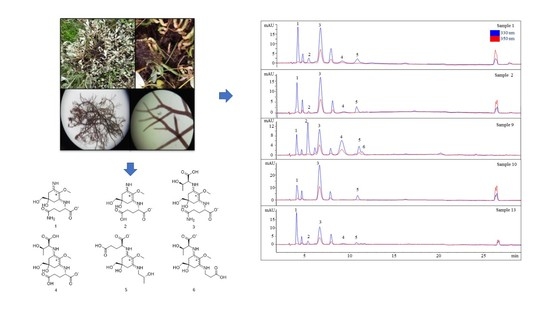
Analysis of the Mycosporine-Like Amino Acid (MAA) Pattern of the Salt Marsh Red Alga Bostrychia scorpioides
Abstract
1. Introduction
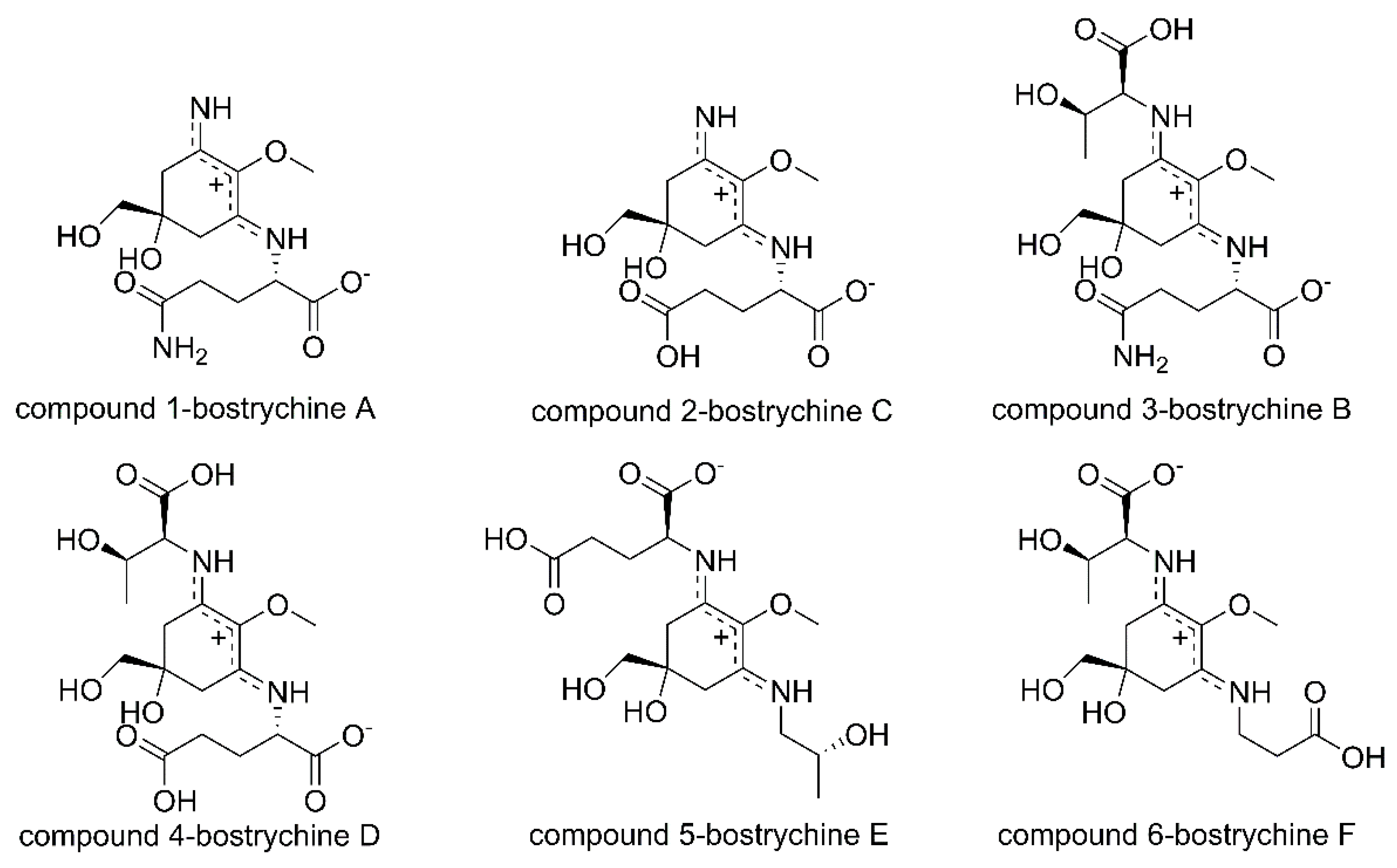
2. Results
2.1. Method Development
2.1.1. Sample Preparation
2.1.2. HPLC-DAD
2.2. Method Validation
2.3. Sample Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Chemicals
4.3. MAA Isolation
4.4. HPLC Sample Preparation
4.5. HPLC-DAD Analysis
4.6. Calibration and Method Validation
4.6.1. Linearity, Limit of Detection (LOD), and Limit of Quantification (LOQ)
4.6.2. Precision and Accuracy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruiz-Nieto, M.; Fernández, J.A.; Niell, F.X.; Carmona, R. Mechanisms of Inorganic Carbon Acquisition in Two Estuarine Rhodophyceans: Bostrychia scorpioides (Hudson) Ex Kützing Montagne and Catenella caespitosa (Withering) L. M. Irvine. Photosynth. Res. 2014, 121, 277–284. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; World-Wide Electronic Publication, National University of Ireland: Galway, Ireland, 2020; Available online: http://www.algaebase.org (accessed on 24 March 2020).
- Van Reine, W.F.P.; Sluiman, H.J. Red Algae Found on European Salt-Marshes. I. Bostrychia scorpioides (Rhodomelaceae). Aquat. Bot. 1980, 9, 323–342. [Google Scholar] [CrossRef]
- Karsten, U.; Kirst, G.O. Incomplete Turgor Pressure Regulation in the “Terrestial” Red Alga, Bostrychia scorpioides (Huds.) Mont. Plant Sci. 1989, 61, 29–36. [Google Scholar] [CrossRef]
- Karsten, U.; King, R.; Kirst, G. The Distribution of D-Sorbitol and D-Dulcitol in the Red Algal Genera Bostrychia and Stictosiphonia (Rhodomelaceae, Rhodophyta): A Re-Evaluation. Br. Phycol. Soc. 1990, 25, 363–366. [Google Scholar] [CrossRef]
- Orfanoudaki, M.; Hartmann, A.; Miladinovic, H.; Nguyen, H.N.; Karsten, U.; Ganzera, M. Bostrychines A–F, Six Novel Mycosporine-Like Amino-Acids and a Novel Betaine from the Red Alga Bostrychia scorpioides. Mar. Drugs 2019, 17, 356. [Google Scholar] [CrossRef]
- Oren, A.; Gunde-Cimerman, N. Mycosporines and Mycosporine-like Amino Acids: UV Protectants or Multipurpose Secondary Metabolites? FEMS Microbiol. Lett. 2007, 269, 1–10. [Google Scholar] [CrossRef]
