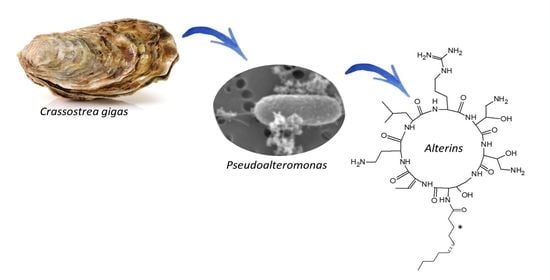
Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity
Abstract
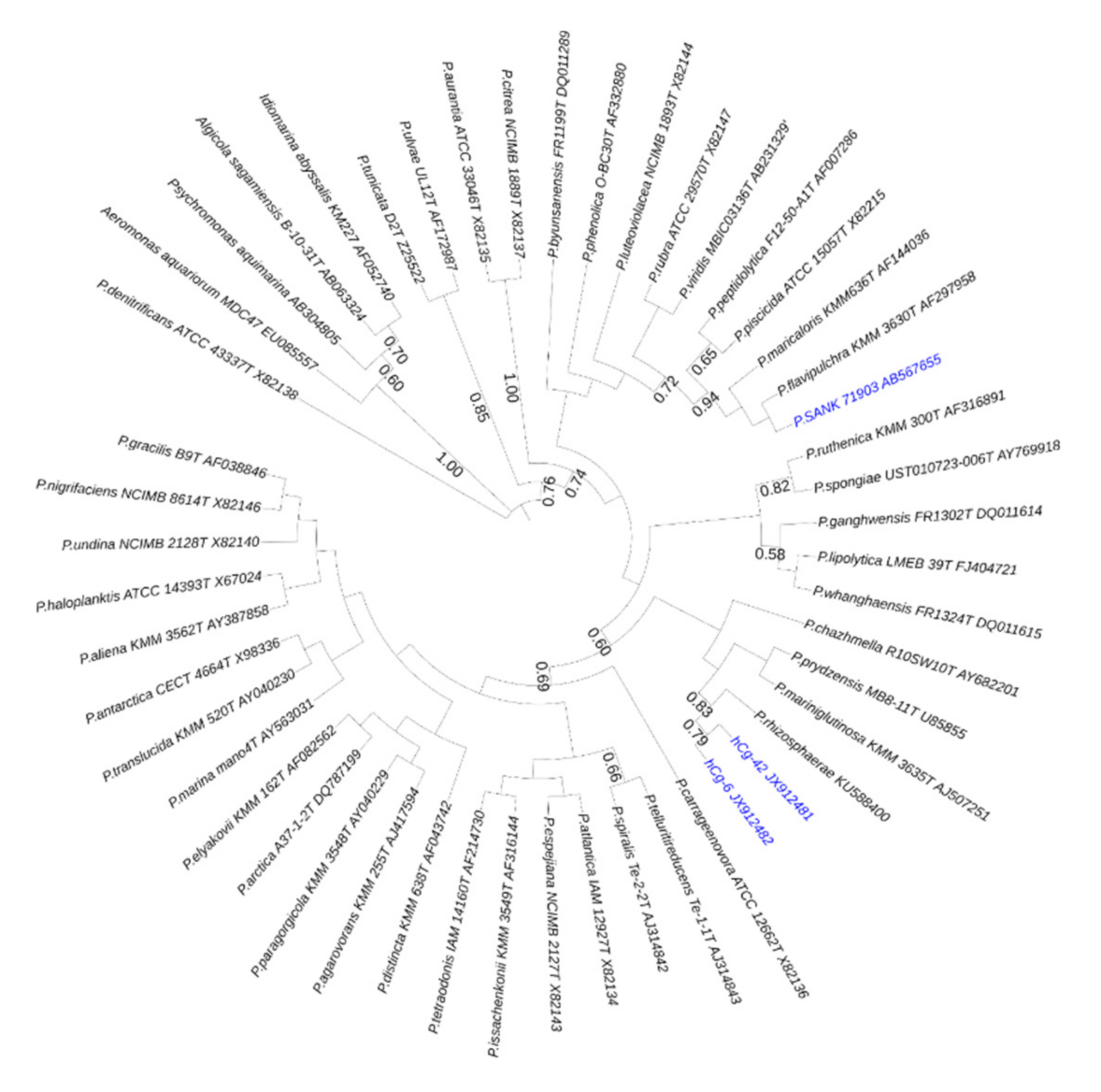
1. Introduction
2. Results
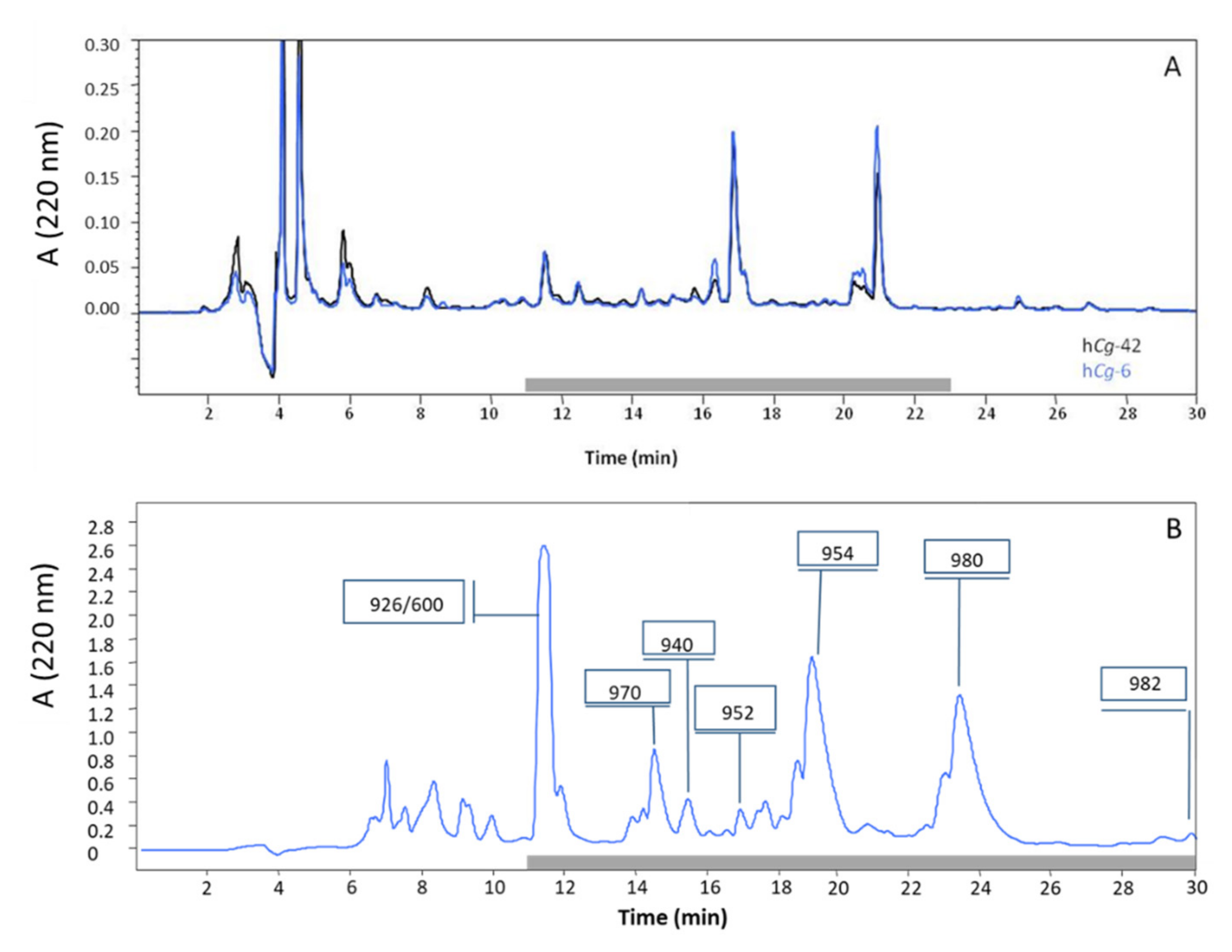
2.1. Purification of Antibacterial Peptides
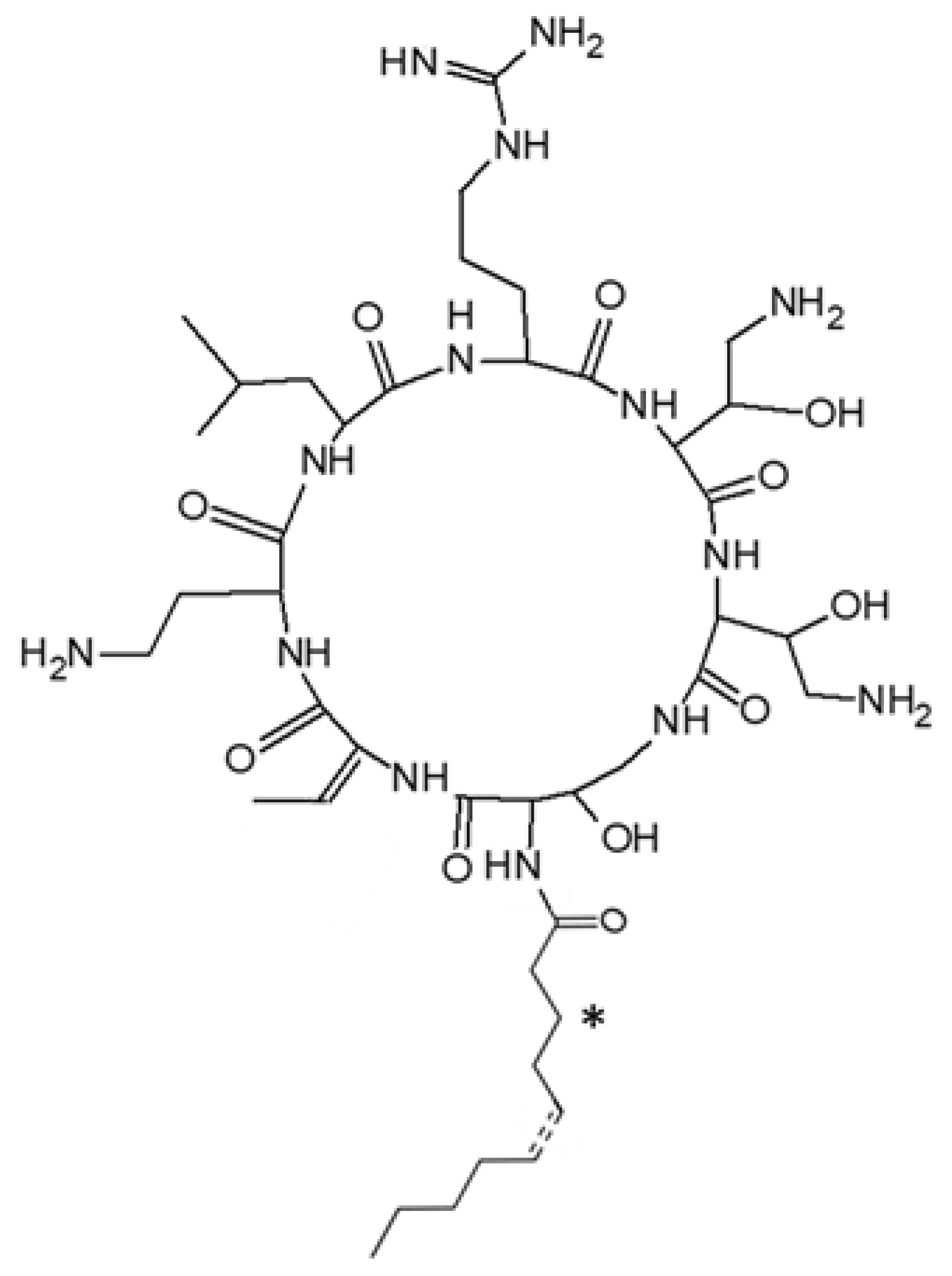
2.2. Structural Characterization of Active Peptides
2.3. Antibacterial Activity
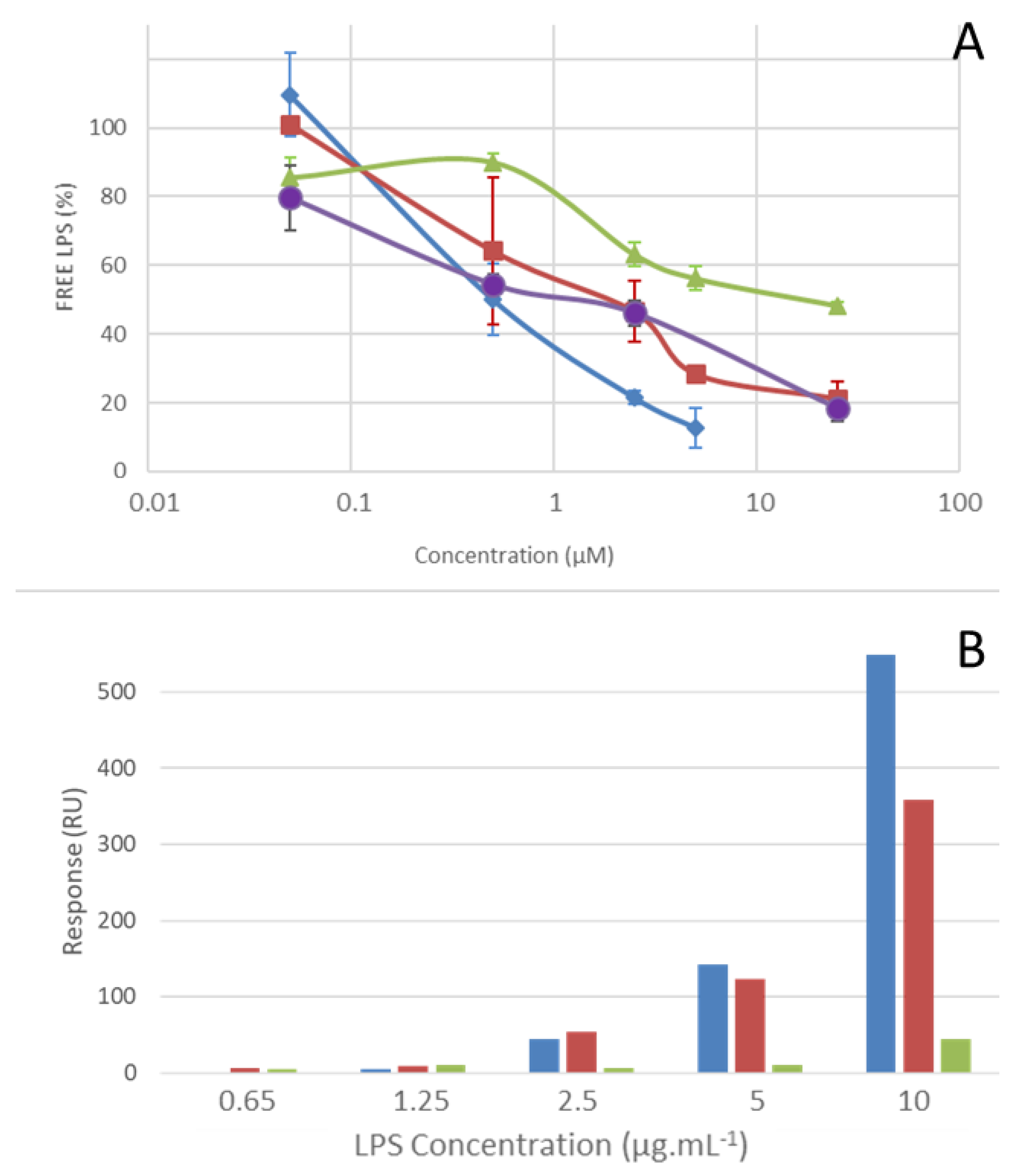
2.4. LPS Binding Properties
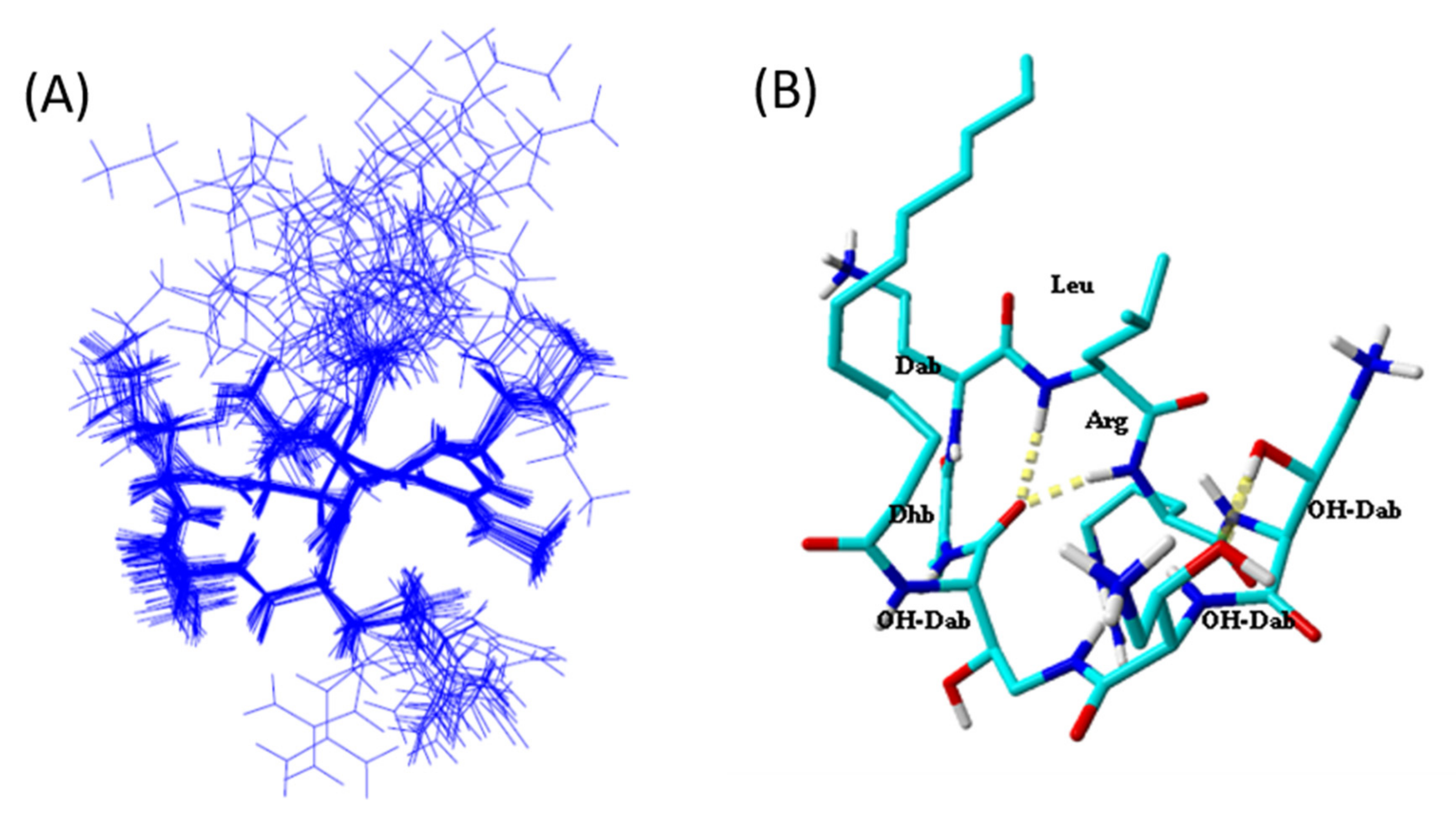
2.5. Tridimensional Structure in LPS Micelles
2.6. Membrane Effects and Bactericidal Activity of Alterins
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Antibacterial Compounds Isolation
4.3. NMR Experiments
4.4. Structure Calculations
4.5. Minimal Inhibitory Concentration (MIC)
4.6. Effect of Alterins on Vibrio Tasmaniensis Viability
4.7. Flow Cytometry
4.8. LPS Binding Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Patents
References
- Moran, M.A. The global ocean microbiome. Science 2015, 350, aac8455. [Google Scholar] [CrossRef]
- Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007, 5, 355–362. [Google Scholar] [CrossRef]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Bosch, T.C.G.; Cryan, J.F.; Gilbert, S.F.; Goodnight, C.J.; Lloyd, E.A.; et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems 2016, 1, e00028-16. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.R.; Easson, C.; Astudillo-García, C.; Olson, J.B.; Erwin, P.M.; López-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef]
- Webster, N.S.; Thomas, T. The Sponge Hologenome. mBio 2016, 7. [Google Scholar] [CrossRef]
