Abstract
The present study aimed to identify mycotoxins in edible tissues of Atlantic salmon (Salmo salar) using liquid chromatography coupled to hybrid quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS). After using a non-targeted screening approach and a home-made spectral library, 233 mycotoxins were analyzed. Moreover, the occurrence of mycotoxins in fish filets was evaluated, and their potential toxicity was predicted by in silico methods. According to the obtained results, forty mycotoxins were identified in analyzed salmon samples, the predominant mycotoxins being enniatins (also rugulosin and 17 ophiobolins), commonly found in cereals and their by-products. Thus, mycotoxin carry-over can occur from feed to organs and edible tissues of cultivated fish. Moreover, the toxicity of detected mycotoxins was predicted by the in silico webserver ProTox-II, highlighting that special attention must be paid to some less reported mycotoxins due to their toxic predicted properties.
1. Introduction
Mycotoxins are natural contaminants commonly found in plant-derived foodstuffs, mainly cereals and their by-products. Since these raw materials are added as ingredients in feed formulation for different animal species, including cultivated fish, the risk of mycotoxin contamination in feed for aquaculture has increased, thus introducing contaminants (i.e., mycotoxins), which were not previously identified in fish tissues []. Diverse studies reported mycotoxin contents in a wide range of randomly sampled feedstuffs and raw materials intended for terrestrial animals [,,,,,]. However, studies focused on feedstuffs intended for aquaculture fish are still scarce, although recently, some studies developed feasible analytical approaches for mycotoxin detection in aquafeeds [,]. The carry-over of mycotoxins from feed into edible portions of fish indicate that mycotoxins and their metabolites present in raw materials and feed for aquaculture fish can be fixed in edible portions and organs [,,].
In addition, mycotoxins have the ability to enter into the food chain through the intake of animal derived products such as milk, meat and eggs from livestock and poultry fed with contaminated feed. Some studies stated that the exposure risk to humans by consumption of these animal derived products can be considered as negligible due to lower contents reported in most cases [,]. However, it should be highlighted that mycotoxins or their metabolites can be considered an additional risk to human health, since they are part of the diet in combination with other chemical contaminants. Moreover, the exposure risk derived from the consumption of these animal by-products also depends on other factors, such as the considered diet, different groups of consumers with different metabolic profiles and their health status.
Mycotoxins have an important impact on aquaculture farming. However, there is a lack of information regarding the consequences for reared fish species, especially compared to that on terrestrial species []. Therefore, due to growing expansion of aquaculture feedstuffs, there is a need to control mycotoxin occurrence in fish produced by this production sector, since more data are required to carry out an adequate risk assessment for human consumption.
Atlantic salmon (Salmo salar) is among the most important farmed fish in Europe, together with other species such as rainbow trout (Oncorhynchus mykiss), sea bass (Dicentrarchus labrax) and gilthead sea bream (Sparus aurata). These species have been the key to producing an increase in the demand and fish consumption and production, thus converting their capture into their aquaculture farming []. The European Commission established a maximum level (ML) for aflatoxin M1 (AFM1) in milk []. However, no maximum levels (MLs) have been set for other mycotoxins in animal source foods (ASF), due to the scarce information on their occurrence in these foodstuffs [,]. Nevertheless, MLs have been set for AFB1 in feed and raw materials, while for deoxynivalenol (DON), zearalenone (ZEA), ochratoxin A (OTA), fumonisins (FB1 and FB2), HT-2 and T-2 toxins, maximum thresholds have been recommended in European legislation. However, according to Bernhoft et al. (2018) [], the maximum recommended level of DON is inappropriate as in their study, the calculated Non-Observed Adverse Effect Level (NOAEL) was lower than the guidance value.
Within this context, multi-mycotoxin methods have been developed in order to investigate the mycotoxin levels in feedstuffs and thus to assess the carry-over from feed to edible tissues [,]. These methods employing mass spectrometric (MS) detection can provide both qualitative and quantitative information at the same time on the assessment of undesirable substances in food and feed [].
