Transcriptome Profiling of the Pacific Oyster Crassostrea gigas Visceral Ganglia over a Reproduction Cycle Identifies Novel Regulatory Peptides
Abstract
1. Introduction
2. Results and Discussion
2.1. VG Transcriptome Sequencing and Assembly
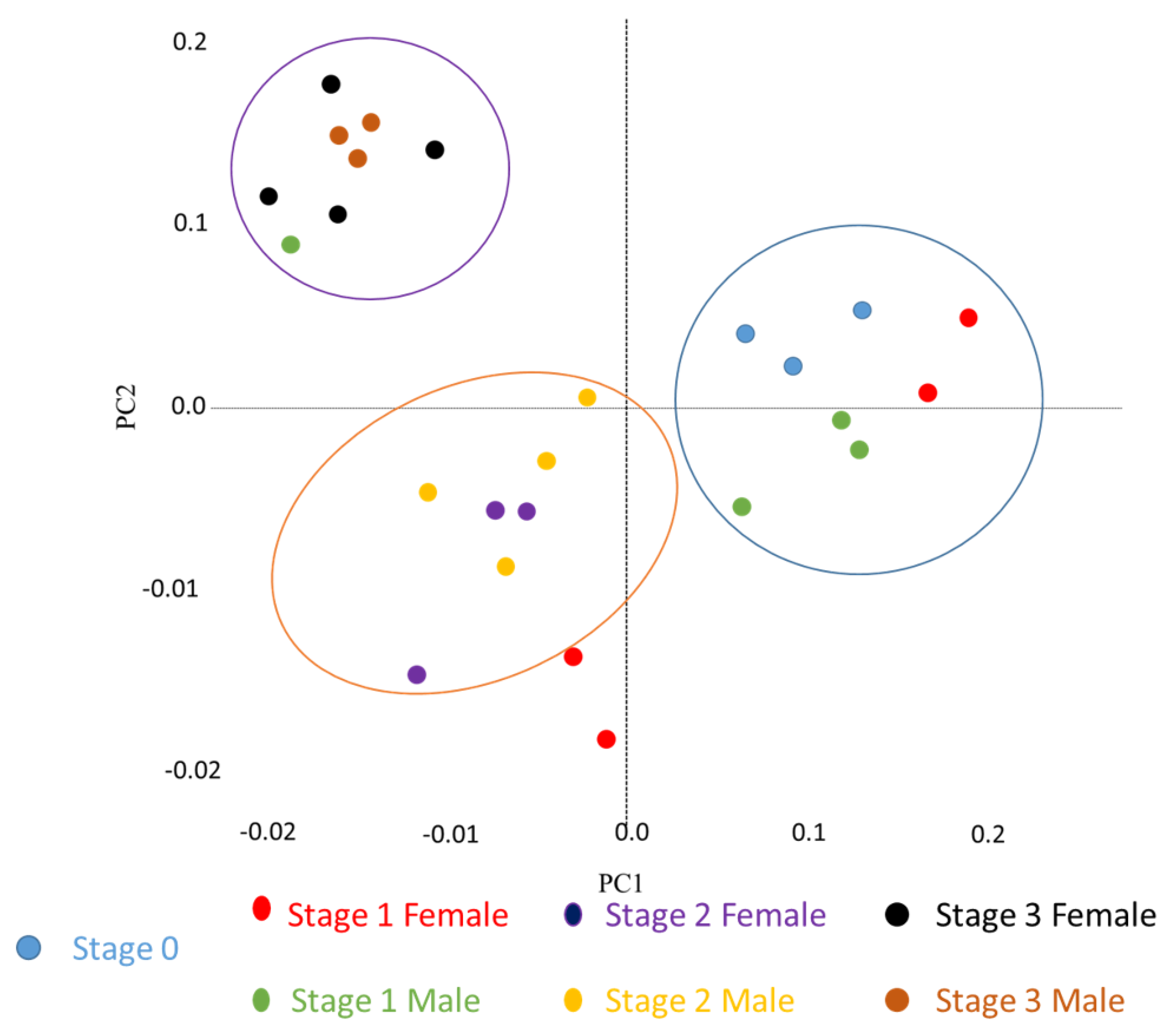
2.2. General Patterns of Transcript Expression in Oyster VG during Gametogenesis
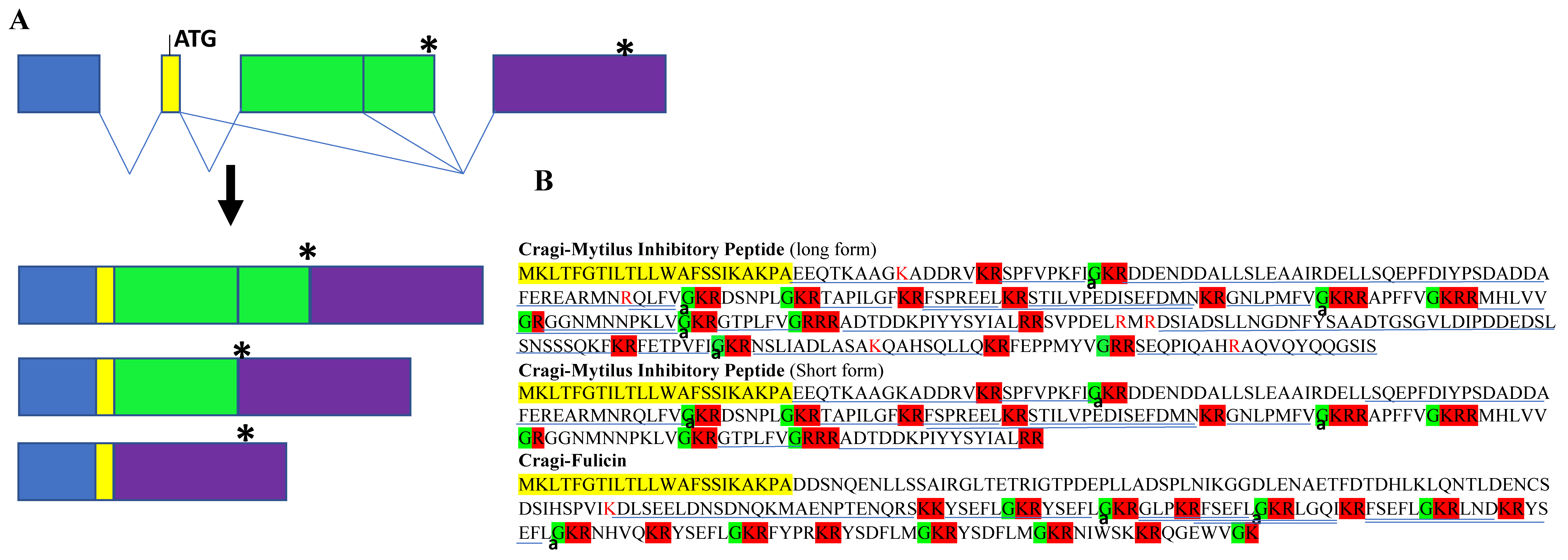
2.3. Identification of New NPP Transcripts
2.4. Bilaterian Families
2.4.1. Cragi-IL17/CFSH (Interleukin 17/Crustacean Female Sex Hormone)
2.4.2. Cragi-EH (Eclosion Hormone)
2.4.3. Cragi-Nesfatin
2.4.4. Cragi-Neuroparsin/IGFBP
2.4.5. Cragi-Urotensin
2.4.6. Cragi-Prokineticin
2.4.7. Cragi-Trunk/PTTH
2.5. Protostome Families
2.5.1. Cragi-Fulicin
2.5.2. Cragi-GNQQN
2.5.3. Cragi-Pleurin
2.5.4. Cragi-Prohormone 3
2.5.5. Cragi-Prohormone-4
2.5.6. Cragi-PXXXamide and QSamide Precursors
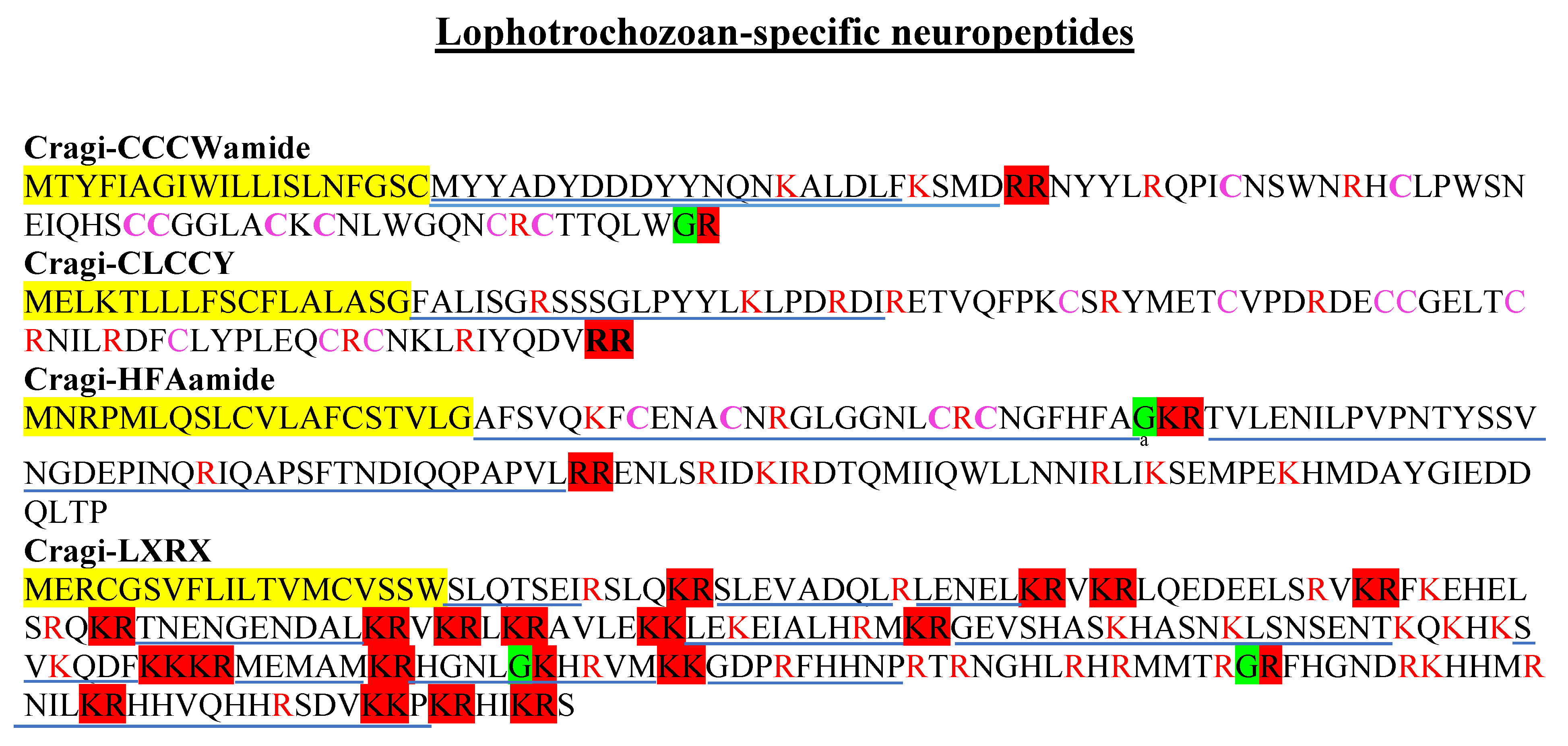
2.6. Lophotrochozoan Families
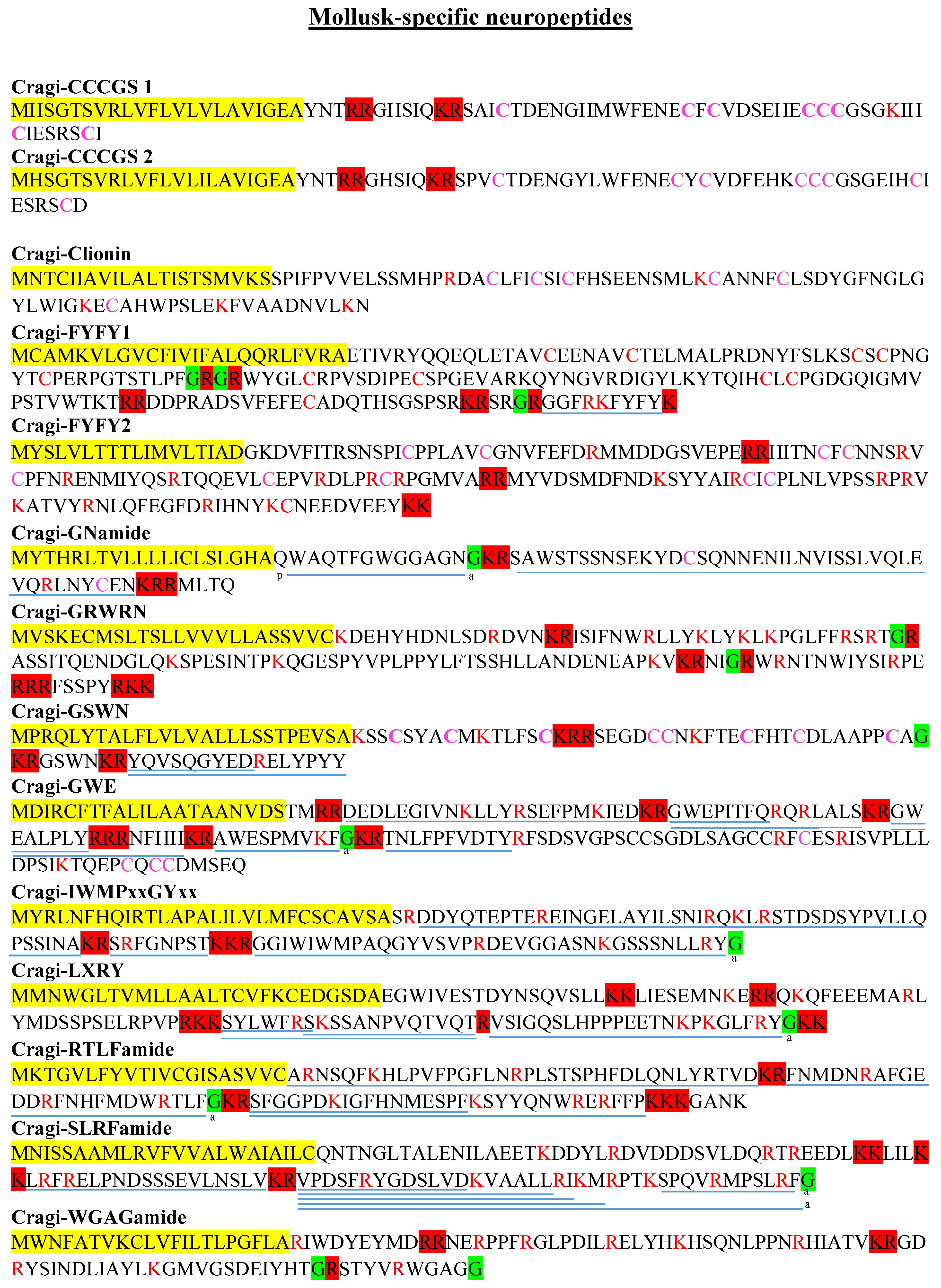
2.7. Mollusk Families
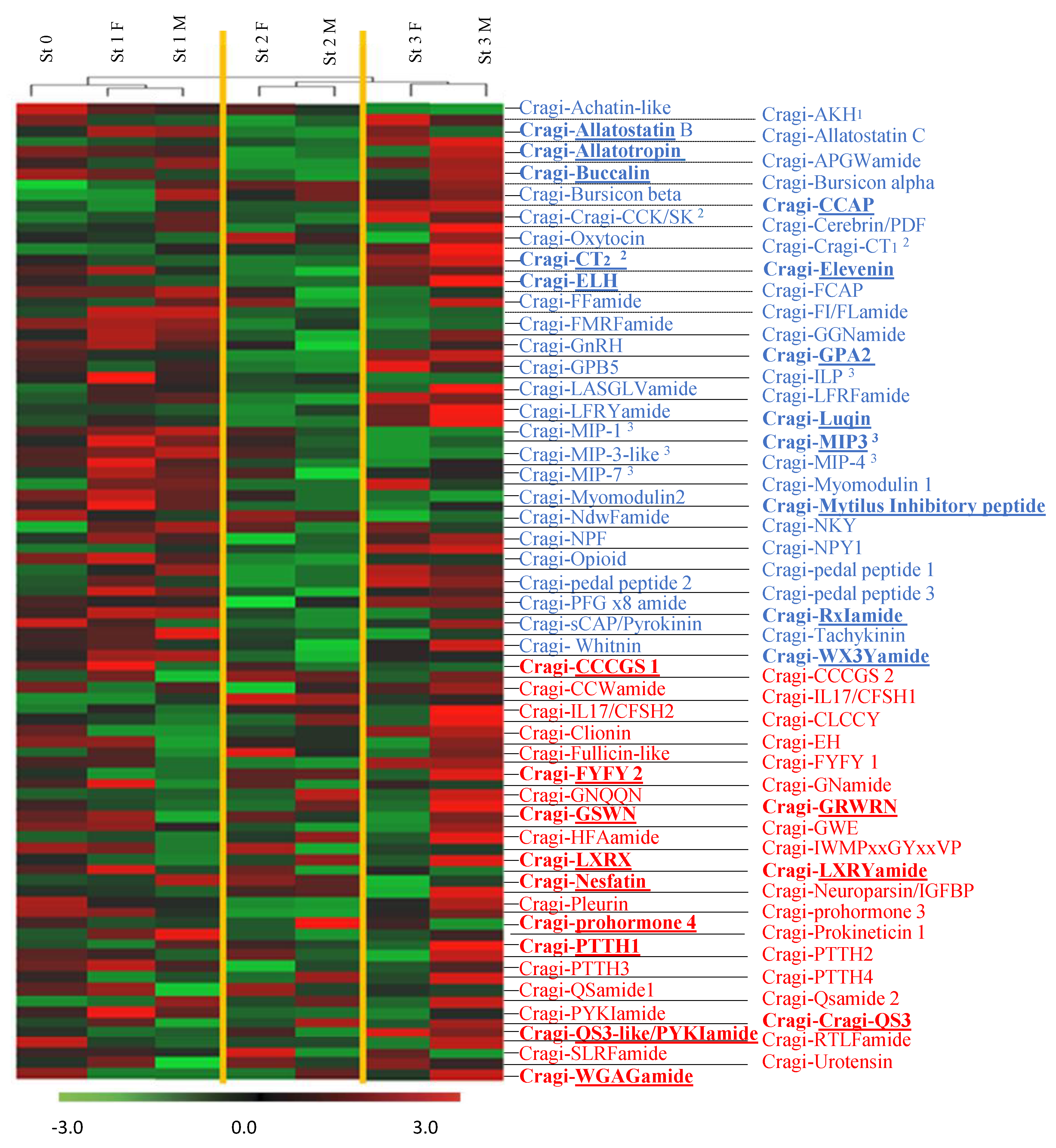
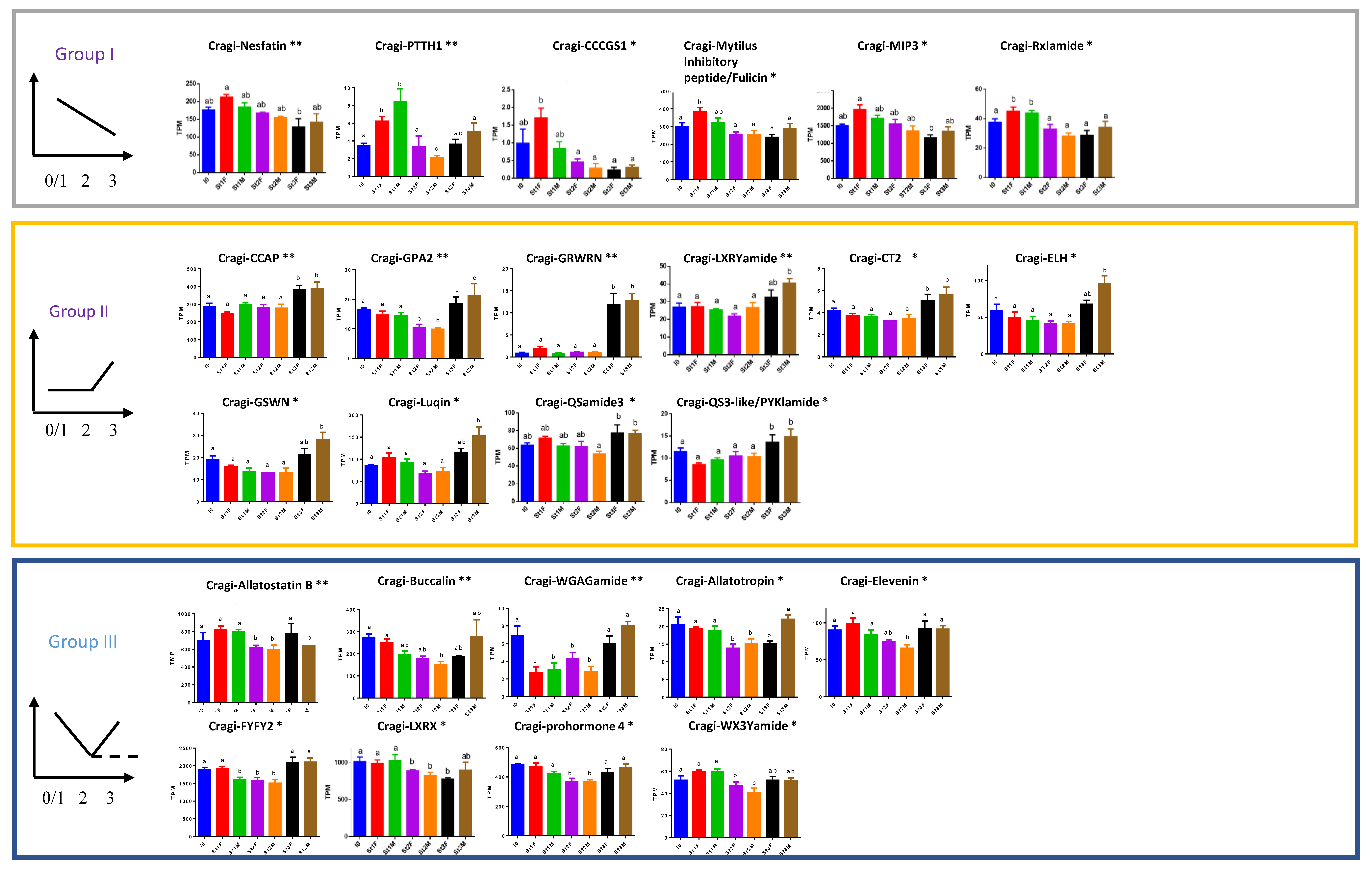
2.8. Differentially Expressed NPP Transcripts during a Reproductive Cycle
2.9. NPPs Differentially Expressed in the First Stages of Gametogenesis
2.10. NPPs Differentially Expressed in the Mature Spawning Stage
2.11. NPPs Expressed in the First Stages of Gametogenesis and in the Mature Spawning Stage
3. Materials and Methods
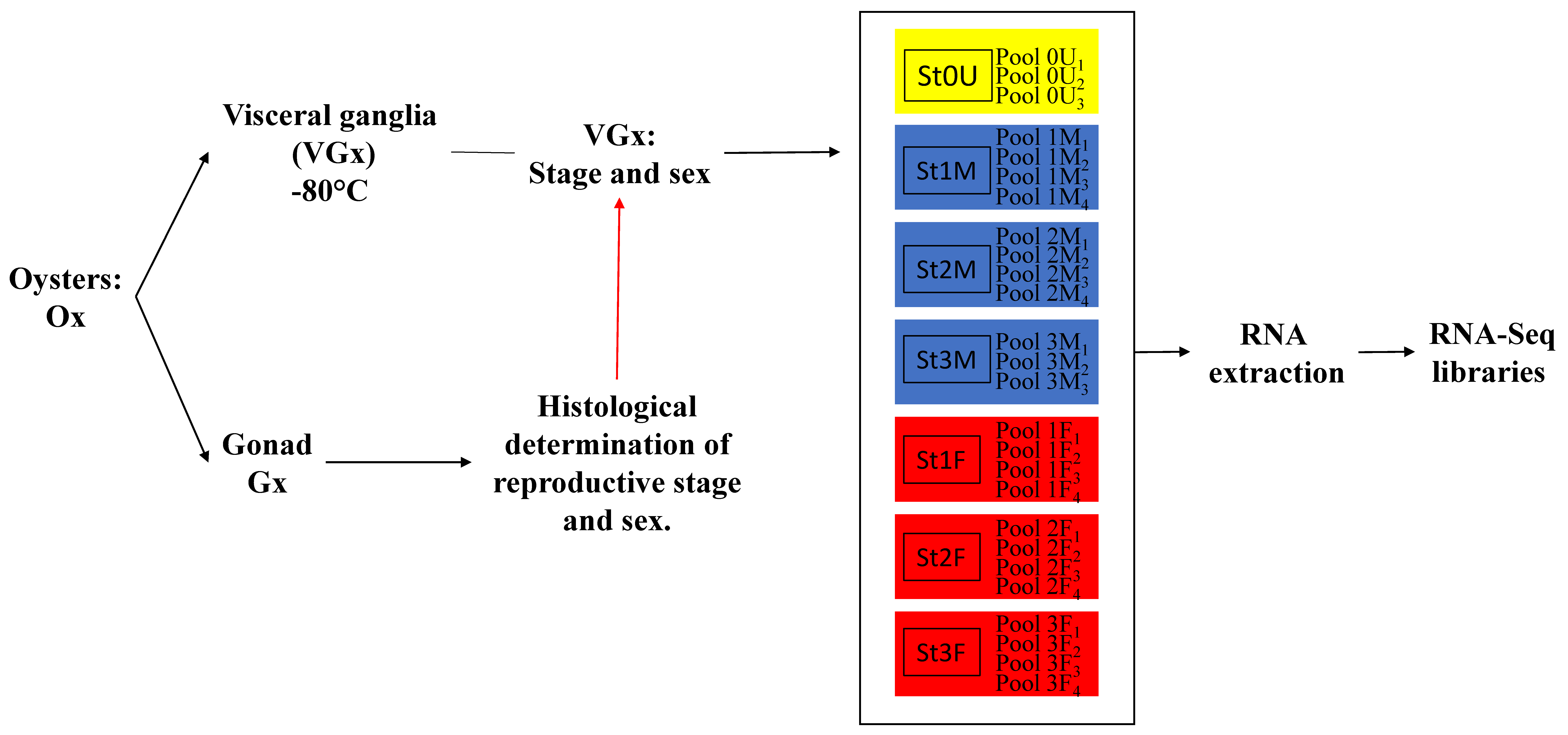
3.1. Animal and Tissue Sampling
3.2. RNA Extraction
3.3. Illumina Sequencing
3.4. Mass Spectrometry Analysis
3.4.1. Sample Preparation for Mass Spectrometry Analysis
3.4.2. Mass Spectrometry Analysis
3.4.3. Peptide Sequencing and Protein Precursor Identification
3.5. In Silico Studies
3.5.1. De Novo RNA-Seq Data Assembly
3.5.2. Global Analysis of the Transcriptome
3.5.3. Neuropeptide Precursor Searches
3.5.4. Search for Differentially Expressed Transcripts during a Reproduction Cycle
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galtsoff, P.S. The american oyster Crassostrea virginica Gmelin. Fish Bull. Fish Wildl. Serv. 1964, 64, 457. [Google Scholar]
- Guo, X.; He, Y.; Zhang, L.; Lelong, C.; Jouaux, A. Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immunol. 2015, 46, 107–119. [Google Scholar] [CrossRef]
- Zhang, G.G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef]
- Schoofs, L.; De Loof, A.; Van Hiel, M.B. Neuropeptides as regulators of behavior in insects. Annu. Rev. Entomol. 2017, 62, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Marder, E. Overview Neuromodulation of Neuronal Circuits: Back to the Future. Neuron 2012, 76, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, M.; Wang, W.; Wang, L.; Liu, Z.; Sun, J.; Wang, K.; Song, L. The immunomodulatory function of invertebrate specific neuropeptide FMRFamide in oyster Crassostrea gigas. Fish Shellfish Immunol. 2019, 88, 480–488. [Google Scholar] [CrossRef]
- Rowe, M.L.; Achhala, S.; Elphick, M.R. Neuropeptides and polypeptide hormones in echinoderms: New insights from analysis of the transcriptome of the sea cucumber Apostichopus japonicus. Gen. Comp. Endocrinol. 2014, 197, 43–55. [Google Scholar] [CrossRef]
- Semmens, D.C.; Mirabeau, O.; Moghul, I.; Pancholi, M.R.; Wurm, Y.; Elphick, M.R. Transcriptomic identification of starfish neuropeptide precursors yields new insights into neuropeptide evolution. Open Biol. 2016, 6, 150224. [Google Scholar] [CrossRef]
- Suwansa-ard, S.; Chaiyamoon, A.; Talarovicova, A.; Tinikul, R.; Tinikul, Y.; Poomtong, T.; Elphick, M.R.; Cummins, S.F. Peptides Transcriptomic discovery and comparative analysis of neuropeptide precursors in sea cucumbers (Holothuroidea). Peptides 2018, 99, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Conzelmann, M.; Williams, E.A.; Krug, K.; Franz-wachtel, M.; Macek, B. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genom. 2013, 14, 906. [Google Scholar] [CrossRef] [PubMed]
- Toullec, J.; Corre, E.; Thorne, M.; Cascella, K.; Ollivaux, C.; Henry, J.; Clark, M. Transcriptome and Peptidome Characterisation of the Main Neuropeptides and Peptidic Hormones of a Euphausiid: The Ice Krill, Euphausia crystallorophias. PLoS ONE 2013, 8, e71609. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, J.A. Neuropeptide evolution: Neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen. Comp. Endocrinol. 2011, 171, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, J.A. The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front. Physiol. 2014, 5, 1–22. [Google Scholar] [CrossRef]