- Gardères, J.; Bedoux, G.; Koutsouveli, V.; Crequer, S.; Desriac, F.; Pennec, G.L. Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Mar. Drugs 2015, 13, 4985–5006. [Google Scholar] [CrossRef] [PubMed]
- Trabal Fernández, N.; Mazón-Suástegui, J.M.; Vázquez-Juárez, R.; Ascencio-Valle, F.; Romero, J. Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol. Ecol. 2014, 88, 69–83. [Google Scholar] [CrossRef] [PubMed]
- King, G.M.; Judd, C.; Kuske, C.R.; Smith, C. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS ONE 2012, 7, e51475. [Google Scholar] [CrossRef]
- Lokmer, A.; Wegner, K.M. Hemolymph microbiome of pacific oysters in response to temperature, temperature stress and infection. ISME J. 2015, 9, 670–682. [Google Scholar] [CrossRef]
- Lokmer, A.; Kuenzel, S.; Baines, J.F.; Wegner, K.M. The role of tissue-specific microbiota in initial establishment success of Pacific oysters. Environ. Microbiol. 2016, 18, 970–987. [Google Scholar] [CrossRef] [PubMed]
- Defer, D.; Desriac, F.; Henry, J.; Bourgougnon, N.; Baudy-Floc’h, M.; Brillet, B.; Le Chevalier, P.; Fleury, Y. Antimicrobial peptides in oyster hemolymph: The bacterial connection. Fish Shellfish Immunol. 2013, 34, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Desriac, F.; Le Chevalier, P.; Brillet, B.; Leguerinel, I.; Thuillier, B.; Paillard, C.; Fleury, Y. Exploring the hologenome concept in marine bivalvia: Haemolymph microbiota as a pertinent source of probiotics for aquaculture. FEMS Microbiol. Lett. 2014, 350, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK; New York, NY, USA, 2000; ISBN 978-0-19-513585-5. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pantel, L.; Florin, T.; Dobosz-Bartoszek, M.; Racine, E.; Sarciaux, M.; Serri, M.; Houard, J.; Campagne, J.-M.; de Figueiredo, R.M.; Midrier, C.; et al. Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site. Mol. Cell 2018, 70, 83–94.e7. [Google Scholar] [CrossRef]