- Wada, N.; Sakamoto, T.; Matsugo, S. Mycosporine-Like Amino Acids and Their Derivatives as Natural Antioxidants. Antioxidants 2015, 4, 603–646. [Google Scholar] [CrossRef]
- Lawrence, K.P.; Long, P.F.; Young, A.R. Mycosporine-Like Amino Acids for Skin Photoprotection. Curr. Med. Chem. 2018, 25, 5512–5527. [Google Scholar] [CrossRef]
- Zuccarello, G.; Muangmai, N.; Preuss, M.; Sanchez, L. The Bostrychia tenella Species Complex: Morphospecies and Genetic Cryptic Species with Resurrection of B. binderi. Phycologia 2015, 54, 261–270. [Google Scholar] [CrossRef]
- Orfanoudaki, M.; Hartmann, A.; Ngoc, H.N.; Gelbrich, T.; West, J.; Karsten, U.; Ganzera, M. Mycosporine-like Amino Acids, Brominated and Sulphated Phenols: Suitable Chemotaxonomic Markers for the Reassessment of Classification of Bostrychia calliptera (Ceramiales, Rhodophyta). Phytochemistry 2020, 174, 112344. [Google Scholar] [CrossRef]
- Orfanoudaki, M.; Hartmann, A.; Kamiya, M.; West, J.; Ganzera, M. Chemotaxonomic Study of Bostrychia Spp. (Ceramiales, Rhodophyta) Based on Their Mycosporine-Like Amino Acid Content. Molecules 2020, 25, 3273. [Google Scholar] [CrossRef]
- Mori, C.C.; Bagatini, I.L.; Garcia da Silva, T.; Parrish, C.C.; Henriques Vieira, A.A. Use of Fatty Acids in the Chemotaxonomy of the Family Selenastraceae (Sphaeropleales, Chlorophyceae). Phytochemistry 2018, 151, 9–16. [Google Scholar] [CrossRef]
- Falshaw, R.; Furneaux, R.H. Chemotaxonomy of New Zealand Red Algae in the Family Gigartinaceae (Rhodophyta) Based on Galactan Structures from the Tetrasporophyte Life-Stage. Carbohydr. Res. 2009, 344, 210–216. [Google Scholar] [CrossRef]
- Amico, V. Marine Brown Algae of Family Cystoseiraceae: Chemistry and Chemotaxonomy. Phytochemistry 1995, 39, 1257–1279. [Google Scholar] [CrossRef]
- Xu, K.; Guo, S.; Jia, X.; Li, X.; Shi, D. Phytochemical and Chemotaxonomic Study on Leathesia nana (Chordariaceae). Biochem. Syst. Ecol. 2018, 81, 42–44. [Google Scholar] [CrossRef]
- Jeffrey, S.; Mactavish, H.; Dunlap, W.; Vesk, M.; Groenewoud, K. Occurrence of UVA-and UVB-Absorbing Compounds in 152 Species (206 Strains) of Marine Microalgae. Mar. Ecol. Prog. Ser. 1999, 189, 35–51. [Google Scholar] [CrossRef]