- Veenstra, J.A.; Rombauts, S.; Grbić, M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem. Mol. Biol. 2012, 42, 277–295. [Google Scholar] [CrossRef]
- Adamson, K.J.; Wang, T.; Zhao, M.; Bell, F.; Kuballa, A.V.; Storey, K.B.; Cummins, S.F. Molecular insights into land snail neuropeptides through transcriptome and comparative gene analysis. BMC Genom. 2015, 16, 308. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Martin, R.; Rao, S.; Choi, M.-Y. Neuropeptides predicted from the transcriptome analysis of the gray garden slug Deroceras reticulatum. Peptides 2017, 93, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Suwansa-ard, S.; Maikaeo, L.; Motti, C.A.; Hall, M.R.; Cummins, S.F. Peptides Neuropeptides encoded within a neural transcriptome of the giant triton snail Charonia tritonis, a Crown-of-Thorns Starfish predator. Peptides 2017, 98, 3–14. [Google Scholar] [CrossRef]
- Veenstra, J.A. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen. Comp. Endocrinol. 2010, 167, 86–103. [Google Scholar] [CrossRef]
- Zatylny-Gaudin, C.; Cornet, V.; Leduc, A.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Bernay, B.; Garderes, J.; Kraut, A.; Couté, Y.; et al. Neuropeptidome of the cephalopod Sepia officinalis: Identification, tissue mapping, and expression pattern of neuropeptides and neurohormones during egg laying. J. Proteome Res. 2016, 15, 48–67. [Google Scholar] [CrossRef]
- Stewart, M.J.; Favrel, P.; Rotgans, B.A.; Wang, T.; Zhao, M.; Sohail, M.; O’Connor, W.A.; Elizur, A.; Henry, J.; Cummins, S.F.; et al. Neuropeptides encoded by the genomes of the Akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: A bioinformatic and peptidomic survey. BMC Genom. 2014, 15, 840. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Li, Y.; Li, W.; Li, R.; Xie, X.; Wang, S.; Hu, X.; Zhang, L.; Bao, Z. Identification and Characterization of Neuropeptides by Transcriptome and Proteome Analyses in a Bivalve Mollusc Patinopecten yessoensis. Front. Genet. 2018, 9, 197. [Google Scholar] [CrossRef]
- De Oliveira, A.L.; Calcino, A.; Wanninger, A. Extensive conservation of the proneuropeptide and peptide prohormone complement in mollusks. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Cherif-Feildel, M.; Berthelin, C.H.; Rivière, G.; Favrel, P.; Kellner, K. Data for evolutive analysis of insulin related peptides in bilaterian species. Data Br. 2019, 22, 546–550. [Google Scholar] [CrossRef]
- Cherif-Feildel, M.; Heude Berthelin, C.; Adeline, B.; Rivière, G.; Favrel, P.; Kellner, K. Molecular evolution and functional characterisation of insulin related peptides in molluscs: Contributions of Crassostrea gigas genomic and transcriptomic-wide screening. Gen. Comp. Endocrinol. 2019, 271, 15–29. [Google Scholar] [CrossRef]
- Dubos, M.-P.; Bernay, B.; Favrel, P. Molecular characterization of an adipokinetic hormone-related neuropeptide (AKH) from a mollusk. Gen. Comp. Endocrinol. 2017, 243, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Bigot, L.; Zatylny-Gaudin, C.; Rodet, F.; Bernay, B.; Boudry, P.; Favrel, P. Characterization of GnRH-related peptides from the Pacific oyster Crassostrea gigas. Peptides 2012, 34, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Dubos, M.-P.P.; Zels, S.; Schwartz, J.; Pasquier, J.; Schoofs, L.; Favrel, P. Characterization of a tachykinin signalling system in the bivalve mollusc Crassostrea gigas. Gen. Comp. Endocrinol. 2018, 266, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.; Dubos, M.-P.; Pasquier, J.; Zatylny-Gaudin, C.; Favrel, P. Emergence of a cholecystokinin/sulfakinin signalling system in Lophotrochozoa. Sci. Rep. 2018, 8, 16424. [Google Scholar] [CrossRef] [PubMed]
- Realis-Doyelle, E.; Schwartz, J.; Dubos, M.-P.; Favrel, P. Molecular and physiological characterization of a crustacean cardioactive signaling system in a lophotrochozoan—The pacific oyster (Crassostrea gigas): A role in reproduction and salinity acclimation. J. Exp. Biol. 2021, 224, jeb241588. [Google Scholar] [CrossRef]