- Hirota-Takahata, Y.; Kozuma, S.; Kuraya, N.; Fukuda, D.; Nakajima, M.; Takatsu, T.; Ando, O. Ogipeptins, novel inhibitors of LPS: Physicochemical properties and structural elucidation. J. Antibiot. 2016, 70, 84–89. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Gonzalez, M.; Gueguen, Y.; Destoumieux-Garzón, D.; Romestand, B.; Fievet, J.; Pugnière, M.; Roquet, F.; Escoubas, J.-M.; Vandenbulcke, F.; Levy, O.; et al. Evidence of a bactericidal permeability increasing protein in an invertebrate, the Crassostrea gigas Cg-BPI. Proc. Natl. Acad. Sci. USA 2007, 104, 17759–17764. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Gronenborn, A.M. Theory and applications of the transferred nuclear overhauser effect to the study of the conformations of small ligands bound to proteins. J. Magn. Reson. (1969) 1982, 48, 402–417. [Google Scholar] [CrossRef]
- Bhattacharjya, S. NMR Structures and Interactions of Antimicrobial Peptides with Lipopolysaccharide: Connecting Structures to Functions. Curr. Top. Med. Chem. 2016, 16, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963. [Google Scholar] [CrossRef]
- Lebaron, P.; Servais, P.; Baudoux, A.-C.; Bourrain, M.; Parthuisot, N. Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 2002, 28, 131–140. [Google Scholar] [CrossRef]
- Bouvier, T.; Del Giorgio, P.A.; Gasol, J.M. A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 2007, 9, 2050–2066. [Google Scholar] [CrossRef]