Mycotoxin toxicity must be evaluated to carry out an adequate risk assessment. In this field, some in silico approaches can provide precise information on the toxicokinetics and the toxicity of some less studied mycotoxins in both food and feed. Thus, in the present study, the oral toxicity and other toxicological endpoints of identified mycotoxins were predicted by using the in silico webserver ProTox-II []. Within this context, the aim of this study was to determine the mycotoxin occurrence in edible tissues of Atlantic salmon (Salmo salar) using a multianalyte method consisting in liquid chromatography coupled to hybrid quadrupole time-of-flight mass spectrometry (LC/Q-TOF MS) and also to predict the potential toxicity of the identified mycotoxins by in silico approaches.
2. Results and Discussion
2.1. Mycotoxin Identification by Non-Target Screening
In this study, LC/Q-TOF-MS was used for structural elucidation, identification, characterization and confirmation of the chemical formulas of mycotoxins due to its improved full-scan sensitivity, mass accuracy and resolving power compared to other equipment such as quadrupole mass spectrometers [,,,,].
TOF analyzer allowed us to investigate the presence of 233 mycotoxins available in a wide list of validated compounds found in a homemade spectral library showing the presence of forty mycotoxins in analyzed salmon fillets (Table 1). To the best of our knowledge, this is the first study reporting the presence of these mycotoxins in fish from aquaculture farming directly purchased from supermarkets.

Table 1.
Identified mycotoxins in salmon samples.
Although the presence of these fungal metabolites has been scarcely reported in feedstuffs and animal derived products, some of them are common contaminants of cereal-based foodstuffs from wheat and corn [], such as enniatins (ENNA, ENNA1, ENNB, ENNB1 and ENNB2) (Figure 1) and fusaproliferin (FUS).
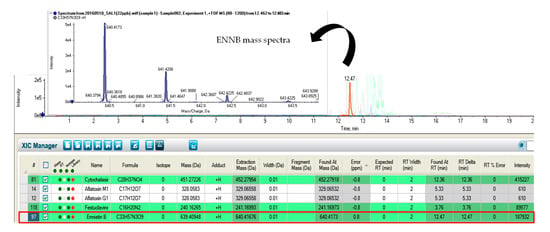
Figure 1.
Chromatogram showing enniatin B (ENNB) identification by LC-Q-TOF-MS.
On the other hand, other less reported mycotoxins in feedstuffs were detected, mainly anisomycin, cytochalasin J (CJ), mycophenolic acid (MPA), ophiobolin A (OA) and B (OB), rugulosin and penicillic acid (PA), among others.
Some of the mycotoxins identified in this study, namely chanoclavine, sulochrin, festuclavine, MPA, FB2 and ENNs, have been reported mainly in bread samples [,], while other mycotoxins have been also identified in feed and raw materials used in feed manufacture, such as MPA, cyclopiazonic acid, PA, radicicol, rugulosin and CJ, as evidenced by Streit et al. []. For instance, the method developed by Rundberget and Wilkins [] allowed the simultaneous determination of MPA together with other less reported mycotoxins in both food and feed, while Sulyok et al. [] were able to detect 15 mycotoxins in wheat and maize kernels similar to those found in this study. Moreover, Zhao et al. [], reported that mycotoxin contamination in feed directly influences the presence of mycotoxins in animal derived products, as they can be retained in organs and edible tissues after metabolization and can be also excreted in some by-products. These results allow us to conclude that these mycotoxins could be present in edible tissues of animals who consume those contaminated feedstuffs, as observed in our study [].
Recent surveys have revealed that diverse fish species in European aquaculture are commonly exposed to Fusarium mycotoxins in feed []. Emerging Fusarium mycotoxins were previously detected by our research team [], and diverse studies have identified mainly AFB1 and/or its metabolites in different organs and tissues from exposed fish [,,,]. Nácher-Mestre et al. (2013) [] applied a screening method to feed and fish fillets performed by UHPLC/Q-TOF-MS, confirming the presence of FB2 and ZEA in feed samples; however, no mycotoxin contamination was detected in fish fillets.