- Schwartz, J.; Réalis-Doyelle, E.; Dubos, M.-P.; Lefranc, B.; Leprince, J.; Favrel, P. Characterization of an evolutionarily conserved calcitonin signaling system in a lophotrochozoan, the Pacific oyster (Crassostrea gigas). J. Exp. Biol. 2019, 222, jeb.201319. [Google Scholar] [CrossRef]
- Bigot, L.; Beets, I.; Dubos, M.-P.; Boudry, P.; Schoofs, L.; Favrel, P. Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J. Exp. Biol. 2014, 217, 2974–2982. [Google Scholar] [CrossRef]
- Yurchenko, O.V.; Skiteva, O.I.; Voronezhskaya, E.E.; Dyachuk, V.A. Nervous system development in the Pacific oyster, Crassostrea gigas (Mollusca: Bivalvia). Front. Zool. 2018, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Kim, B.M.; Hwang, I.J.; Lee, J.S.; Choi, I.Y.; Kim, Y.J.; Rhee, J.S. Thermal stress induces a distinct transcriptome profile in the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 19, 62–70. [Google Scholar] [CrossRef]
- Sussarellu, R.; Huvet, A.; Lapègue, S.; Quillen, V.; Lelong, C.; Cornette, F.; Fast Jensen, L.; Bierne, N.; Boudry, P. Additive transcriptomic variation associated with reproductive traits suggest local adaptation in a recently settled population of the Pacific oyster, Crassostrea gigas. BMC Genom. 2015, 19, 808. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Li, Q.; Yu, H. Gonad Transcriptome Analysis of the Pacific Oyster Crassostrea gigas Identifies Potential Genes Regulating the Sex Determination and Differentiation Process. Mar. Biotechnol. 2018, 20, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Dheilly, N.M.; Lelong, C.; Huvet, A.; Kellner, K.; Dubos, M.-P.; Riviere, G.; Boudry, P.; Favrel, P. Gametogenesis in the Pacific oyster Crassostrea gigas: A microarrays-based analysis identifies sex and stage specific genes. PLoS ONE 2012, 7, e36353. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Manousaki, T.; Tsakogiannis, A.; Lagnel, J.; Sarropoulou, E.; Xiang, J.Z.; Papandroulakis, N.; Mylonas, C.C.; Tsigenopoulos, C.S. The sex-specific transcriptome of the hermaphrodite sparid sharpsnout seabream (Diplodus puntazzo). BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, A.; Manousaki, T.; Lagnel, J.; Papanikolaou, N.; Papandroulakis, N.; Mylonas, C.C.; Tsigenopoulos, C.S. The gene toolkit implicated in functional sex in Sparidae hermaphrodites: Inferences from comparative transcriptomics. Front. Genet. 2019, 10, 749. [Google Scholar] [CrossRef]
- Riviere, G.; Klopp, C.; Ibouniyamine, N.; Huvet, A.; Boudry, P.; Favrel, P. GigaTON: An extensive publicly searchable database providing a new reference transcriptome in the pacific oyster Crassostrea gigas. BMC Bioinform. 2015, 16, 401. [Google Scholar] [CrossRef] [PubMed]
- Fleury, E.; Huvet, A.; Lelong, C.; Lorgeril, J.; De Boulo, V.; Gueguen, Y.; Bachère, E.; Tanguy, A.; Moraga, D.; Fabioux, C.; et al. Generation and analysis of a 29,745 unique Expressed Sequence Tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: The GigasDatabase. BMC Genom. 2009, 15, 1–15. [Google Scholar] [CrossRef]
- Zmora, N.; Chung, J.S. A Novel Hormone Is Required for the Development of Reproductive Phenotypes in Adult Female Crabs. Endocrinology 2014, 155, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, J.A. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocrinol. 2015, 224, 84–95. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Zhang, Y.; Xiang, Z.; Tong, Y.; Qu, F.; Yu, Z. Genomic characterization and expression analysis of five novel IL-17 genes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2014, 40, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhang, H.; Zhang, R.; Li, H.; Wang, W.; Wang, L.; Wang, H.; Qiu, L.; Song, L. CgIL17-5, an ancient inflammatory cytokine in Crassostrea gigas exhibiting the heterogeneity functions compared with vertebrate interleukin17 molecules. Dev. Comp. Immunol. 2015, 53, 339–348. [Google Scholar] [CrossRef]
- Truman, J.W. Hormonal control of ecdysis. In Comprehensive Insect Physiology, Biochemistry and Pharmacology; Kerkut, G.A., Gilbert, L.I., Eds.; Pergamon: New York, NY, USA, 1985; pp. 413–440. [Google Scholar]
- Holman, G.M.; Nachman, R.J.; Wright, M.S. Insect neuropeptides. Annu. Rev. Entomol. 1990, 35, 201–217. [Google Scholar] [CrossRef]
