In-Depth In Silico Search for Cuttlefish (Sepia officinalis) Antimicrobial Peptides Following Bacterial Challenge of Haemocytes
Abstract
1. Introduction
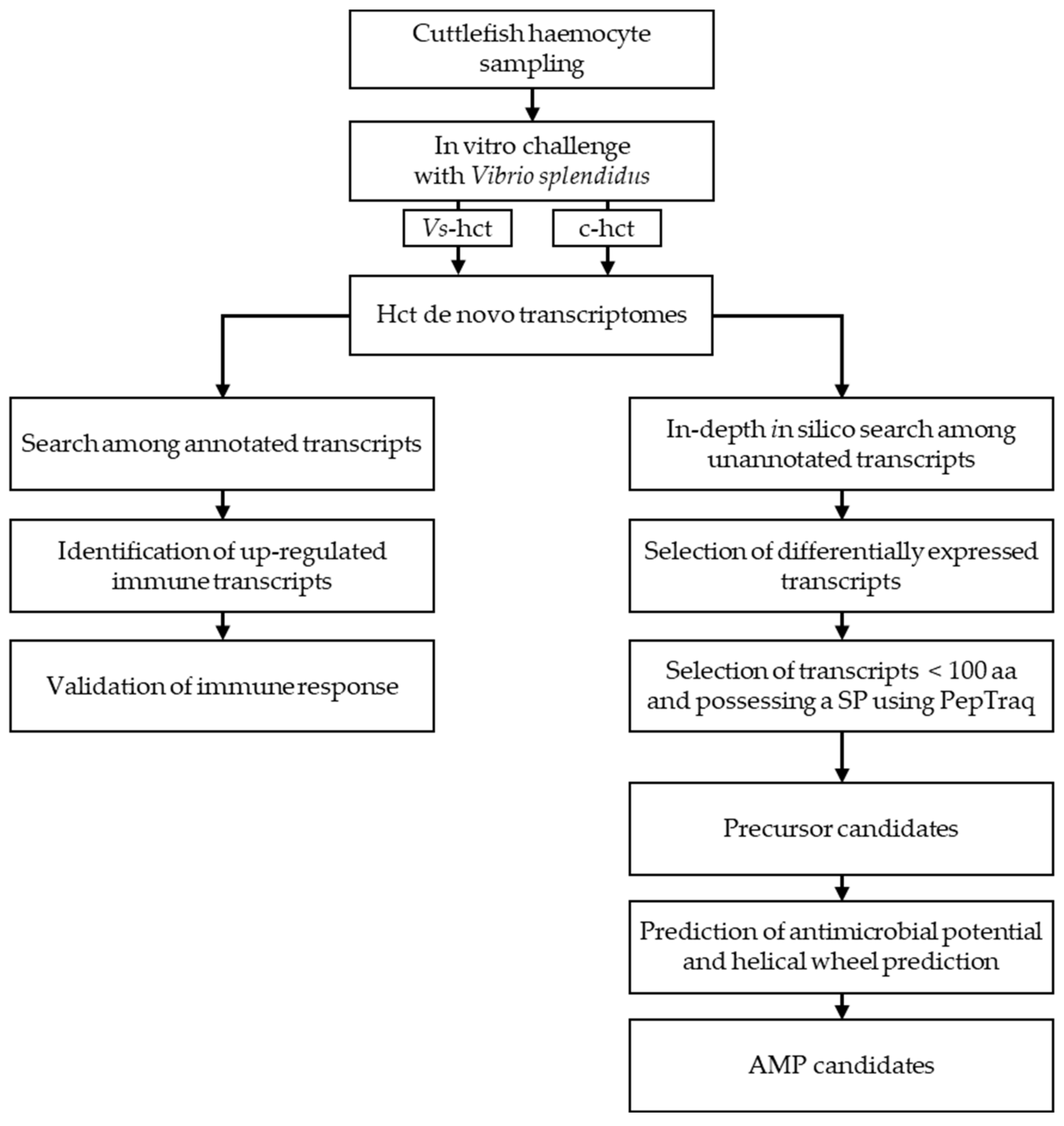
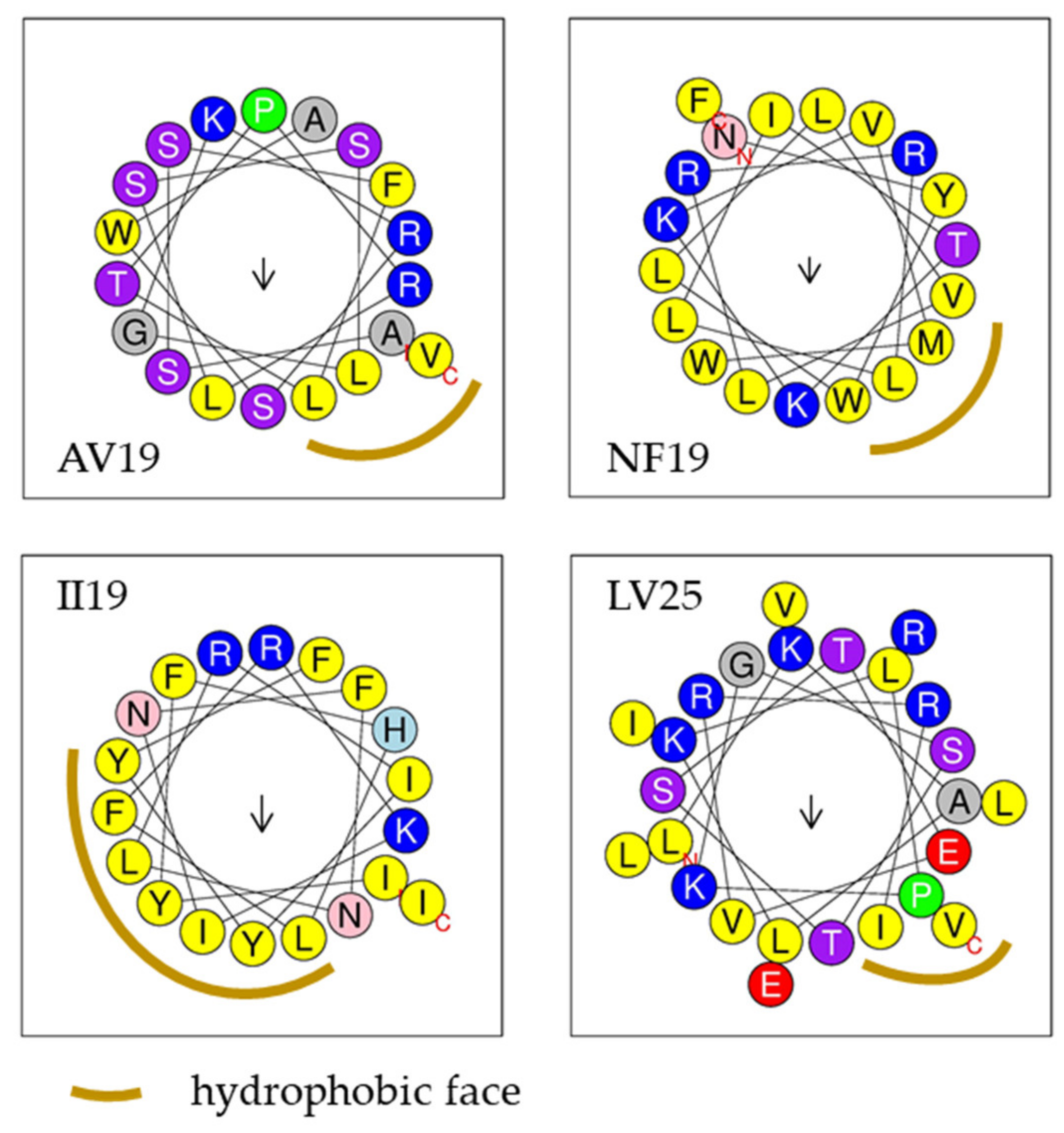
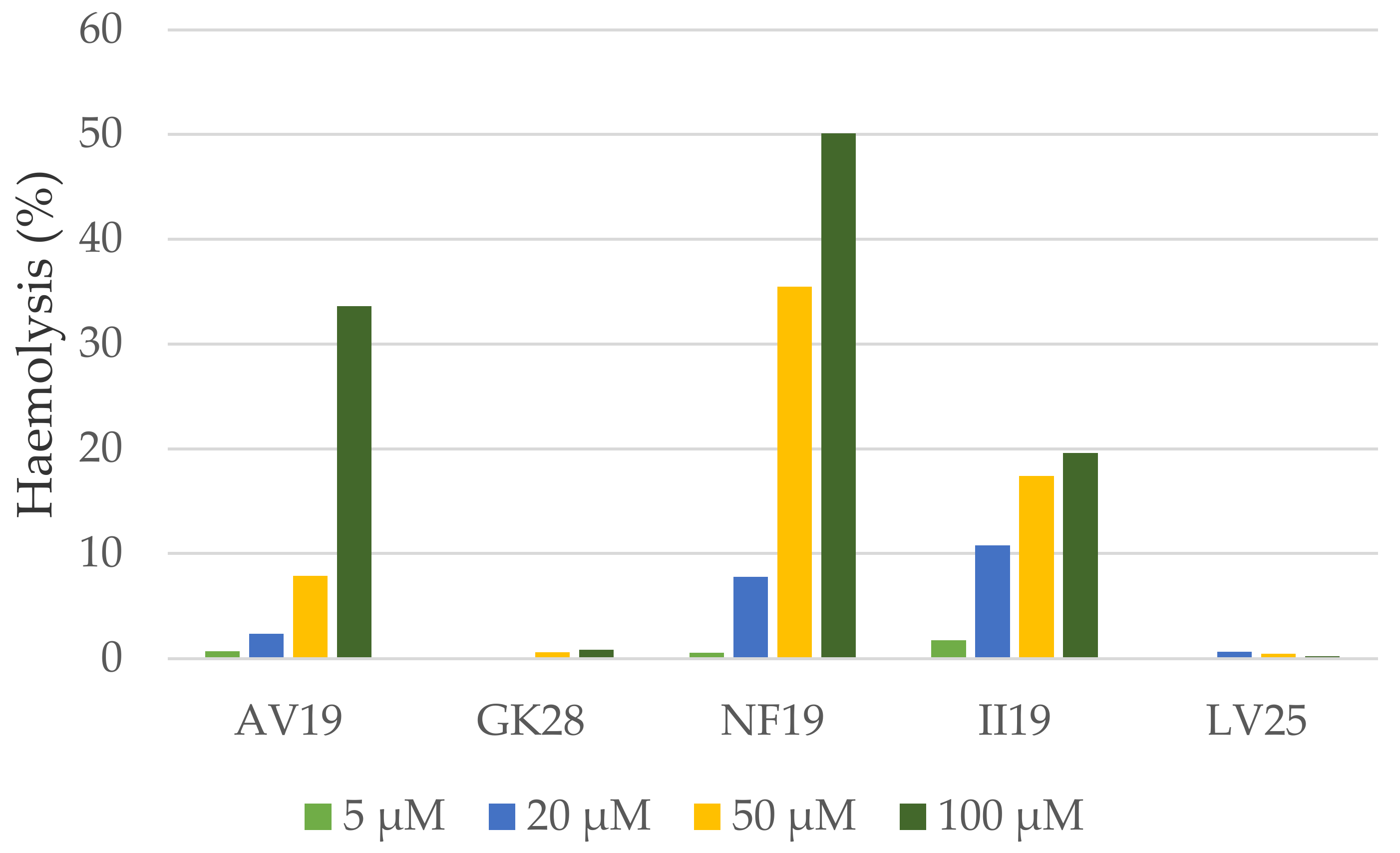
2. Results—Discussion
3. Materials and Methods
3.1. Animals
3.2. Ethical Statement
3.3. Haemolymph Collection and Challenge with Heat-Killed Bacteria
3.4. Illumina Sequencing
3.5. Bioinformatic Analysis
3.5.1. Transcriptome Assembly
3.5.2. Transcriptome Annotation
3.5.3. PepTraq
3.5.4. In-Depth In Silico Search
3.6. Peptide Synthesis
3.7. Antimicrobial Assay
3.8. Haemolytic Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitta, G.; Hubert, F.; Noël, T.; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta BBA Proteins Proteom. 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Gueguen, Y.; Herpin, A.; Aumelas, A.; Garnier, J.; Fievet, J.; Escoubas, J.-M.; Bulet, P.; Gonzalez, M.; Lelong, C.; Favrel, P.; et al. Characterization of a Defensin from the Oyster Crassostrea gigas. J. Biol. Chem. 2006, 281, 313–323. [Google Scholar] [CrossRef]
- Robinson, W.E.; McDougall, B.; Tran, D.; Selsted, M.E. Anti-HIV-1 activity of indolicidin, an antimicrobial peptide from neutrophils. J. Leukoc. Biol. 1998, 63, 94–100. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; de Lima, L.M.P.; Migliolo, L.; Mendes, G.D.S.; de Jesus, M.G.; Franco, O.L.; Silva, P.A. Linear antimicrobial peptides with activity against herpes simplex virus 1 and Aichi virus. Pept. Sci. 2017, 108, e22871. [Google Scholar] [CrossRef] [PubMed]
- Fehlbaum, P.; Bulet, P.; Michaut, L.; Lagueux, M.; Broekaert, W.F.; Hetru, C.; Hoffmann, J.A. Insect immunity: Septic injury of drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 1994, 269, 33159–33163. [Google Scholar] [PubMed]
- Benincasa, M.; Scocchi, M.; Pacor, S.; Tossi, A.; Nobili, D.; Basaglia, G.; Busetti, M.; Gennaro, R. Fungicidal activity of five cathelicidin peptides against clinically isolated yeasts. J. Antimicrob. Chemother. 2006, 58, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, S.E.; Miletti, L.C.; Steindel, M.; Bachère, E.; Barracco, M.A. Trypanocidal and leishmanicidal activities of different antimicrobial peptides (AMPs) isolated from aquatic animals. Exp. Parasitol. 2008, 118, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Brand, G.D.; Leite, J.R.S.A.; Silva, L.P.; Albuquerque, S.; Prates, M.V.; Azevedo, R.B.; Carregaro, V.; Silva, J.S.; Sá, V.C.L.; Brandão, R.A.; et al. Dermaseptins from Phyllomedusa oreades and Phyllomedusa distincta. Anti-Trypanosoma cruzi activity without cytotoxicity to mammalian cells. J. Biol. Chem. 2002, 277, 49332–49340. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.; Phoenix, D. Anionic Antimicrobial Peptides from Eukaryotic Organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Waller, F.; Wang, K. Innate Immunity in Plants: The Role of Antimicrobial Peptides. In Antimicrobial Peptides and Innate Immunity; Springer: Basel, Switzerland, 2013; pp. 29–51. ISBN 9783034805414. [Google Scholar]
