aspergillus Iizukae. Mar. Drugs 2018, 16, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.Z.; Peng, S.; Yang, J.; Cong, Z.W.; Lin, X.P.; Liao, S.R.; Yang, B.; Zhou, X.F.; Zhou, X.J.; Liu, Y.H.; et al. Structurally Diverse Sesquiterpenoids and Polyketides from a Sponge-Associated Fungus Aspergillus Sydowii SCSI041301. Fitoterapia 2019, 135, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tang, X.L.; Luo, X.C.; Sun, C.X.; Liu, K.C.; Zhang, Y.; Li, P.L.; Li, G.Q. Isolation and Identification of Three New Sterigmatocystin Derivatives from the Fungus Aspergillus Versicolor Guided by Molecular Networking Approach. Chem. Biodivers. 2020, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Ouyang, M.A.; Tan, Q.W. New Asperxanthone and Asperbiphenyl from the Marine Fungus Aspergillus sp. Pest Manag. Sci. 2009, 65, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Yang, B.; Liu, J.; Xun, T.; Liu, Y.; Zhou, X. Penicillixanthone A, a Marine-Derived Dual-Coreceptor Antagonist as Anti-HIV-1 Agent. Nat. Prod. Res. 2019, 33, 1467–1471. [Google Scholar] [CrossRef]
- Wang, J.F.; Zhou, L.M.; Chen, S.T.; Yang, B.; Liao, S.R.; Kong, F.D.; Lin, X.P.; Wang, F.Z.; Zhou, X.F.; Liu, Y.H. New Chlorinated Diphenyl Ethers and Xanthones from a Deep-Sea-Derived I Fungus Penicillium Chrysogenum SCSIO 41001. Fitoterapia 2018, 125, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Gan, Q.; Lin, C.Y.; Lu, C.J.; Chang, Y.M.; Che, Q.; Zhang, G.J.; Zhu, T.J.; Gu, Q.Q.; Wu, Z.Q.; Li, M.Y.; et al. Staprexanthones, Xanthone-Type Stimulators of Pancreatic Beta-Cell Proliferation from a Mangrove Endophytic Fungus. J. Nat. Prod. 2020, 83, 2996–3003. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Huang, Z.H.; Ma, X.; Zheng, Z.H.; Zhang, X.X.; Lu, X.H.; Qi, S.H. Mycotoxins as Inhibitors of Protein Tyrosine Phosphatases from the Deep-Sea-Derived Fungus Aspergillus Puniceus SCSIO Z021. Bioorganic Chem. 2021, 107, 104571. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; He, F.; Yu, J.H.; Zhai, H.J.; Cheng, Z.Q.; Jiang, C.S.; Zhang, Y.Y.; Zhang, Y.; Zhang, X.Y.; Chen, G.Y.; et al. New Chromones from a Marine-Derived Fungus, Arthrinium sp., and Their Biological Activity. Molecules 2018, 23, 1982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.H.; Liu, D.; Xu, Y.; Chen, J.L.; Lin, W.H. Antioxidant Xanthones and Anthraquinones Isolated from a Marine-Derived Fungus Aspergillus Versicolor. Chin. J. Nat. Med. 2018, 16, 219–224. [Google Scholar] [CrossRef]
- Du, X.W.; Liu, D.; Huang, J.; Zhang, C.J.; Proksch, P.; Lin, W.H. Polyketide Derivatives from the Sponge Associated Fungus Aspergillus Europaeus with Antioxidant and NO Inhibitory Activities. Fitoterapia 2018, 130, 190–197. [Google Scholar] [CrossRef]
- Wang, J.F.; Xu, F.Q.; Wang, Z.; Lu, X.; Wan, J.T.; Yang, B.; Zhou, X.F.; Zhang, T.Y.; Tu, Z.C.; Liu, Y.H. A New Naphthalene Glycoside from the Sponge-Derived Fungus Arthrinium sp. ZSDS1-F3. Nat. Prod. Res. 2014, 28, 1070–1074. [Google Scholar] [CrossRef]
- Liu, H.J.; Chen, S.H.; Liu, W.Y.; Liu, Y.Y.; Huang, X.S.; She, Z.G. Polyketides with Immunosuppressive Activities from Mangrove Endophytic Fungus Penicillium sp. ZJ-SY2. Mar. Drugs 2016, 14, 217. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Huang, J.X.; Zhou, X.F.; Lin, X.P.; Liu, J.; Liao, S.R.; Wang, J.F.; Liu, F.A.; Tao, H.M.; Liu, Y.H. The Fungal Metabolites with Potential Antiplasmodial Activity. Curr. Med. Chem. 2018, 25, 3796–3825. [Google Scholar] [CrossRef] [PubMed]
- Tempone, A.G.; de Oliveira, C.M.; Berlinck, R.G.S. Current Approaches to Discover Marine Antileishmanial Natural Products. Planta Med. 2011, 77, 572–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, A.; Zhang, X.W.; Zhang, M.; Li, W.; Ma, Z.Y.; Zhu, H.J.; Cao, F. Aspergixanthones I–K, New Anti-Vibrio Prenylxanthones from the Marine-Derived Fungus Aspergillus sp. ZA-01. Mar. Drugs 2018, 16, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.Q.; Li, X.M.; Xu, G.M.; Li, X.; An, C.Y.; Wang, B.G. Antibacterial Anthraquinone Derivatives Isolated from a Mangrove-Derived Endophytic Fungus Aspergillus Nidulans by Ethanol Stress Strategy. J. Antibiot. 2018, 71, 778–784. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).