- Zhou, L.; Li, S.; Wang, Z.; Li, F.; Xiang, J. An eclosion hormone-like gene participates in the molting process of Palaemonid shrimp Exopalaemon carinicauda. Dev. Genes Evol. 2017, 227, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Troestscher, R.; Kramer, J.; Schooley, D.A. Isolation and primary structure of the eclosion hormone of the tobacco hornworm, Manduca sexta. Biochem. Biophys. Res. Commun. 1987, 146, 746–750. [Google Scholar] [CrossRef]
- Zandawala, M.; Moghul, I.; Guerra, L.A.Y.; Delroisse, J.; Hara, T.D.O.; Abylkassimova, N.; Hugall, A.F.; O’Hara, T.D.; Elphick, M.R. Discovery of novel representatives of bilaterian neuropeptide families and reconstruction of neuropeptide precursor evolution in ophiuroid echinodems. Open Biol. 2017, 7, 170129. [Google Scholar] [CrossRef]
- De Oliveira, A.L.; Calcino, A.; Wanninger, A. Ancient origins of arthropod moulting pathway components. eLife 2019, 8, 1–15. [Google Scholar] [CrossRef]
- Clark, A.C.; Del Campo, M.L.; Ewer, J. Neuroendocrine control of larval ecdysis behavior in Drosophila: Complex regulation by partially redundant neuropeptides. J. Neurosci. 2004, 24, 4283–4292. [Google Scholar] [CrossRef]
- Zieger, E.; Robert, N.S.M.; Calcino, A.; Wanninger, A. Ancestral role of ecdysis-related neuropeptides in animal life cycle transitions. Curr. Biol. 2021, 31, 207–213.e4. [Google Scholar] [CrossRef]
- Oh-I, S.; Shimizu, H.; Satoh, T.; Okada, S.; Adachi, S.; Inoue, K.; Eguchi, H.; Yamamoto, M.; Imaki, T.; Hashimoto, K.; et al. Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature 2006, 443, 709–712. [Google Scholar] [CrossRef]
- Gonzalez, R.; Perry, R.L.S.; Gao, X.; Gaidhu, M.P.; Tsushima, R.G.; Ceddia, R.B.; Unniappan, S. Nutrient responsive nesfatin-1 regulates energy balance and induces glucose-stimulated insulin secretion in rats. Endocrinology 2011, 152, 3628–3637. [Google Scholar] [CrossRef]
- Gonzalez, R.; Shepperd, E.; Thiruppugazh, V.; Lohan, S.; Grey, C.L.; Chang, J.P.; Unniappan, S. Nesfatin-1 regulates the hypothalamo-pituitary-ovarian axis of fish. Biol. Reprod. 2012, 87, 1–10. [Google Scholar] [CrossRef]
- Otte, S.; Barnikol-Watanabe, S.; Vorbrüggen, G.; Hilschmann, N. NUCB1, the Drosophila melanogaster homolog of the mammalian EF-hand proteins NEFA and nucleobindin. Mech. Dev. 1999, 86, 155–158. [Google Scholar] [CrossRef]
- Shimizu, H.; Oh-I, S.; Hashimoto, K.; Nakata, M.; Yamamoto, S.; Yoshida, N.; Eguchi, H.; Kato, I.; Inoue, K.; Satoh, T.; et al. Peripheral administration of nesfatin-1 reduces food intake in mice: The leptin-independent mechanism. Endocrinology 2009, 150, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Badisco, L.; Claeys, I.; Van Loy, T.; Van Hiel, M.; Franssens, V.; Simonet, G.; Broeck, J. Vanden Neuroparsins, a family of conserved arthropod neuropeptides. Gen. Comp. Endocrinol. 2007, 153, 64–71. [Google Scholar] [CrossRef]
- Girardie, J.; Girardie, A.; Huet, J.C.; Pernollet, J.C. Amino acid sequence of locust neuroparsins. FEBS Lett. 1989, 245, 4–8. [Google Scholar] [CrossRef]
- Veenstra, J.A. What the loss of the hormone neuroparsin in the melanogaster subgroup of Drosophila can tell us about its function. Insect Biochem. Mol. Biol. 2010, 40, 354–361. [Google Scholar] [CrossRef]
- Amankwah, B.K.; Wang, C.; Sun, C.; Wang, W.; Chan, S. Crustacean neuroparsins—A mini-review. Gene 2020, 732, 14361. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Go, W.; Fritz, M.; Mann, K. Perlustrin, a Haliotis laevigata (Abalone) Nacre Protein, is Homologous to the Insulin-like Growth Factor Binding Protein N-Terminal Module of Vertebrates. Gene 2001, 249, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Pearson, D.; Shively, J.E.; Clark, B.R.; Geschwind, I.I.; Barkley, M.; Nishioka, R.S.; Bern, H.A. Urotensin II, a somatostatin-like peptide in the caudal neurosecretory system of fishes. Proc. Natl. Acad. Sci. USA 1980, 77, 5021–5024. [Google Scholar] [CrossRef]
- Vaudry, H.; Do Rego, J.C.; Le Mevel, J.C.; Chatenet, D.; Tostivint, H.; Fournier, A.; Tonon, M.C.; Pelletier, G.; Michael Conlon, J.; Leprince, J. Urotensin II, from fish to human. Ann. N. Y. Acad. Sci. 2010, 1200, 53–66. [Google Scholar] [CrossRef]
- Romanova, E.V.; Sasaki, K.; Alexeeva, V.; Vilim, F.S.; Jing, J.; Richmond, T.A.; Weiss, K.R.; Sweedler, J.V. Urotensin II in Invertebrates: From Structure to Function in Aplysia californica. PLoS ONE 2012, 7, e48764. [Google Scholar] [CrossRef] [PubMed]