- Offret, C.; Jégou, C.; Mounier, J.; Fleury, Y.; Chevalier, P.L. New insights into the haemo- and coelo-microbiota with antimicrobial activities from Echinodermata and Mollusca. J. Appl. Microbiol. 2019, 126, 1023–1031. [Google Scholar] [CrossRef]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; David, S.A.; Mathan, V.I.; Balaram, P. Polymyxin B nonapeptide: Conformations in water and in the lipopolysaccharide-bound state determined by two-dimensional NMR and molecular dynamics. Biopolymers 1997, 41, 251–265. [Google Scholar] [CrossRef]
- Pristovsek, P.; Kidric, J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: An NMR and molecular modeling study. J. Med. Chem. 1999, 42, 4604–4613. [Google Scholar] [CrossRef]
- Mares, J.; Kumaran, S.; Gobbo, M.; Zerbe, O. Interactions of Lipopolysaccharide and Polymyxin Studied by NMR Spectroscopy. J. Biol. Chem. 2009, 284, 11498–11506. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, J.D.; Karas, J.A.; Li, J.; Velkov, T. Structure of micelle bound cationic peptides by NMR spectroscopy using a lanthanide shift reagent. Chem. Commun. 2020, 56, 2897–2900. [Google Scholar] [CrossRef] [PubMed]
- Kadar, B.; Kocsis, B.; Nagy, K.; Szabo, D. The renaissance of polymyxins. Curr. Med. Chem. 2013, 20, 3759–3773. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Daugelavicius, R.; Bamford, D.H. Polymyxin B induces lysis of marine pseudoalteromonads. Antimicrob. Agents Chemother. 2007, 51, 3908–3914. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benedict, R.G.; Langlykke, A.F. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 1947, 54, 24. [Google Scholar]
- Kozuma, S.; Hirota-Takahata, Y.; Fukuda, D.; Kuraya, N.; Nakajima, M.; Ando, O. Identification and biological activity of ogipeptins, novel LPS inhibitors produced by marine bacterium. J. Antibiot. 2016, 70. [Google Scholar] [CrossRef]