- Paiva, A.D.; Breukink, E. Antimicrobial Peptides Produced by Microorganisms. In Antimicrobial Peptides and Innate Immunity; Springer: Basel, Switzerland, 2013; pp. 53–95. ISBN 9783034805414. [Google Scholar]
- Sun, D.; Eccleston, E.D.; Fallon, A.M. Cloning and expression of three cecropin cDNAs from a mosquito cell line. FEBS Lett. 1999, 454, 147–151. [Google Scholar] [CrossRef]
- Destoumieux, D.; Bulet, P.; Loew, D.; Van Dorsselaer, A.; Rodriguez, J.; Bachère, E. Penaeidins, a New Family of Antimicrobial Peptides Isolated from the Shrimp Penaeus vannamei (Decapoda). J. Biol. Chem. 1997, 272, 28398–28406. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and Functional Characterization of Three Chicken Cathelicidins with Potent Antimicrobial Activity. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar] [CrossRef]
- Douglas, S.E.; Gallant, J.W.; Liebscher, R.S.; Dacanay, A.; Tsoi, S.C.M. Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev. Comp. Immunol. 2003, 27, 589–601. [Google Scholar] [CrossRef]
- Houyvet, B.; Bouchon-Navaro, Y.; Bouchon, C.; Goux, D.; Bernay, B.; Corre, E.; Zatylny-Gaudin, C. Identification of a moronecidin-like antimicrobial peptide in the venomous fish Pterois volitans: Functional and structural study of pteroicidin-α. Fish Shellfish Immunol. 2017, 72, 318–324. [Google Scholar] [CrossRef]
- Leoni, G.; De Poli, A.; Mardirossian, M.; Gambato, S.; Florian, F.; Venier, P.; Wilson, D.; Tossi, A.; Pallavicini, A.; Gerdol, M. Myticalins: A Novel Multigenic Family of Linear, Cationic Antimicrobial Peptides from Marine Mussels (Mytilus spp.). Mar. Drugs 2017, 15, 261. [Google Scholar] [CrossRef]
- Gerdol, M.; Gomez-Chiarri, M.; Castillo, M.G.; Figueras, A.; Fiorito, G.; Moreira, R.; Novoa, B.; Pallavicini, A.; Ponte, G.; Roumbedakis, K.; et al. Immunity in Molluscs: Recognition and Effector Mechanisms, with a Focus on Bivalvia. In Advances in Comparative Immunology; Springer International Publishing: Cham, Switzerland, 2018; pp. 225–341. ISBN 9781461490654. [Google Scholar]
- Gonzalez, M.; Gueguen, Y.; Desserre, G.; de Lorgeril, J.; Romestand, B.; Bachère, E. Molecular characterization of two isoforms of defensin from hemocytes of the oyster Crassostrea gigas. Dev. Comp. Immunol. 2007, 31, 332–339. [Google Scholar] [CrossRef]
- Gueguen, Y.; Bernard, R.; Julie, F.; Paulina, S.; Delphine, D.-G.; Franck, V.; Philippe, B.; Evelyne, B. Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol. Immunol. 2009, 46, 516–522. [Google Scholar] [CrossRef]
- Mitta, G.; Vandenbulcke, F.; Hubert, F.; Roch, P. Mussel defensins are synthesised and processed in granulocytes then released into the plasma after bacterial challenge. J. Cell Sci. 1999, 112, 4233–4242. [Google Scholar]