- Negri, L.; Lattanzi, R.; Giannini, E.; Melchiorri, P. Bv8/Prokineticin proteins and their receptors. Life Sci. 2007, 81, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Söderhäll, I.; Kim, Y.-A.; Jiravanichpaisal, P.; Lee, S.-Y.; Söderhäll, K. An ancient role for a prokineticin domain in invertebrate hematopoiesis. J. Immunol. 2005, 174, 6153–6160. [Google Scholar] [CrossRef]
- Hsiao, C.Y.; Song, Y.L. A long form of shrimp astakine transcript: Molecular cloning, characterization and functional elucidation in promoting hematopoiesis. Fish Shellfish Immunol. 2010, 28, 77–86. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, S.; Li, M.; Xin, L.; Wang, L.; Wang, H.; Qiu, L.; Song, L. A cytokine-like factor astakine accelerates the hemocyte production in Pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 2016, 55, 179–187. [Google Scholar] [CrossRef]
- Monnier, J.; Samson, M. Cytokine properties of prokineticins. FEBS J. 2008, 275, 4014–4021. [Google Scholar] [CrossRef]
- Bullock, C.; Li, J.-D.; Zhou, Q.-Y. Structural Determinants Required for the Bioactivities of Prokineticins and Identification of Prokineticin Receptor Antagonists. Mol. Pharmacol. 2004, 65, 582–588. [Google Scholar] [CrossRef]
- Mirabeau, O.; Joly, J. Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. USA 2013, 110, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kim, Y.-A.; Lee, B.; Söderhäll, K.; Söderhäll, I. Identification and properties of a receptor for the invertebrate cytokine astakine, involved in hematopoiesis. Exp. Cell Res. 2009, 315, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, H.; Xin, L.; Xu, J.; Jia, Z.; Wang, L.; Song, L. The immunological capacity in the larvae of Pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 2016, 49, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Nagasawa, H.; Isogai, A.; Ishizaki, H.; Suzuki, A. Prothoracicotropic hormone of the silkworm, Bombyx mori: Amino acid sequence and dimeric structure. Agric. Biol. Chem. 1991, 55, 73–86. [Google Scholar] [PubMed]
- Jékely, G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 8702–8707. [Google Scholar] [CrossRef]
- Grillo, M.; Furriols, M.; De Miguel, C.; Franch-Marro, X.; Casanova, J. Conserved and divergent elements in Torso RTK activation in Drosophila development. Sci. Rep. 2012, 2, 1–7. [Google Scholar] [CrossRef]
- Rewitz, K.; Yamanaka, N.; Gilbert, L.; O’Connor, M. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 2009, 326, 1403–1405. [Google Scholar] [CrossRef]
- Jun, J.; Han, G.; Yun, H.; Lee, G.; Hyun, S. Torso, a Drosophila receptor tyrosine kinase, plays a novel role in the larval fat body in regulating insulin signaling and body growth. J. Comp. Physiol. Biochem. Syst. Environ. Physiol. 2016, 186, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Mineo, A.; Furriols, M.; Casanova, J. The trigger (and the restriction) of Torso RTK activation. Open Biol. 2018, 8, 180180. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.; Furriols, C.; McCormick, C.; Struhl, G. Similarities between trunk and spätzle, putative extracellular ligands specifying body pattern in Drosophila. Genes Dev. 1995, 9, 2539–2544. [Google Scholar] [CrossRef]
- Ohta, N.; Kubota, I.; Takao, T.; Shimonishi, Y.; Yasuda-Kamatani, Y.; Minakata, H.; Nomoto, K.; Muneoka, Y.; Kobayashi, M. Fulicin, a novel neuropeptide containing a D-amino acid residue isolated from the ganglia of Achatina fulica. Biochem. Biophys. Res. Commun. 1991, 178, 486–493. [Google Scholar] [CrossRef]
- Hirata, T.; Kubota, I.; Iwasawa, N.; Takabatake, I.; Ikeda, T.; Muneoka, Y. Structures and actions of Mytilus Inhibitory Peptides. Biochem. Biophys. Res. Commun. 1988, 152, 1376–1382. [Google Scholar] [CrossRef]
- Bauknecht, P.; Jékely, G. Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Rep. 2015, 12, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Van Sinay, E.; Mirabeau, O.; Depuydt, G.; Van Hiel, M.B.; Peymen, K.; Watteyne, J.; Zels, S.; Schoofs, L.; Beets, I. Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2017, 114, E4065–E4074. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, Y.; Masuda, K.; Minakata, H. Fulicin regulates the female reproductive organs of the snail, Achatina fulica. Peptides 2000, 21, 1203–1208. [Google Scholar] [CrossRef]