- Hirota-Takahata, Y.; Kozuma, S.; Kuraya, N.; Fukuda, D.; Nakajima, M.; Ando, O. Pedopeptins, novel inhibitors of LPS: Taxonomy of producing organism, fermentation, isolation, physicochemical properties and structural elucidation. J. Antibiot. 2014, 67, 243–251. [Google Scholar] [CrossRef]
- Kozuma, S.; Hirota-Takahata, Y.; Fukuda, D.; Kuraya, N.; Nakajima, M.; Ando, O. Screening and biological activities of pedopeptins, novel inhibitors of LPS produced by soil bacteria. J. Antibiot. 2014, 67, 237–242. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure—Activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef]
- Jerala, R. Structural biology of the LPS recognition. Int. J. Med. Microbiol. 2007, 297, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Inácio, R.G.; Raimundo, J.M.; Martins, M.; Castanho, M.A.R.B.; Santos, N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers 2012, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, P.; Rosa, R.D.; Destoumieux-Garzón, D. An intimate link between antimicrobial peptide sequence diversity and binding to essential components of bacterial membranes. Biochim. Biophys. Acta BBA Biomembr. 2016, 1858, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Saravanan, R.; Mohanram, H.; Mangoni, M.L.; Bhattacharjya, S. NMR Structures and Interactions of Temporin-1Tl and Temporin-1Tb with Lipopolysaccharide Micelles. J. Biol. Chem. 2011, 286, 24394–24406. [Google Scholar] [CrossRef] [PubMed]
- Larrick, J.W.; Hirata, M.; Balint, R.F.; Lee, J.; Zhong, J.; Wright, S.C. Human CAP18: A novel antimicrobial lipopolysaccharide-binding protein. Infect. Immun. 1995, 63, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ. 2013, 105, 45–55. [Google Scholar] [CrossRef]