- Rosa, R.D.; Santini, A.; Fievet, J.; Bulet, P.; Destoumieux-Garzón, D.; Bachère, E. Big Defensins, a Diverse Family of Antimicrobial Peptides That Follows Different Patterns of Expression in Hemocytes of the Oyster Crassostrea gigas. PLoS ONE 2011, 6, e25594. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, Z.; Zhang, X.; Shi, Q.; Wang, C.; Hu, Z.; Li, H. Identification and characterization of a novel defensin from Asian green mussel Perna viridis. Fish Shellfish Immunol. 2018, 74, 242–249. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liang, H.; Zhu, J.; Fang, X. Separation, identification and gene expression analysis of PmAMP-1 from Pinctada fucata martensii. Fish Shellfish Immunol. 2019, 92, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, Q.; Wang, Q.; Chen, L.; Liu, Y.; Cong, M.; Wu, H.; Li, F.; Ji, C.; Zhao, J. A defensin-like antimicrobial peptide from the manila clam Ruditapes philippinarum: Investigation of the antibacterial activities and mode of action. Fish Shellfish Immunol. 2018, 80, 274–280. [Google Scholar] [CrossRef]
- Li, M.; Zhu, L.; Zhou, C.; Sun, S.; Fan, Y.; Zhuang, Z. Molecular characterization and expression of a novel big defensin (Sb-BDef1) from ark shell, Scapharca broughtonii. Fish Shellfish Immunol. 2012, 33, 1167–1173. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, X.C.; Liu, H.H.; Fan, M.H.; Sun, J.J.; Shen, W. Molecular characterization of a novel antimicrobial peptide from Mytilus coruscus. Fish Shellfish Immunol. 2013, 34, 610–616. [Google Scholar] [CrossRef]
- Charlet, M.; Chernysh, S.; Philippe, H.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Isolation of Several Cystein-rich Antimicrobial Peptides from the Blood of a Mollusc, Mytilus edulis. J. Biol. Chem. 1996, 271, 21808–21813. [Google Scholar] [CrossRef]
- Zhao, J.; Li, C.; Chen, A.; Li, L.; Su, X.; Li, T. Molecular Characterization of a Novel Big Defensin from Clam Venerupis philippinarum. PLoS ONE 2010, 5, e13480. [Google Scholar] [CrossRef]
- Wang, D.; Li, F.; Li, S.; Wen, R.; Xiang, J. Expression profiles of antimicrobial peptides (AMPs) and their regulation by Relish. Chin. J. Oceanol. Limnol. 2012, 30, 611–619. [Google Scholar] [CrossRef]
- Dolashka, P.; Moshtanska, V.; Borisova, V.; Dolashki, A.; Stevanovic, S.; Dimanov, T.; Voelter, W. Antimicrobial proline-rich peptides from the hemolymph of marine snail Rapana venosa. Peptides 2011, 32, 1477–1483. [Google Scholar] [CrossRef]
- De Zoysa, M.; Whang, I.; Lee, Y.; Lee, S.; Lee, J.-S.; Lee, J. Defensin from disk abalone Haliotis discus discus: Molecular cloning, sequence characterization and immune response against bacterial infection. Fish Shellfish Immunol. 2010, 28, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Houyvet, B.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Henry, J.; Zatylny-Gaudin, C. Design of antimicrobial peptides from a cuttlefish database. Amino Acids 2018, 50, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Farto, R.; Armada, S.P.; Montes, M.; Guisande, J.A.; Pérez, M.J.; Nieto, T.P. Vibrio lentus associated with diseased wild octopus (Octopus vulgaris). J. Invertebr. Pathol. 2003, 83, 149–156. [Google Scholar] [CrossRef]
- Prado, S.; Dubert, J.; da Costa, F.; Martínez-Patiño, D.; Barja, J.L. Vibrios in hatchery cultures of the razor clam, Solen marginatus (Pulteney). J. Fish Dis. 2014, 37, 209–217. [Google Scholar] [CrossRef]