In a subsequent study, these authors evaluated the mycotoxin carry-over of aflatoxins (AFs), trichothecenes (TCs) and FBs, from feed to fish fillets in Atlantic salmon (Salmo Salar) and gilthead sea bream (Sparus aurata) [], concluding that no mycotoxin carry-over was found in analyzed samples. Conversely, Guan et al. [] evaluated DON occurrence and described the TC transformation by deacetylation and/or de-epoxidation reactions in different fish species. This fact is in accordance with our findings, where DON was not detected, but deepoxy-deoxynivalenol (DOM-1), obtained from DON de-epoxidation, was present in salmon fillets analyzed (Figure 2). This could be explained because DON is rapidly metabolized and its retention and accumulation in animal tissues is generally low []. These findings were also supported by Tola et al. [], who described that DOM-1 was formed by DON de-epoxidation and deacetylation by microorganisms from the digestive tract in fish species. In addition, other assays have revealed that microbes in the digestive tract of brown bullhead (Ameiurus nebulosus), brown trout (Salmo trutta), pink salmon (Oncorhynchus gorbuscha) and other fish species were capable of transforming DON into DOM-1, while hepatic microsomes in the liver of common carp (Cyprinus carpio) were able to transform DON into deoxynivalenol 3 glucuronide (DON-3-glc). Moreover, according to the study reported by Bernhoft et al. (2017) [], DON was metabolized in the liver of Atlantic salmon (Salmo salar) exposed to DON contaminated feed, resulting in the formation of DON-3glc. In their study, DON residues were detected in all tissues; however, when compared to terrestrial species, it can be observed that in Atlantic salmon the elimination of DON could be considerably slower.
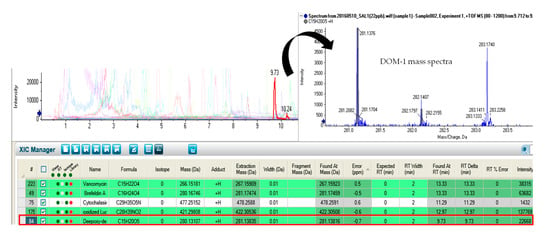
Figure 2.
Chromatogram showing the identification of deepoxy-deoxynivalenol (DOM-1).
Within the identified molecules, some of them corresponded to antibiotics, namely tetracyclines and β-lactams. The presence of these veterinary drug residues in edible tissues can be explained by their use in the treatment of food-producing animals. In animal production, when veterinary drugs are used, it is mandatory to respect a withdrawal period before the slaughter of animals intended for human consumption to avoid the presence of their residues in animal by-products, which can suppose a risk for consumers in terms of allergy and antibioresistance. In 2017, the World Health Organization (WHO) recommended reducing antibiotic use in animals used in the food industry, due to the increasing risk of antibiotic resistant bacteria, concluding that animals that require antibiotics should be treated with antibiotics that pose the smallest risk to human health. Some studies have established connections between antibiotic resistant infections and food-producing animals. Thus, it must be pointed out that antibiotic use in farm animals contributes to the overall problem of antibiotic resistance and thus poses an additional hazard of this animal by-products for consumers.
Furthermore, some compounds from the Penicillin family have been identified. Allergic reactions to penicillins have been commonly reported even at therapeutic doses. This fact highlights the importance of avoiding the presence of these undesirable compounds in animal origin products which can produce serious allergic reactions to consumers.
2.2. In Silico Toxicity Prediction
Most of the identified mycotoxins in the present study have not been commonly reported in scientific literature. Thus, little information on their toxicity is available. For this reason, in silico prediction methods were used in this survey to predict the toxicity of detected and identified mycotoxins.