- Nicolas, J.L.; Basuyaux, O.; Mazuri, J.; Thbault, A. Vibrio carchariae, a pathogen of the abalone Haliotis tuberculata. Dis. Aquat. Organ. 2002, 50, 35–43. [Google Scholar] [CrossRef]
- Destoumieux-Garzón, D.; Rosa, R.D.; Schmitt, P.; Barreto, C.; Vidal-Dupiol, J.; Mitta, G.; Gueguen, Y.; Bachère, E. Antimicrobial peptides in marine invertebrate health and disease. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004, 278, 313–352. [Google Scholar] [CrossRef]
- Nicolas, I.; Bordeau, V.; Bondon, A.; Baudy-Floc’h, M.; Felden, B. Novel antibiotics effective against gram-positive and -negative multi-resistant bacteria with limited resistance. PLoS Biol. 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins Struct. Funct. Bioinforma. 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Marie, D.; Partensky, F.; Jacquet, S.; Vaulot, D. Enumeration and Cell Cycle Analysis of Natural Populations of Marine Picoplankton by Flow Cytometry Using the Nucleic Acid Stain SYBR Green I. Appl. Environ. Microbiol. 1997, 63, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Bouvy, M.; Got, P.; Domaizon, I.; Pagano, M.; Leboulanger, C.; Bouvier, C.; Carré, C.; Roques, C.; Dupuy, C. Plankton communities of the five “Iles Eparses” (Western Indian Ocean) considered to be pristine ecosystems. Acta Oecol. 2016, 72, 9–20. [Google Scholar] [CrossRef]
- Novo, D.J.; Perlmutter, N.G.; Hunt, R.H.; Shapiro, H.M. Multiparameter Flow Cytometric Analysis of Antibiotic Effects on Membrane Potential, Membrane Permeability, and Bacterial Counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 2000, 44, 827–834. [Google Scholar] [CrossRef]