- Geraldes, V.; de Medeiros, L.S.; Jacinavicius, F.R.; Long, P.F.; Pinto, E. Development and Validation of a Rapid LC-MS/MS Method for the Quantification of Mycosporines and Mycosporine-like Amino Acids (MAAs) from Cyanobacteria. Algal Res. 2020, 46, 101796. [Google Scholar] [CrossRef]
- Whitehead, K.; Hedges, J.I. Analysis of Mycosporine-like Amino Acids in Plankton by Liquid Chromatography Electrospray Ionization Mass Spectrometry. Mar. Chem. 2002, 80, 27–39. [Google Scholar] [CrossRef]
- Chaves-Peña, P.; de la Coba, F.; Figueroa, F.L.; Korbee, N. Quantitative and Qualitative HPLC Analysis of Mycosporine-Like Amino Acids Extracted in Distilled Water for Cosmetical Uses in Four Rhodophyta. Mar. Drugs 2019, 18, 27. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Murauer, A.; Ganzera, M. Quantitative Analysis of Mycosporine-like Amino Acids in Marine Algae by Capillary Electrophoresis with Diode-Array Detection. J. Pharm. Biomed. Anal. 2017, 138, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Becker, K.; Karsten, U.; Remias, D.; Ganzera, M. Analysis of Mycosporine-Like Amino Acids in Selected Algae and Cyanobacteria by Hydrophilic Interaction Liquid Chromatography and a Novel MAA from the Red Alga Catenella Repens. Mar. Drugs 2015, 13, 6291–6305. [Google Scholar] [CrossRef] [PubMed]
- Carreto, J.I.; Carignan, M.O.; Montoya, N.G. A High-Resolution Reverse-Phase Liquid Chromatography Method for the Analysis of Mycosporine-like Amino Acids (MAAs) in Marine Organisms. Mar. Biol. 2005, 146, 237–252. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, N.; Zhou, J.; Dong, S.; Zhang, X.; Guo, L.; Guo, G. Distribution, Contents, and Types of Mycosporine-Like Amino Acids (MAAs) in Marine Macroalgae and a Database for MAAs Based on These Characteristics. Mar. Drugs 2020, 18, 43. [Google Scholar] [CrossRef]
- Figueroa, F.L.; Korbee, N.; de Clerck, O.; Bárbara, I.; Gall, E.A.R. Characterization of Grateloupia lanceola (Halymeniales, Rhodophyta), an Obscure Foliose Grateloupia from the Iberian Peninsula, Based on Morphology, Comparative Sequence Analysis and Mycosporine-like Amino Acid Composition. Eur. J. Phycol. 2007, 42, 231–242. [Google Scholar] [CrossRef]