- Rojas, R.; Miranda, C.D.; Opazo, R.; Romero, J. Characterization and pathogenicity of Vibrio splendidus strains associated with massive mortalities of commercial hatchery-reared larvae of scallop Argopecten purpuratus (Lamarck, 1819). J. Invertebr. Pathol. 2015, 124, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Pujalte, M.; Sitjà-Bobadilla, A.; Álvarez-Pellitero, P.; Garay, E. Carriage of potentially fish-pathogenic bacteria in Sparus aurata cultured in Mediterranean fish farms. Dis. Aquat. Organ. 2003, 54, 119–126. [Google Scholar] [CrossRef]
- Lasa, A.; Avendaño-Herrera, R.; Estrada, J.M.; Romalde, J.L. Isolation and identification of Vibrio toranzoniae associated with diseased red conger eel (Genypterus chilensis) farmed in Chile. Vet. Microbiol. 2015, 179, 327–331. [Google Scholar] [CrossRef]
- Benoist, L.; Corre, E.; Bernay, B.; Henry, J.; Zatylny-Gaudin, C. -Omic Analysis of the Sepia officinalis White Body: New Insights into Multifunctionality and Haematopoiesis Regulation. J. Proteome Res. 2020, 19, 3072–3087. [Google Scholar] [CrossRef]
- Zatylny-Gaudin, C.; Cornet, V.; Leduc, A.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Bernay, B.; Garderes, J.; Kraut, A.; Couté, Y.; et al. Neuropeptidome of the Cephalopod Sepia officinalis: Identification, Tissue Mapping, and Expression Pattern of Neuropeptides and Neurohormones during Egg Laying. J. Proteome Res. 2016, 15, 48–67. [Google Scholar] [CrossRef]
- Cornet, V.; Henry, J.; Corre, E.; Le Corguille, G.; Zanuttini, B.; Zatylny-Gaudin, C. Dual role of the cuttlefish salivary proteome in defense and predation. J. Proteom. 2014, 108, 209–222. [Google Scholar] [CrossRef]
- Castellanos-Martínez, S.; Arteta, D.; Catarino, S.; Gestal, C. De novo transcriptome sequencing of the Octopus vulgaris hemocytes using illumina RNA-Seq technology: Response to the infection by the gastrointestinal parasite Aggregata octopiana. PLoS ONE 2014, 9, e107873. [Google Scholar] [CrossRef] [PubMed]
- Gestal, C.; Costa, M.; Figueras, A.; Novoa, B. Analysis of differentially expressed genes in response to bacterial stimulation in hemocytes of the carpet-shell clam Ruditapes decussatus: Identification of new antimicrobial peptides. Gene 2007, 406, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Mateo, D.R.; Greenwood, S.J.; Araya, M.T.; Berthe, F.C.J.; Johnson, G.R.; Siah, A. Differential gene expression of γ-actin, Toll-like receptor 2 (TLR-2) and interleukin-1 receptor-associated kinase 4 (IRAK-4) in Mya arenaria haemocytes induced by in vivo infections with two Vibrio splendidus strains. Dev. Comp. Immunol. 2010, 34, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Araya, M.T.; Siah, A.; Mateo, D.R.; Markham, F.; McKenna, P.; Johnson, G.R.; Berthe, F.C.J. Morphological and Molecular Effects of Vibrio splendidus on Hemocytes of Softshell Clams, Mya arenaria. J. Shellfish Res. 2009, 28, 751–758. [Google Scholar] [CrossRef]
- Herath, H.M.L.P.B.; Elvitigala, D.A.S.; Godahewa, G.I.; Whang, I.; Lee, J. Molecular insights into a molluscan transferrin homolog identified from disk abalone (Haliotis discus discus) evidencing its detectable role in host antibacterial defense. Dev. Comp. Immunol. 2015, 53, 222–233. [Google Scholar] [CrossRef]