ProTox-II
The oral toxicity prediction data provided by ProTox-II are based in 2D similarity and the recognition of toxic fragments. Results are expressed as LD50 (mg/kg). In Table 2, the predicted LD50 and the corresponding toxicity class for each identified mycotoxin are shown. In material and methods section, the characteristics to classify the substances within different toxicity groups are described.

Table 2.
Acute Oral Toxicity prediction obtained by using ProTox-II web server.
It should be highlighted that, according to the obtained predictions, ENNB and ENNB2 showed a predicted LD50 of 3 mg/kg, both with a 100% of average similarity and prediction accuracy. Thus, the assigned toxicity class was 1. Therefore, special attention should be paid to these mycotoxins due to their predicted toxicity, which is comparable to that of T-2 Toxin (Table 2), the latter being a toxic fungal metabolite with the lowest tolerable daily intake (TDI) within the Fusarium mycotoxins []. Regarding mycotoxins classified in category 2 (LD50 between 5 and 50 mg/kg), we found DOM-1, which showed a predicted LD50 of 34 mg/kg. In the case of oxidized luol, no prediction results could be obtained due to its chemical structure.
Using the ProTox-II web server, the organ toxicity (hepatotoxicity) can be also predicted, which was evaluated for different identified mycotoxins as the liver is the organ where mycotoxins are metabolized. In Table 3, the results obtained regarding the organ toxicity and the calculated prediction values for diverse toxicological endpoints using the ProTox-II web server are reported.

Table 3.
Organ toxicity and toxicological endpoints predicted activity calculated using the ProTox-II web server.
Regarding the organ toxicity, results obtained showed that cyclopenin, phomopsin A and tetracyclin were predicted as hepatotoxic. On the other hand, regarding the different toxicity endpoints evaluated, some mycotoxins were shown to be carcinogenic, immunotoxic, mutagenic and/or cytotoxic. Both fumigaclavine A and T-2 toxin were predicted as carcinogenic, immunotoxic and mutagenic substances, while curvularin, FB2, ophiobolin B, radicicol, rugulosin and vancomycin were predicted as carcinogenic and immunotoxic.
Within the toxicological endpoints, carcinogenicity and mutagenicity are relevant parameters to evaluate and to assess the toxic potential of different substances []. In this survey, Chanoclavine 56, cyclopenin, DOM-1, dihidrolysergol, festuclavine and methysergide were predicted as mutagenic compounds, while fumigaclavine A and T-2 toxin were predicted as both carcinogenic and mutagenic compounds (Table 3).
ENN B and ENNB2 were predicted as cytotoxic mycotoxins, a fact already reported in different studies performed by in vitro assays in different cell cultures [,]. The same occurs in the case of ophiobolin B (predicted as carcinogenic and immunotoxic), which has been described as toxic to animals in in vivo toxicity assays in mice [].
In Table 4 and Table 5, the prediction results obtained for the toxicological pathways, nuclear receptor signaling pathways and stress response pathways are reported, respectively. According to the Tox21 Consortium, chemical compounds might have the potential to disrupt processes in the human body that may lead to negative health effects []. Regarding the nuclear receptor signaling pathway, seven different pathways were assessed. The computational estimations revealed that curvularin and sulochrin could interact with the estrogen receptor alpha (ER), FK 506 was active to interact with aromatase receptor and methysergide could interact with the aryl hydrocarbon receptor (AhR). Regarding the stress response pathways, five diverse assays were assessed by in silico approaches. Computational predictions indicated that special attention should be paid to curvularin, which showed to be active to interact with the nuclear factor (erythroid-derived 2-like 2/antioxidant responsive element (nrf2/ARE), heat shock response element (HSE), mitochondrial membrane potential (MMP) and phosphoprotein p53 (tumor supressor).

Table 4.
Toxicological pathways: nuclear receptor signaling pathways predicted for detected mycotoxins.

Table 5.
Toxicological pathways: stress response pathways predicted for detected mycotoxins.