- del Giorgio, P.A.; Bouvier, T.C. Linking the physiologic and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 2002, 47, 471–486. [Google Scholar] [CrossRef]
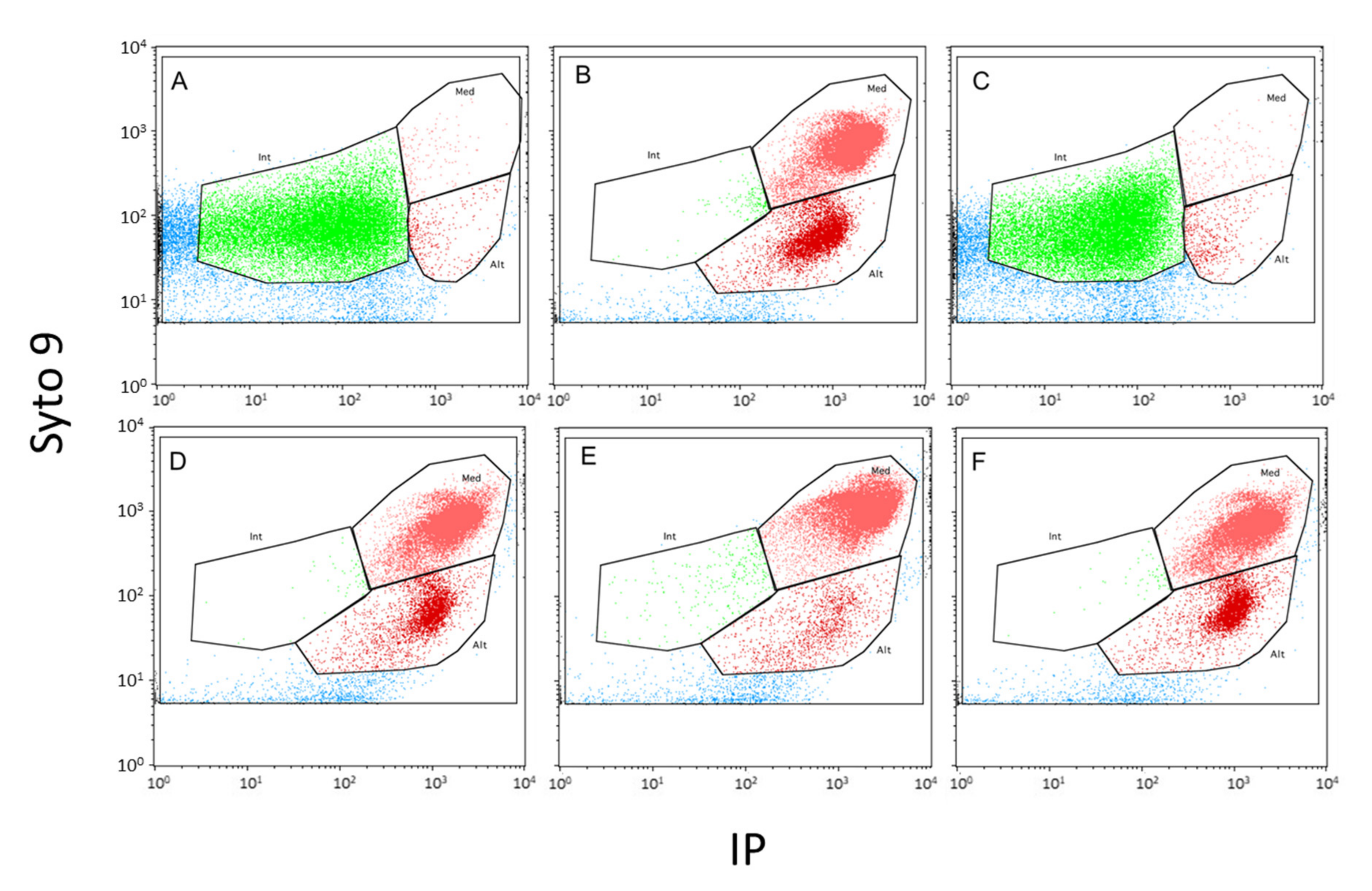
 ) and cells exhibiting an altered membrane with low (
) and cells exhibiting an altered membrane with low ( ) and high (
) and high ( ) nucleic acid content.
) nucleic acid content.
 ) and cells exhibiting an altered membrane with low (
) and cells exhibiting an altered membrane with low ( ) and high (
) and high ( ) nucleic acid content.
) nucleic acid content.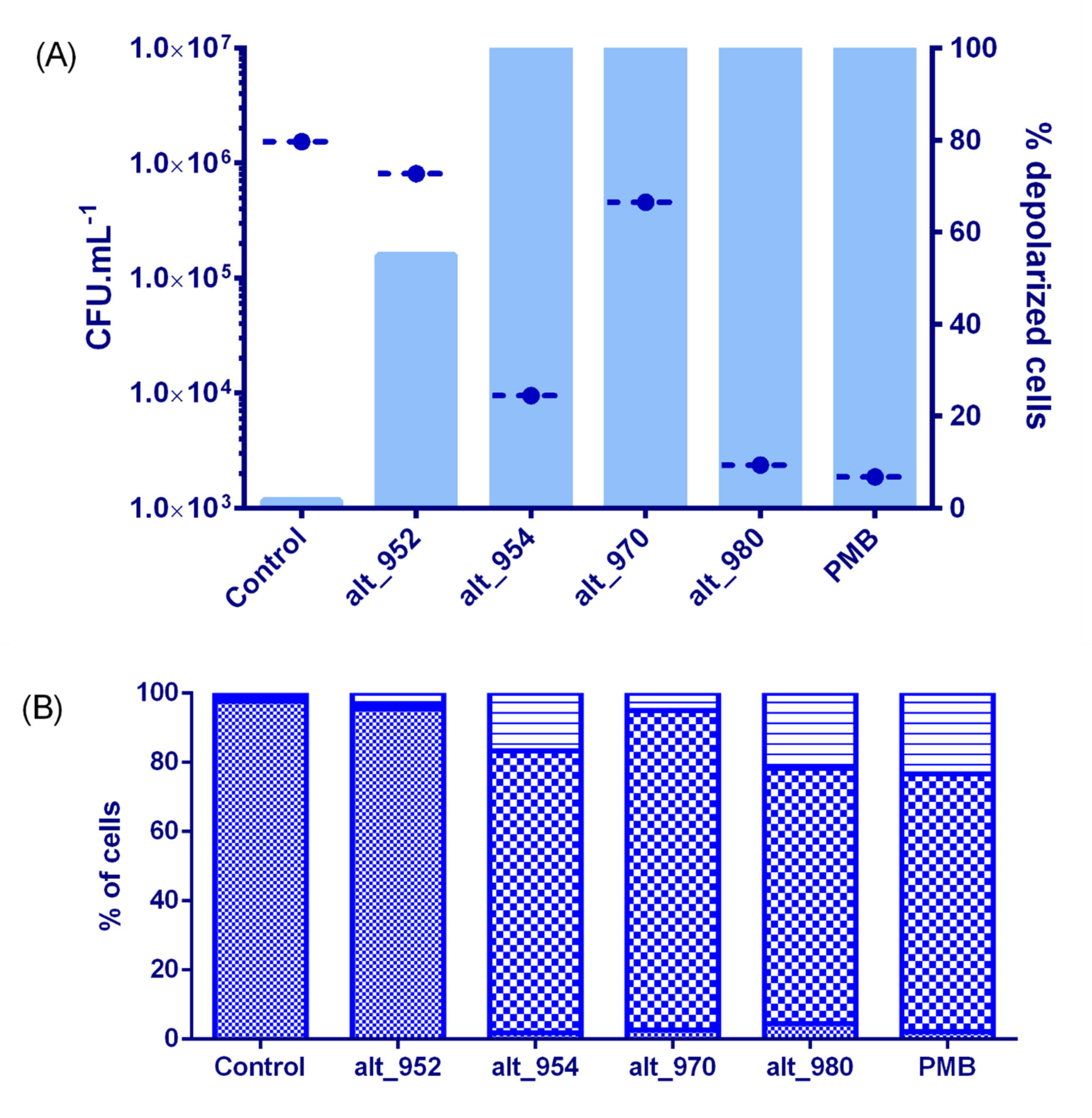
| Denomination | Hydrocarbon Tail | MW (Da) | Pseudoalteromonas Strains | |||
|---|---|---|---|---|---|---|
| hCg-6 | hCg-42 | SANK 71903 | ||||
| alt_926 | C8:0 | 926 | + | + | − | |
| alt_940 | C9:0 | 940 | + | + | − | |
| alt_952 | cis C10:1Δ5 | 952 | + | + | − | |
| alt_954 | C10:0 | 954 | + | + | + | |
| Alterin | alt_970 | 3′-OH C10:0 | 970 | + | + | − |
| alt_980 | cis C12:1 Δ5 | 980 | + | + | + | |
| alt_982 | C12:0 | 982 | + | − | + | |
| alt_1008 | cis C14:1Δ7 | 1008 | − | − | + | |
| MICs (µM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Alterins | PMB. | ||||||||
| MW (Da) | 926 | 940 | 954 | 952 | 970 | 980 | 982 | ||
| Hydrocarbon Tail | C8:0 | C9:0 | C10:0 | C10:1 | 3′OH-C10:0 | C12:1 | C12:0 | ||
| A. caviae | CIP 7616 | 5.5 | ND | 17.6 | 5.8 | ND | 3.1 | ||
| A. hydrophila | CIP 7614 | >100 | 25 | 2.7 | ND | ND | 2.9 | ND | 0.8 |
| E. coli | ML35 | 100 | 50 | 1.4 | 25 | 35 | 1.5 | ND | 3.1 |
| E. coli | SBS363 | 50 | 25 | 0.2 | 0.8 | 0.1 | 0.4 | 6.3 | 0.2 |
| P. aeruginosa | ATCC 27853 | >100 | 50 | 22 | 50 | 35.2 | 23 | ND | 1.6 |
| Pseudoalteromonas | hCg-6 | >100 | >100 | >100 | >100 | >100 | >100 | ND | >100 |