- Richards, R.C.; O’Neil, D.B.; Thibault, P.; Ewart, K.V. Histone H1: An Antimicrobial Protein of Atlantic Salmon (Salmo salar). Biochem. Biophys. Res. Commun. 2001, 284, 549–555. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Cornet, V.; Henry, J.; Corre, E.; Le Corguillé, G.; Zatylny-Gaudin, C. The Toll/NF-κB pathway in cuttlefish symbiotic accessory nidamental gland. Dev. Comp. Immunol. 2015, 53, 42–46. [Google Scholar] [CrossRef]
- Li, Y.-F.; Liu, Y.-Z.; Chen, Y.-W.; Chen, K.; Batista, F.M.; Cardoso, J.C.R.; Chen, Y.-R.; Peng, L.-H.; Zhang, Y.; Zhu, Y.-T.; et al. Two toll-like receptors identified in the mantle of Mytilus coruscus are abundant in haemocytes. Fish Shellfish Immunol. 2019, 90, 134–140. [Google Scholar] [CrossRef]
- Toubiana, M.; Gerdol, M.; Rosani, U.; Pallavicini, A.; Venier, P.; Roch, P. Toll-like receptors and MyD88 adaptors in Mytilus: Complete cds and gene expression levels. Dev. Comp. Immunol. 2013, 40, 158–166. [Google Scholar] [CrossRef]
- Qiu, L.; Song, L.; Xu, W.; Ni, D.; Yu, Y. Molecular cloning and expression of a Toll receptor gene homologue from Zhikong Scallop, Chlamys farreri. Fish Shellfish Immunol. 2007, 22, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Liu, B.; Fan, S.; Li, H.; Chen, M.; Zhang, B.; Su, J.; Meng, Z.; Yu, D. Differentially expressed immune-related genes in hemocytes of the pearl oyster Pinctada fucata against allograft identified by transcriptome analysis. Fish Shellfish Immunol. 2017, 62, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Toubiana, M.; Rosani, U.; Giambelluca, S.; Cammarata, M.; Gerdol, M.; Pallavicini, A.; Venier, P.; Roch, P. Toll signal transduction pathway in bivalves: Complete cds of intermediate elements and related gene transcription levels in hemocytes of immune stimulated Mytilus galloprovincialis. Dev. Comp. Immunol. 2014, 45, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Liu, Z.; Zhou, Z.; Wang, L.; Wang, L.; Yue, F.; Wang, J.; Wang, H.; Song, L. Transcriptional activation and translocation of ancient NOS during immune response. FASEB J. 2016, 30, 3527–3540. [Google Scholar] [CrossRef]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef]
- Ni, D.; Song, L.; Wu, L.; Chang, Y.; Yu, Y.; Qiu, L.; Wang, L. Molecular cloning and mRNA expression of peptidoglycan recognition protein (PGRP) gene in bay scallop (Argopecten irradians, Lamarck 1819). Dev. Comp. Immunol. 2007, 31, 548–558. [Google Scholar] [CrossRef]
- Su, J.; Ni, D.; Song, L.; Zhao, J.; Qiu, L. Molecular cloning and characterization of a short type peptidoglycan recognition protein (CfPGRP-S1) cDNA from Zhikong scallop Chlamys farreri. Fish Shellfish Immunol. 2007, 23, 646–656. [Google Scholar] [CrossRef]
- Gonzalez, M.; Gueguen, Y.; Destoumieux-Garzon, D.; Romestand, B.; Fievet, J.; Pugniere, M.; Roquet, F.; Escoubas, J.-M.; Vandenbulcke, F.; Levy, O.; et al. Evidence of a bactericidal permeability increasing protein in an invertebrate, the Crassostrea gigas Cg-BPI. Proc. Natl. Acad. Sci. USA 2007, 104, 17759–17764. [Google Scholar] [CrossRef]