- Hotter, V.; Glaser, K.; Hartmann, A.; Ganzera, M.; Karsten, U. Polyols and UV-Sunscreens in the Prasiola-Clade (Trebouxiophyceae, Chlorophyta) as Metabolites for Stress Response and Chemotaxonomy. J. Phycol. 2018, 54, 264–274. [Google Scholar] [CrossRef]
- Llewellyn, C.A.; Greig, C.; Silkina, A.; Kultschar, B.; Hitchings, M.D.; Farnham, G. Mycosporine-like Amino Acid and Aromatic Amino Acid Transcriptome Response to UV and Far-Red Light in the Cyanobacterium Chlorogloeopsis fritschii PCC 6912. Sci. Rep. 2020, 10, 20638. [Google Scholar] [CrossRef]
- Portwich, A.; Garcia-Pichel, F. Biosynthetic Pathway of Mycosporines (Mycosporine-like Amino Acids) in the Cyanobacterium Chlorogloeopsis Sp. Strain PCC 6912. Phycologia 2003, 42, 384–392. [Google Scholar] [CrossRef]

| Calibration Data | ||||||
| Substance | Regression Equation a | Coefficient of Determination | Linearity b (μg··mL−1) | LOD c (μg·mL−1) | LOQ d (μg·mL−1) | |
| bostrychine C | y = 22248x + 1.4832 | R² = 0.9996 | 0.06–105.20 | 0.07 | 0.23 | |
| bostrychine B | y = 29657x + 2.0058 | R² = 0.9999 | 0.11–71.40 | 0.04 | 0.13 | |
| bostrychine D | y = 35033x + 8.1483 | R² = 0.9997 | 0.26–83.00 | 0.13 | 0.40 | |
| bostrychine E | y = 42335x + 5.4806 | R² = 0.9999 | 0.04–69.00 | 0.10 | 0.29 | |
| bbostrychine F | y = 27415x + 3.7719 | R² = 0.9999 | 0.10–63.60 | 0.22 | 0.65 | |
| Accuracy and Precision | ||||||
| Precision | Accuracy g | |||||
| Substance | Intraday e | Interday f | Substance | Low | Medium | High |
| bostrychine A | 2.45 | 2.91 | bostrychine C | 95.15 ± 1.03 | 103.06 ± 0.18 | 95.39 ± 0.11 |
| bostrychine C | 3.49 | 4.44 | bostrychine B | 95.53 ± 0.52 | 95.15 ± 2.99 | 97.75 ± 0.16 |
| bostrychine B | 1.97 | 2.06 | bostrychine F | 102.77 ± 0.40 | 95.15 ± 0.72 | 103.78 ± 0.85 |
| bostrychine D | 3.78 | 3.60 | ||||
| bostrychine E | 4.31 | 4.81 | ||||
| Sample | Bostrychine A (mg·g−1) (Srel% a) | Bostrychine C (mg·g−1) (Srel%) | Bostrychine B (mg·g−1) (Srel%) | Bostrychine D (mg·g−1) (Srel%) | Bostrychine E (mg·g−1) (Srel%) | Bostrychine F (mg·g−1) (Srel%) |
|---|---|---|---|---|---|---|
| 1 | 2.04 (2.91) | 0.40 (4.43) | 3.30 (2.06) | 0.20 (3.60) | 0.31 (4.81) | |
| 2 | 1.8 (1.10) | 0.14 (2.80) | 3.15 (0.75) | <LOQ b | 0.28 (4.98) | |