- Bogdanov, Y.D.; Balaban, P.M.; Poteryaev, D.A.; Zakharov, I.S. Putative neuropeptides and an EF-hand motif region are encoded by a novel gene expressed in the four giant interneurons of the terrestrial snail. Neuroscience 1998, 85, 637–647. [Google Scholar] [CrossRef]
- Moroz, L.L.; Edwards, J.R.; Puthanveettil, S.V.; Kohn, A.B.; Ha, T.; Heyland, A.; Knudsen, B.; Sahni, A.; Yu, F.; Liu, L.; et al. Neuronal transcriptome of Aplysia: Neuronal compartments and circuitry. Cell 2006, 127, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Zatylny-Gaudin, C.; Favrel, P. Diversity of the RFamide peptide family in mollusks. Front. Endocrinol. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hummon, A.B.; Richmond, T.A.; Verleyen, P.; Baggerman, G.; Huybrechts, J.; Ewing, M.A.; Vierstraete, E.; Rodriguez-Zas, S.L.; Schoofs, L.; Robinson, G.E.; et al. From the genome to the proteome: Uncovering peptides in the Apis brain. Science 2006, 314, 647–649. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Cummins, S.F.; Fitzgibbon, Q.; Battaglene, S.; Elizur, A. Analysis of the central nervous system transcriptome of the Eastern rock lobster Sagmariasus verreauxi reveals its putative neuropeptidome. PLoS ONE 2014, 9, e97323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Brandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.; Purcell, A.; Norton, R.; Safavi-Hemami, H. Hormone-like peptides in the venoms of marine cone snails. Gen. Comp. Endocrinol. 2017, 244, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, A.; Annangudi, S.P.; Richmond, T.A.; Ament, S.A.; Xie, F.; Southey, B.R.; Rodriguez-Zas, S.R.; Robinson, G.E.; Sweedler, J.V. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc. Natl. Acad. Sci. USA 2009, 106, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, B.; Cardoen, D.; Bogaerts, A.; Landuyt, B.; Schoofs, L.; Verleyen, P. Mass spectrometric profiling of (neuro)-peptides in the worker honeybee, Apis mellifera. Neuropharmacology 2010, 58, 248–258. [Google Scholar] [CrossRef]
- Han, B.; Fang, Y.; Feng, M.; Hu, H.; Qi, Y.; Huo, X.; Meng, L.; Wu, B.; Li, J. Quantitative Neuropeptidome Analysis Reveals Neuropeptides Are Correlated with Social Behavior Regulation of the Honeybee Workers. J. Proteome Res. 2015, 14, 4382–4393. [Google Scholar] [CrossRef]
- Xie, J.; Sang, M.; Song, X.; Zhang, S.; Kim, D.; Veenstra, J.A.; Park, Y.; Li, B. A new neuropeptide insect parathyroid hormone iPTH in the red flour beetle Tribolium castaneum. PLoS Genet. 2020, 16, 1–20. [Google Scholar] [CrossRef]
- Dheilly, N.M.; Jouaux, A.; Boudry, P.; Favrel, P.; Lelong, C. Transcriptomic profiling of gametogenesis in triploid Pacific Oysters Crassostrea gigas: Towards an understanding of partial sterility associated with triploidy. PLoS ONE 2014, 9, e112094. [Google Scholar]
- Dimarcq, J.-L.; Bulet, P.; Hetru, C.; Hoffmann, J. Cysteine-Rich Antimicrobial Peptides in Invertebrates. Biopolymers 1998, 47, 465–477. [Google Scholar] [CrossRef]
- Gueguen, Y.; Herpin, A.; Aumelas, A.; Garnier, J.; Fievet, J.; Escoubas, J.-M.M.; Bulet, P.; Gonzalez, M.; Lelong, C.; Favrel, P.; et al. Characterization of a defensin from the oyster Crassostrea gigas: Recombinant production, folding, solution structure, antimicrobial activities and gene expression. J. Biol. Chem. 2006, 281, 313–323. [Google Scholar] [CrossRef]
- Fainzilber, M.; Smit, A.B.; Syed, N.I.; Wildering, W.C.; Hermann, P.M.; van der Schors, R.C.; Jimenez, C.; Li, K.W.; van Minnen, J.; Bulloch, A.G.; et al. CRNF, a molluscan neurotrophic factor that interacts with the p75 neurotrophin receptor. Science 1996, 274, 1540–1543. [Google Scholar] [CrossRef]
- Jouaux, A.; Franco, A.; Heude-Berthelin, C.; Sourdaine, P.; Blin, J.L.; Mathieu, M.; Kellner, K. Identification of Ras, Pten and p70S6K homologs in the Pacific oyster Crassostrea gigas and diet control of insulin pathway. Gen. Comp. Endocrinol. 2012, 176, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-J.; Drummond-Barbosa, D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl. Acad. Sci. USA 2009, 106, 1117–1121. [Google Scholar] [CrossRef]