- Gorbushin, A.M.; Borisova, E.A. Lectin-like molecules in transcriptome of Littorina littorea hemocytes. Dev. Comp. Immunol. 2015, 48, 210–220. [Google Scholar] [CrossRef]
- Kumar, P.; Kannan, M.; ArunPrasanna, V.; Vaseeharan, B.; Vijayakumar, S. Proteomics analysis of crude squid ink isolated from Sepia esculenta for their antimicrobial, antibiofilm and cytotoxic properties. Microb. Pathog. 2018, 116, 345–350. [Google Scholar] [CrossRef]
- Ezquerra-Brauer, J.M.; Miranda, J.M.; Chan-Higuera, J.E.; Barros-Velázquez, J.; Aubourg, S.P. New icing media for quality enhancement of chilled hake (Merluccius merluccius) using a jumbo squid (Dosidicus gigas) skin extract. J. Sci. Food Agric. 2017, 97, 3412–3419. [Google Scholar] [CrossRef] [PubMed]
- Abdelmalek, B.E.; Sila, A.; Krichen, F.; Karoud, W.; Martinez-Alvarez, O.; Ellouz-Chaabouni, S.; Ayadi, M.A.; Bougatef, A. Sulfated polysaccharides from Loligo vulgaris skin: Potential biological activities and partial purification. Int. J. Biol. Macromol. 2015, 72, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Halverson, T.; Basir, Y.J.; Knoop, F.C.; Conlon, J.M. Purification and characterization of antimicrobial peptides from the skin of the North American green frog Rana clamitans. Peptides 2000, 21, 469–476. [Google Scholar] [CrossRef]
- Conlon, J.M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Kolodziejek, J.; Nowotny, N.; Bevier, C.R.; Moler, P.E. Peptidomic analysis of skin secretions from Rana heckscheri and Rana okaloosae provides insight into phylogenetic relationships among frogs of the Aquarana species group. Regul. Pept. 2007, 138, 87–93. [Google Scholar] [CrossRef]
- Gao, B.; Zhu, S. Mesobuthus Venom-Derived Antimicrobial Peptides Possess Intrinsic Multifunctionality and Differential Potential as Drugs. Front. Microbiol. 2018, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Oyama, L.B.; Girdwood, S.E.; Cookson, A.R.; Fernandez-Fuentes, N.; Privé, F.; Vallin, H.E.; Wilkinson, T.J.; Golyshin, P.N.; Golyshina, O.V.; Mikut, R.; et al. The rumen microbiome: An underexplored resource for novel antimicrobial discovery. npj Biofilms Microbiomes 2017, 3, 33. [Google Scholar] [CrossRef]
- Azkargorta, M.; Bregón-Villahoz, M.; Escobes, I.; Ibáñez-Pérez, J.; Iloro, I.; Iglesias, M.; Diez-Zapirain, M.; Rabanal, A.; Prieto, B.; Moragues, M.-D.; et al. In-depth proteomics and natural peptidomics analyses reveal antibacterial peptides in human endometrial fluid. J. Proteom. 2020, 216, 103652. [Google Scholar] [CrossRef]
- Sangster, C.R.; Smolowitz, R.M. Description of Vibrio alginolyticus Infection in Cultured Sepia officinalis, Sepia apama, and Sepia pharaonis. Biol. Bull. 2003, 205, 233–234. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Menanteau-Ledouble, S.; Kumar, G.; Saleh, M.; El-Matbouli, M. Aeromonas salmonicida: Updates on an old acquaintance. Dis. Aquat. Organ. 2016, 120, 49–68. [Google Scholar] [CrossRef]