| Pseudoalteromonas | hCg-42 | ND | ND | >100 | >100 | >100 | >100 | ND | >100 |
| S. enterica | CIP 8297 | 12.5 | 6.25 | 0.7 | 6.25 | 1.1 | 1.4 | ND | 0.8 |
| V. harveyi | ORM4 | 50 | 6.25 | 1.4 | 12.5 | ND | 1.4 | ND | 25 |
| V. parahaemolyticus | 13-028A/3 | ND | ND | 6.25 | ND | 6.25 | 6.25 | ND | >100 |
| V. tapetis | CECT 4600T | ND | ND | >100 | >100 | >100 | >100 | ND | >100 |
| V. tasmaniensis | LGP 32 | 50 | 6.25 | 2.7 | 50 | 8.8 | 2.9 | ND | 3 |
| Y. ruckeri | ATCC 29473 | 50 | 6.25 | 0.3 | 0.4 | 1.1 | 0.4 | 12.5 | 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desriac, F.; El Harras, A.; Simon, M.; Bondon, A.; Brillet, B.; Le Chevalier, P.; Pugnière, M.; Got, P.; Destoumieux-Garzón, D.; Fleury, Y. Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity. Mar. Drugs 2020, 18, 630. https://doi.org/10.3390/md18120630
Desriac F, El Harras A, Simon M, Bondon A, Brillet B, Le Chevalier P, Pugnière M, Got P, Destoumieux-Garzón D, Fleury Y. Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity. Marine Drugs. 2020; 18(12):630. https://doi.org/10.3390/md18120630
Chicago/Turabian StyleDesriac, Florie, Abderrafek El Harras, Matthieu Simon, Arnaud Bondon, Benjamin Brillet, Patrick Le Chevalier, Martine Pugnière, Patrice Got, Delphine Destoumieux-Garzón, and Yannick Fleury. 2020. "Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity" Marine Drugs 18, no. 12: 630. https://doi.org/10.3390/md18120630
APA StyleDesriac, F., El Harras, A., Simon, M., Bondon, A., Brillet, B., Le Chevalier, P., Pugnière, M., Got, P., Destoumieux-Garzón, D., & Fleury, Y. (2020). Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity. Marine Drugs, 18(12), 630. https://doi.org/10.3390/md18120630