- Michaelson, D.; Korta, D.Z.; Capua, Y.; Hubbard, E.J.A. Insulin signaling promotes germline proliferation in C. elegans. Development 2010, 137, 671–680. [Google Scholar] [PubMed]
- Smit, A.B.; Van Kesteren, R.E.; Spijker, S.; Van Minnen, J.; Van Golen, F.A.; Jiménez, C.R.; Li, K.W. Peptidergic modulation of male sexual behavior in Lymnaea stagnalis: Structural and functional characterization of -FVamide neuropeptides. J. Neurochem. 2003, 87, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Van In, V.; Ntalamagka, N.; O’Connor, W.; Wang, T.; Powell, D.; Cummins, S.F.; Elizur, A. Reproductive neuropeptides that stimulate spawning in the Sydney Rock Oyster (Saccostrea glomerata). Peptides 2016, 82, 109–119. [Google Scholar]
- Endress, M.; Zatylny-Gaudin, C.; Corre, E.; Le Corguillé, G.; Benoist, L.; Leprince, J.; Lefranc, B.; Bernay, B.; Leduc, A.; Rangama, J.; et al. Crustacean cardioactive peptides: Expression, localization, structure, and a possible involvement in regulation of egg-laying in the cuttlefish Sepia officinalis. Gen. Comp. Endocrinol. 2018, 260, 67–79. [Google Scholar] [CrossRef]
- Chiu, A.Y.; Hunkapiller, M.W.; Heller, E.; Stuart, D.K.; Hood, L.E.; Strumwasser, F. Purification and primary structure of the neuropeptide egg-laying hormone of Aplysia californica. Proc. Natl. Acad. Sci. USA 1979, 76, 6656–6660. [Google Scholar] [CrossRef]
- Ebberink, R.H.M.; Van Loenhout, H.; Geraerts, W.P.M.; Joosse, J. Purification and amino acid sequence of the ovulation neurohormone of Lymnaea stagnalis. Proc. Natl. Acad. Sci. USA 1985, 82, 7767–7771. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Matsumi, H.; Bhalla, A.; Bae, J.; Mosselman, S.; Hsu, S.Y.; Hsueh, A.J.W. Thyrostimulin, a heterodimer of two new human glycoprotein hormone subunits, activates the thyroid-stimulating hormone receptor. J. Clin. Investig. 2002, 109, 1445–1452. [Google Scholar] [CrossRef]
- Paluzzi, J.P.; Vanderveken, M.; O’Donnell, M.J. The heterodimeric glycoprotein hormone, GPA2/GPB5, regulates ion transport across the hindgut of the adult mosquito, Aedes aegypti. PLoS ONE 2014, 9, e86386. [Google Scholar] [CrossRef]
- Heyland, A.; Plachetzki, D.; Donelly, E.; Gunaratne, D.; Bobkova, Y.; Jacobson, J.; Kohn, A.B.; Moroz, L.L. Distinct expression patterns of glycoprotein hormone subunits in the lophotrochozoan Aplysia: Implications for the evolution of neuroendocrine systems in animals. Endocrinology 2012, 153, 5440–5451. [Google Scholar] [CrossRef] [PubMed]
- Vandersmissen, H.P.; Van Hiel, M.B.; Van Loy, T.; Vleugels, R.; Vanden Broeck, J.; Silencing, D. melanogaster lgr1 impairs transition from larval to pupal stage. Gen. Comp. Endocrinol. 2014, 209, 135–147. [Google Scholar] [CrossRef]
- Cho, S.; Rogers, K.W.; Fay, D.S. The C. elegans glycopeptide hormone receptor ortholog, FSHR-1, regulates germline differentiation and survival. Curr. Biol. 2007, 17, 203–212. [Google Scholar] [CrossRef]
- Rocco, D.A.; Garcia, A.S.G.; Scudeler, E.L.; Dos Santos, D.C.; Nóbrega, R.H.; Paluzzi, J.P.V. Glycoprotein hormone receptor knockdown leads to reduced reproductive success in male Aedes aegypti. Front. Physiol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Rocco, D.A.; Paluzzi, J.P.V. Expression profiling, downstream signaling, and inter-subunit interactions of GPA2/GPB5 in the adult mosquito Aedes aegypti. Front. Endocrinol. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Tensen, C.P.; Cox, K.J.; Smit, A.B.; van der Schors, R.C.; Meyerhof, W.; Richter, D.; Planta, R.J.; Hermann, P.M.; van Minnen, J.; Geraerts, W.P.; et al. The lymnaea cardioexcitatory peptide (LyCEP) receptor: A G-protein-coupled receptor for a novel member of the RFamide neuropeptide family. J. Neurosci. 1998, 18, 9812–9821. [Google Scholar] [CrossRef]
- Giardino, N.D.; Aloyz, R.S.; Zollinger, M.; Miller, M.W.; DesGroseillers, L. L5-67 and LUQ-1 peptide precursors of Aplysia californica: Distribution and localization of immunoreactivity in the central nervous system and in peripheral tissues. J. Comp. Neurol. 1996, 374, 230–245. [Google Scholar] [CrossRef]
- Williams, E.A. Function and Distribution of the Wamide Neuropeptide Superfamily in Metazoans. Front. Endocrinol. 2020, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Bartalska, K.; Audsley, N.; Yamanaka, N.; Yapici, N.; Lee, J.-Y.; Kim, Y.-C.; Markovic, M.; Isaac, E.; Tanaka, Y.; et al. MIPs are ancestral ligands for the sex peptide receptor. Proc. Natl. Acad. Sci. USA 2010, 107, 6520–6525. [Google Scholar] [CrossRef] [PubMed]