- Fichi, G.; Cardeti, G.; Perrucci, S.; Vanni, A.; Cersini, A.; Lenzi, C.; De Wolf, T.; Fronte, B.; Guarducci, M.; Susini, F. Skin lesion-associated pathogens from Octopus vulgaris: First detection of Photobacterium swingsii, Lactococcus garvieae and betanodavirus. Dis. Aquat. Organ. 2015, 115, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Lozano-Leon, A.; DePaola, A.; Ishibashi, M.; Shimada, K.; Nishibuchi, M.; Liebana, E. Characterization of Pathogenic Vibrio parahaemolyticus Isolates from Clinical Sources in Spain and Comparison with Asian and North American Pandemic Isolates. J. Clin. Microbiol. 2004, 42, 4672–4678. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Cellura, C.; Toubiana, M.; Parrinello, N.; Roch, P. Specific expression of antimicrobial peptide and HSP70 genes in response to heat-shock and several bacterial challenges in mussels. Fish Shellfish Immunol. 2007, 22, 340–350. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Philip, D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Macmanes, M.D.; et al. De novo transcript sequence recostruction from RNA-Seq: Reference generation and analysis with Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Hiller, K.; Grote, A.; Scheer, M.; Munch, R.; Jahn, D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004, 32, W375–W379. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMP R3: A database on sequences, structures and signatures of antimicrobial peptides: Table 1. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific -helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Hetru, C.; Bulet, P. Strategies for the isolation and characterization of antimicrobial peptides of invertebrates. Methods Mol. Biol. 1997, 78, 35–49. [Google Scholar] [CrossRef]
- Duval, E.; Zatylny, C.; Laurencin, M.; Baudy-Floc’h, M.; Henry, J. KKKKPLFGLFFGLF: A cationic peptide designed to exert antibacterial activity. Peptides 2009, 30, 1608–1612. [Google Scholar] [CrossRef]

| Transcript | Name | Transcript Length (nt) | Protein Length (aa) | Expression (TPM) | Fold Change | |
|---|---|---|---|---|---|---|
| c-hct | Vs-hct | |||||
| TR5906|c0_g1_i2 | TLRγ | 2861 | 833 | 3.55 | 4.58 | 1.29 |
| TR31922|c0_g2_i3 | TLR-h (partial) | 1741 | 536 | 0.31 | 4.71 | 15.11 |
| TR20767|c0_g1_i1 | MyD88 | 1938 | 338 | 12.13 | 14.50 | 1.20 |
| TR42010|c1_g1_i1 | IRAK4 | 1939 | 319 | 8.56 | 10.63 | 1.24 |
| TR37884|c0_g2_i2 | TRAF6 | 2364 | 547 | 2.53 | 3.05 | 1.21 |
| TR17535|c0_g1_i1 | IκB | 2609 | 337 | 233.96 | 230.08 | 0.98 |
| TR34670|c0_g1_i1 | IKK-h | 2724 | 476 | 10.64 | 12.29 | 1.16 |
| TR41212|c6_g1_i2 | Rel/NF-κB-h | 2614 | 491 | 1.85 | 2.56 | 1.39 |
| TR24628|c0_g1_i1 | iNOS-h (partial) | 4137 | 1106 | 0.00 | 0.52 | - |
| TR41722|c24_g56_i1 | PGRP-h1 | 873 | 213 | 254.12 | 466.71 | 1.84 |
| TR14900|c0_g1_i1 | PGRP-h2 | 956 | 204 | 87.28 | 154.73 | 1.77 |
| TR40338|c3_g2_i1 | BPI/LPB | 2342 | 537 | 134.38 | 149.31 | 1.11 |
| TR32087|c0_g2_i1 | Gal-2 (partial) | 817 | 236 | 86.15 | 120.87 | 1.40 |
| Transcript Name | Length (aa) | Expression (TPM) | Fold Change | SP | AMP Name | |
|---|---|---|---|---|---|---|
| c-hct | Vs-hct | |||||
| TR42258|c1_g1_i1 | 54 | 8.92 | 7.57 | 0.85 | Yes | AV19 |