3. Materials and Methods
3.1. Samples
Norwegian Atlantic salmon (Salmo salar) (10 samples) from aquaculture farming were acquired from different supermarkets located in the metropolitan area of Valencia (Spain) and analyzed for mycotoxin content determination. Samples were acquired in individual packages at different markets within one month in 2016, and they came from different producers and batches. All samples were stored in a dark and dry place at −20 °C until analysis. After their packages had been opened, they were analyzed within the same day. These samples were first analyzed by LC-MS/MS LIT, and results showing ENN contents were reported in a previous study []. The results showed some unidentified peaks; thus, those samples were analyzed by LC-Q-TOF-MS in order to identify those compounds by exact mass.
3.2. Mycotoxin Extraction and LC-Q-TOF-MS Analysis
The mycotoxin extraction used was carried out according to the method previously reported by Tolosa et al. []. For chromatographic separation, an Agilent 1290 HPLC system (Agilent, Santa Clara, CA, USA) with an Acquity UHPLC BEH C18 analytical column (50 × 2.1 mm and 1.7 μm particle size) (Waters) at a flow rate of 350 μL/min was employed. The column temperature was set to 60 °C, and the injected volume was 10 µL, while the mobile phase consisted in water (0.15 mM ammonium formate) and MeOH 0.1% formic acid. The percentage of organic modifier (B) was changed linearly as follows: 0 min, 5%; 2 min, 25%; 13 min, 100%; 15 min, 100%; 15.1 min, 5%; 25 min, 5%.
A hybrid quadrupole-orthogonal acceleration-TOF mass spectrometer (AB SCIEX TripleTOF™ 5600 LC/MS/MS System, Ontario, Canada), with an orthogonal Z-spray-ESI interface operating in positive ion mode, was used. The data acquisition was performed in positive mode, and mode mass spectra were acquired in a scan range from 100 to 1000 m/z, with a resolving power of 10,000 full width at half maximum (FWHM) mass resolution at m/z 556.2771. For automated accurate mass measurement, an external calibrant delivery system (CDS) which infuses calibration solution was used prior to sample injection. The MS was carried out using an IDA acquisition method with the survey scan type (TOF-MS) and the dependent scan type (product ion) using 50V of collision energy (CE). Data were qualitatively evaluated using the PeakViewTM software (AB Sciex, Ontario, Canada).
Ion source parameters were as follows: cone voltage 25 V, capillary voltage 3.5 kV, desolvation temperature 500 °C, interface temperature 450 °C and source temperature 120 °C. Ion Spray Voltage (ISVF) was 5500 and declustering potential, 120 V. The Ion source gas 1 (GC1) and 2 (GC2) were 40 psi.
To promote ion-source fragmentation in MS2 experiments, an acquisition function with medium CE of 50 V was applied using argon as the collision gas (99.995%; Praxair, Madrid, Spain).
3.3. Non-Targeted Suspect Screening (TOF)
Mass spectrometry (MS) is among the most employed methods for structure elucidation, and high resolution MS is the method of choice for the identification of unknown masked mycotoxins in processed or unprocessed food []. In the non-target screening carried out in the present study, the compounds were identified by the exact m/z ion in chromatograms by searching in a database containing the empirical formula, the RT, isotopic abundance, number of double bonds and product ion mass spectra.
3.4. In Silico Prediction Methods
To carry out the prediction by in silico methods, the ProTox-II platform was used [,]. The only essential information to carry out the prediction is the chemical structure or the Pubchem-name of the molecule. The ProTox-II platform is divided into a five different classification steps: (1) acute toxicity (oral toxicity model with six different toxicity classes); (2) organ toxicity (1 model); (3) toxicological endpoints (4 models); (4) toxicological pathways (12 models) and (5) toxicity targets (15 models).
ProTox-II incorporates molecular similarity, fragment propensities, most frequent features (fragment similarity-based CLUSTER cross-validation) and machine-learning, based a total of 33 models for the prediction of various toxicity endpoints such as acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity, adverse outcomes pathways (Tox21) and toxicity targets.