- Abdel-latief, M.; Meyering-Vos, M.; Hoffmann, K.H. Expression and localization of the Spodoptera frugiperda allatotropin (Spofr-AT) and allatostatin (Spofr-AS) genes. Arch. Insect Biochem. Physiol. 2004, 55, 188–199. [Google Scholar] [CrossRef]
- Taussig, R.; Kaldany, R.R.; Scheller, R.H. A cDNA clone encoding neuropeptides isolated from Aplysia neuron L11. Proc. Natl. Acad. Sci. USA 1984, 81, 4988–4992. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, H.; Maehara, S.; Ohta, H.; Seki, T.; Tanaka, Y. Elevenin regulates the body color through a G protein-coupled receptor NlA42 in the brown planthopper Nilaparvata lugens. Gen. Comp. Endocrinol. 2018, 258, 33–38. [Google Scholar] [CrossRef]
- Rodet, F.; Lelong, C.; Dubos, M.-P.; Costil, K.; Favrel, P. Molecular cloning of a molluscan gonadotropin-releasing hormone receptor orthologue specifically expressed in the gonad. Biochim. Biophys. Acta 2005, 1730, 187–195. [Google Scholar] [CrossRef]
- Lubet, P. Recherches sur le cycle sexuel et l’émission des gamètes chez les mytilidés et les Pectinidés. Revue des Travaux de l’Institut des Pêches Maritimes 1959, 23, 387–548. [Google Scholar]
- Cabau, C.; Escudié, F.; Djari, A.; Guiguen, Y.; Bobe, J.; Klopp, C. Compacting and correcting Trinity and Oases RNA-Seq de novo assemblies. PeerJ 2017, 5, e2988. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hunter, S.; Jones, P.; Mitchell, A.; Apweiler, R.; Attwood, T.K.; Bateman, A.; Bernard, T.; Binns, D.; Bork, P.; Burge, S.; et al. InterPro in 2011: New developments in the family and domain prediction database. Nucleic Acids Res. 2012, 40, 306–312. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Andrey Sivachenko, K.C.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; DePristo, M.A. The Genome Analysis Toolkit: A MapReduceframework for analyzing next-generation DNAsequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Mariette, J.; Noirot, C.; Nabihoudine, I.; Bardou, P.; Hoede, C.; Djari, A.; Cabau, C.; Klopp, C. RNAbrowse: RNA-Seq de novo assembly results browser. PLoS ONE 2014, 9, 1–7. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Ruotti, V.; Stewart, R.M.; Thomson, J.A.; Dewey, C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2009, 26, 493–500. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Peñaloza, C.; Gutierrez, A.P.; Eöry, L.; Wang, S.; Guo, X.; Archibald, A.L.; Bean, T.P.; Houston, R.D. A chromosome-level genome assembly for the Pacific oyster Crassostrea gigas. Gigascience 2021, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L.; et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef] [PubMed]
- Müllner, D. Fastcluster: Fast Hierarchical, Agglomerative. J. Stat. Softw. 2013, 53, 1–18. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Réalis-Doyelle, E.; Schwartz, J.; Cabau, C.; Le Franc, L.; Bernay, B.; Rivière, G.; Klopp, C.; Favrel, P. Transcriptome Profiling of the Pacific Oyster Crassostrea gigas Visceral Ganglia over a Reproduction Cycle Identifies Novel Regulatory Peptides. Mar. Drugs 2021, 19, 452. https://doi.org/10.3390/md19080452
Réalis-Doyelle E, Schwartz J, Cabau C, Le Franc L, Bernay B, Rivière G, Klopp C, Favrel P. Transcriptome Profiling of the Pacific Oyster Crassostrea gigas Visceral Ganglia over a Reproduction Cycle Identifies Novel Regulatory Peptides. Marine Drugs. 2021; 19(8):452. https://doi.org/10.3390/md19080452
Chicago/Turabian StyleRéalis-Doyelle, Emilie, Julie Schwartz, Cédric Cabau, Lorane Le Franc, Benoit Bernay, Guillaume Rivière, Christophe Klopp, and Pascal Favrel. 2021. "Transcriptome Profiling of the Pacific Oyster Crassostrea gigas Visceral Ganglia over a Reproduction Cycle Identifies Novel Regulatory Peptides" Marine Drugs 19, no. 8: 452. https://doi.org/10.3390/md19080452
APA StyleRéalis-Doyelle, E., Schwartz, J., Cabau, C., Le Franc, L., Bernay, B., Rivière, G., Klopp, C., & Favrel, P. (2021). Transcriptome Profiling of the Pacific Oyster Crassostrea gigas Visceral Ganglia over a Reproduction Cycle Identifies Novel Regulatory Peptides. Marine Drugs, 19(8), 452. https://doi.org/10.3390/md19080452