| TR27534|c0_g1_i1 | 48 | 17.39 | 23.79 | 1.37 | Yes | GK28 |
| TR36613|c0_g1_i1 | 47 | 20.28 | 17.01 | 0.84 | Yes | NF19 |
| TR42563|c7_g3_i1 | 40 | 1.99 | 0.78 | 0.39 | Yes | II19 |
| TR5654|c0_g1_i1 | 45 | 9.77 | 17.33 | 1.77 | Yes | LV25 |
| Name | Sequence | Length (aa) | MW | C | HR | α-Helix Prediction and NRH | CAMP Algorithms | |||
|---|---|---|---|---|---|---|---|---|---|---|
| SVM | RFC | ANN | DAC | |||||||
| AV19 | ASSFLTPRLSSLGKRSWAV | 19 | 2063.38 | +3 | 42% | Yes-2 | 0.75 | 0.8 | AMP | 0.91 |
| GK28 | GFCNFMHLKPISRELRRELYGRTRRRRK | 28 | 3577.25 | +8 | 28% | No | 0.69 | 0.31 | AMP | 0.8 |
| NF19 | NYWLLVLRRLLITKKVMWF | 19 | 2493.14 | +4 | 63% | Yes-7 | 0.43 | 0.71 | AMP | 0.65 |
| II19 | IYFHLFRKINFNLRIYYFI | 19 | 2581.1 | +3 | 52% | Yes-6 | 0.13 | 0.65 | AMP | 0.87 |
| LV25 | LKALKLPKGSTSTEVRRILVLEIRV | 25 | 2820.45 | +4 | 44% | Yes-4 | 0.68 | 0.88 | NAMP | 0.84 |
| Name | Gram Negative | Gram Positive | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vibrio alginolyticus | Vibrio splendidus | Vibrio aestueranius | Vibrio anguillarum | Vibrio parahaemo-lyticus | Escherichia coli | Halomonas aquamarina | Aeromonas salmonicida | Enterococcus faecalis | Lactococcus garvieae | |||||||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| AV19 | - | - | 10–20 | 10–20 | - | - | - | - | 10–20 | 20–50 | - | - | - | - | - | - | - | - | - | - |
| GK28 | ≤5 | 20–50 | ≤5 | ≤5 | 10–20 | >50 | 10–20 | 20–50 | ≤5 | 10–20 | ≤5 | 20–50 | - | - | - | - | - | - | - | - |
| NF19 | - | - | 5–10 | 10–20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| II19 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| LV25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| SYN | 10–20 | >20 | 5–10 | 10–20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Ox | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | ≤5 | >10 | - | ≤5 | ≤5 | ≤5 | 5–10 | ≤5 | > 10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benoist, L.; Houyvet, B.; Henry, J.; Corre, E.; Zanuttini, B.; Zatylny-Gaudin, C. In-Depth In Silico Search for Cuttlefish (Sepia officinalis) Antimicrobial Peptides Following Bacterial Challenge of Haemocytes. Mar. Drugs 2020, 18, 439. https://doi.org/10.3390/md18090439
Benoist L, Houyvet B, Henry J, Corre E, Zanuttini B, Zatylny-Gaudin C. In-Depth In Silico Search for Cuttlefish (Sepia officinalis) Antimicrobial Peptides Following Bacterial Challenge of Haemocytes. Marine Drugs. 2020; 18(9):439. https://doi.org/10.3390/md18090439
Chicago/Turabian StyleBenoist, Louis, Baptiste Houyvet, Joël Henry, Erwan Corre, Bruno Zanuttini, and Céline Zatylny-Gaudin. 2020. "In-Depth In Silico Search for Cuttlefish (Sepia officinalis) Antimicrobial Peptides Following Bacterial Challenge of Haemocytes" Marine Drugs 18, no. 9: 439. https://doi.org/10.3390/md18090439
APA StyleBenoist, L., Houyvet, B., Henry, J., Corre, E., Zanuttini, B., & Zatylny-Gaudin, C. (2020). In-Depth In Silico Search for Cuttlefish (Sepia officinalis) Antimicrobial Peptides Following Bacterial Challenge of Haemocytes. Marine Drugs, 18(9), 439. https://doi.org/10.3390/md18090439