3.4.1. Acute Oral Toxicity Prediction
Substances are classified into different toxicity classes, depending on the LD50 (mg/kg body weight), which are defined according to the globally harmonized system of classification in labelling of chemicals (GHS):
- Class I: fatal if swallowed (LD50 ≤ 5 mg/kg);
- Class II: fatal if swallowed (5 mg/kg < LD50 ≤ 50 mg/kg);
- Class III: toxic if swallowed (50 mg/kg < LD50 ≤ 300 mg/kg);
- Class IV: harmful if swallowed (300 mg/kg < LD50 ≤ 2000 mg/kg);
- Class V: may be harmful if swallowed (2000 mg/kg < LD50 ≤ 5000 mg/kg).
3.4.2. Toxicity Endpoint and Organ Toxicity Prediction
The same in silico prediction tool (ProTox-II) was employed for the prediction of various toxicity endpoints; namely hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity and immunotoxicity. The predictive models are based on data from both in vitro (e.g., Tox21 assays, Ames bacterial mutation assays, hepG2 cytotoxicity assays and immunotoxicity assays) and in vivo assays (e.g., carcinogenicity, hepatotoxicity).
3.4.3. Toxicological Pathways
Two types of target-pathway-based models are implemented In ProTox-II: (i) Nuclear Receptor Signaling Pathways (7 pathway assays shown in Table 4) and (ii) Stress Response Pathways (5 pathway assays shown in Table 5) [].
This approach is based in the fact that a chemical compound can activate or inhibit a receptor or an enzyme when it interacts with them, resulting in a perturbation in diverse biological pathways, thereby disrupting the cellular process and causing cell death. The main purpose of the initiative is to prioritize substances for further in-depth toxicological evaluation as well as to identify some mechanisms for further investigation such as disease-associated pathways. Thus, by applying this computational prediction tool, it is possible to test quickly and efficiently whether certain chemical compounds have the potential to disrupt processes in the human body that may lead to adverse health effects.
4. Conclusions
From the results obtained, it is possible to conclude that the use of a multiclass screening methodology was shown to be effective for the identification of 40 mycotoxins in edible salmon tissues from aquaculture using a homemade database with 233 compounds. Screening selectivity was supported by accurate mass measurements provided by the Q-TOF-MS technique. It is the first time that these 40 mycotoxins have been identified and documented in farmed fish, as they had previously only been found in different cereal samples. The explanation for the appearance of these mycotoxins in farmed fish is the inclusion of cereals with mycotoxins as raw material in the feed during the processing and manufacturing processes, which results in the carryover of the feed to the edible parts of the fish. Furthermore, a metabolite formed through de-epoxidation of DON (DOM-1) was detected in salmon tissues. Therefore, it is necessary to ensure that farmed fish for human consumption is free of contaminants or contains concentrations below the maximum limits established for legislated mycotoxins. In light of these findings, the potential health risk associated with eating mycotoxin-contaminated fish should attract the public’s attention, as these products are an important part of the daily diet in combination with other foods. These results are supported by the fact that some of the detected mycotoxins showed a low LD50 using in silico approaches. However, the next purpose is to confirm these findings achieved through in silico predictions with in vitro techniques to corroborate the results obtained.