| 3 | 3.35 (0.54) | <LOQ b | 6.30 (0.66) | 0.47 (4.96) | ||
| 4 | 3.16 (0.78) | 0.22 (3.33) | 4.03 (0.62) | 0.16 (4.47) | 0.21 (4.52) | <LOQ b |
| 5 | 2.49 (1.63) | 0.25 (1.83) | 3.04 (2.80) | 0.18 (2.01) | 0.21 (2.35) | <LOQ b |
| 6 | 2.84 (1.28) | 0.07 (2.59) | 3.85 (0.20) | 0.02 (3.17) | 0.24 (4.24) | |
| 7 | 2.17 (0.36) | 0.16 (4.38) | 3.49 (0.49) | 0.11 (3.62) | 0.17 (2.10) | |
| 8 | 1.87 (0.29) | <LOQ | 2.35 (0.42) | 0.14 (2.15) | ||
| 9 | 0.88 (2.31) | 1.76 (3.00) | 1.10 (3.37) | 0.73 (4.47) | 0.33 (4.26) | <LOQ b |
| 10 | 1.63 (0.50) | 3.14 (0.41) | 0.28 (5.16) | |||
| 11 | 1.46 (1.66) | 0.11 (4.17) | 1.95 (2.50) | 0.16 (4.42) | ||
| 12 | 8.05 (0.47) | <LOQ b | 2.04 (0.26) | 0.11 (4.86) | ||
| 13 | 1.96 (0.96) | 0.30 (3.95) | 1.14 (1.03) | <LOQ b | 0.08 (4.90) | |
| 14 | 4.42 (1.73) | 6.44 (0.74) | ||||
| 15 | 1.53 (1.31) | 0.14 (2.41) | 1.11 (0.65) | <LOQ b | 0.09 (3.44) | |
| 16 | 2.49 (0.61) | 5.66 (0.27) | 0.18 (3.40) | |||
| 17 | 1.42 (1.29) | 3.94 (0.52) | 0.39 (1.73) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orfanoudaki, M.; Hartmann, A.; Mayr, J.; Figueroa, F.L.; Vega, J.; West, J.; Bermejo, R.; Maggs, C.; Ganzera, M. Analysis of the Mycosporine-Like Amino Acid (MAA) Pattern of the Salt Marsh Red Alga Bostrychia scorpioides. Mar. Drugs 2021, 19, 321. https://doi.org/10.3390/md19060321
Orfanoudaki M, Hartmann A, Mayr J, Figueroa FL, Vega J, West J, Bermejo R, Maggs C, Ganzera M. Analysis of the Mycosporine-Like Amino Acid (MAA) Pattern of the Salt Marsh Red Alga Bostrychia scorpioides. Marine Drugs. 2021; 19(6):321. https://doi.org/10.3390/md19060321
Chicago/Turabian StyleOrfanoudaki, Maria, Anja Hartmann, Julia Mayr, Félix L. Figueroa, Julia Vega, John West, Ricardo Bermejo, Christine Maggs, and Markus Ganzera. 2021. "Analysis of the Mycosporine-Like Amino Acid (MAA) Pattern of the Salt Marsh Red Alga Bostrychia scorpioides" Marine Drugs 19, no. 6: 321. https://doi.org/10.3390/md19060321
APA StyleOrfanoudaki, M., Hartmann, A., Mayr, J., Figueroa, F. L., Vega, J., West, J., Bermejo, R., Maggs, C., & Ganzera, M. (2021). Analysis of the Mycosporine-Like Amino Acid (MAA) Pattern of the Salt Marsh Red Alga Bostrychia scorpioides. Marine Drugs, 19(6), 321. https://doi.org/10.3390/md19060321