Author Contributions
Conceptualization, F.J.B. and E.F.; methodology, E.F.; software, J.T. and N.P.; validation, J.T. and N.P.; formal analysis, J.T. and N.P.; investigation, J.T. and N.P.; resources, J.T. and N.P.; data curation, J.T. and N.P.; writing—original draft preparation, J.T. and N.P.; writing—review and editing, J.T., N.P. and E.F.; visualization, F.J.B.; supervision, E.F. and F.J.B.; project administration, E.F.; funding acquisition, E.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the EU Commission and BBI-JU Horizon H2020, through AQUABIOPRO-FIT project (Aquaculture and agriculture biomass side stream proteins and bioactives for feed, fitness and health promoting nutritional supplements, grant number 790956) and the projects AGL2016/77610/R (Economy and Competitiveness Spanish Ministry) as well as OTR2013-11518INVES/CPI-16-143/APOTIP 2016-A-040 (Universitat de València).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nizza, A.; Piccolo, G. Chemical-nutritional characteristics of diets in aquaculture. Veter. Res. Commun. 2009, 33, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Binder, E.M. Managing the risk of mycotoxins in modern feed production. Anim. Feed. Sci. Technol. 2007, 133, 149–166. [Google Scholar] [CrossRef]
- Binder, E.; Tan, L.; Chin, L.; Handl, J.; Richard, J. Worldwide occurrence of mycotoxins in commodities, feeds and feed ingredients. Anim. Feed. Sci. Technol. 2007, 137, 265–282. [Google Scholar] [CrossRef]
- Zinedine, A.; Mañes, J. Occurrence and legislation of mycotoxins in food and feed from Morocco. Food Control. 2009, 20, 334–344. [Google Scholar] [CrossRef]
- Rodrigues, I.; Naehrer, K. A three-year survey on the worldwide occurrence of mycotoxins in feedstuffs and feed. Toxins 2012, 4, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Streit, E.; Schatzmayr, G.; Tassis, P.; Tzika, E.D.; Marin, D.E.; Taranu, I.; Tabuc, C.; Nicolau, A.I.; Aprodu, I.; Puel, O.; et al. Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins 2012, 4, 788–809. [Google Scholar] [CrossRef]
- Streit, E.; Schwab, C.; Sulyok, M.; Naehrer, K.; Krska, R.; Schatzmayr, G. Multi-mycotoxin screening reveals the occurrence of 139 different secondary metabolites in feed and feed ingredients. Toxins 2013, 5, 504–523. [Google Scholar] [CrossRef]
- Nácher-Mestre, J.; Serrano, R.; Beltran, E.; Pérez-Sánchez, J.; Silva, J.; Karalazos, V.; Hernández, F.; Berntssen, M. Occurrence and potential transfer of mycotoxins in gilthead sea bream and Atlantic salmon by use of novel alternative feed ingredients. Chemosphere 2015, 128, 314–320. [Google Scholar] [CrossRef]
- Tolosa, J.; Font, G.; Mañes, J.; Ferrer, E. Natural occurrence of emerging fusarium mycotoxins in feed and fish from aquaculture. J. Agric. Food Chem. 2014, 62, 12462–12470. [Google Scholar] [CrossRef]
- Nomura, H.; Ogiso, M.; Yamashita, M.; Takaku, H.; Kimura, A.; Chikasou, M.; Nakamura, Y.; Fujii, S.; Watai, M.; Yamada, H. Uptake by dietary exposure and elimination of aflatoxins in muscle and liver of rainbow trout (Oncorhynchus mykiss). J. Agric. Food Chem. 2011, 59, 5150–5158. [Google Scholar] [CrossRef]
- Bernhoft, A.; Høgåsen, H.R.; Rosenlund, G.; Moldal, T.; Grove, S.; Berntssen, M.; Thoresen, S.I.; Alexander, J. Effects of dietary deoxynivalenol or ochratoxin A on performance and selected health indices in Atlantic salmon (Salmo salar). Food Chem. Toxicol. 2018, 121, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Bernhoft, A.; Høgåsen, H.R.; Rosenlund, G.; Ivanova, L.; Berntssen, M.H.G.; Alexander, J.; Eriksen, G.S.; Fæste, C.K. Tissue distribution and elimination of deoxynivalenol and ochratoxin A in dietary-exposed Atlantic salmon (Salmo salar). Food Addit. Contam. Part A 2017, 34, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, T.; Young, J.C.; Boland, G.J.; Scott, P.M. Chemical and biological transformations for detoxification of trichothecene mycotoxins in human and animal food chains: A review. Trends Food Sci. Technol. 2010, 21, 67–76. [Google Scholar] [CrossRef]
- Caruso, D.; Talamond, P.; Moreau, Y. Mycotoxinesetpisciculture: Un risqueoublié? CAH Agric. 2013, 22, 165–173. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2016. Contributing to Food Security and Nutrition for All; FAO: Rome, Italy, 2016; p. 200. ISBN 978-92-5-109185-2. Available online: http://www.fao.org/3/a-i5555e.pdf (accessed on 2 March 2019).
- European Commission. Commission Regulation (EC) No. 1881/2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs. Off. J. Eur. Union 2006, 364, 5–24. [Google Scholar]
- Zhao, Z.; Liu, N.; Yang, L.; Deng, Y.; Wang, J.; Song, S.; Lin, S.; Wu, A.; Zhou, Z.; Hou, J.-F. Multi-mycotoxin analysis of animal feed and animal-derived food using LC–MS/MS system with timed and highly selective reaction monitoring. Anal. Bioanal. Chem. 2015, 407, 7359–7368. [Google Scholar] [CrossRef]
- Abdallah, M.F.; Girgin, G.; Baydar, T.; Krska, R.; Sulyok, M. Occurrence of multiple mycotoxins and other fungal metabolites in animal feed and maize samples from Egypt using LC-MS/MS. J. Sci. Food Agric. 2017, 97, 4419–4428. [Google Scholar] [CrossRef]
- Åberg, A.T.; Solyakov, A.; Bondesson, U. Development and in-house validation of an LC-MS/MS method for the quantification of the mycotoxins deoxynivalenol, zearalenone, T-2 and HT-2 toxin, ochratoxin A and fumonisin B1 and B2 in vegetable animal feed. Food Addit. Contam. Part A 2013, 30, 541–549. [Google Scholar] [CrossRef]
- Mol, H.G.J.; Plaza-Bolanños, P.; Zomer, P.; De Rijk, T.C.; Stolker, A.A.M.; Mulder, P.P.J. Toward a generic extraction method for simultaneous determination of pesticides, mycotoxins, plant toxins, and veterinary drugs in feed and food matrixes. Anal. Chem. 2008, 80, 9450–9459. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [Google Scholar] [CrossRef]
- Quesada, S.P.; Paschoal, J.A.R.; Reyes, F.G.R. A simple method for the determination of fluoroquinolone residues in tilapia (Oreochromis niloticus) and pacu (Piaractus mesopotamicus) employing LC-MS/MS QToF. Food Addit. Contam. Part. A 2013, 30, 813–825. [Google Scholar] [CrossRef]
- Liao, C.-D.; Wong, J.W.; Zhang, K.; Yang, P.; Wittenberg, J.B.; Trucksess, M.W.; Hayward, D.G.; Lee, N.S.; Chang, J.S. Multi-mycotoxin analysis of finished grain and nut products using ultrahigh-performance liquid chromatography and positive electrospray ionization–quadrupole orbital ion trap high-resolution mass spectrometry. J. Agric. Food Chem. 2015, 63, 8314–8332. [Google Scholar] [CrossRef]
- Cirlini, M.; Dall’Asta, C.; Galaverna, G. Hyphenated chromatographic techniques for structural characterization and determination of masked mycotoxins. J. Chromatogr. A 2012, 1255, 145–152. [Google Scholar] [CrossRef]
- Nácher-Mestre, J.; Ibáñez, M.V.; Serrano, R.; Pérez-Sánchez, J.; Hernández, F. Qualitative screening of undesirable compounds from feeds to fish by liquid chromatography coupled to mass spectrometry. J. Agric. Food Chem. 2013, 61, 2077–2087. [Google Scholar] [CrossRef]
- Aresta, A.; Cioffi, N.; Palmisano, F.; Zambonin, C.G. Simultaneous determination of ochratoxin A and cyclopiazonic, mycophenolic, and tenuazonic acids in cornflakes by solid-phase microextraction coupled to high-performance liquid chromatography. J. Agric. Food Chem. 2003, 51, 5232–5237. [Google Scholar] [CrossRef]
- Sulyok, M.; Krska, R.; Schuhmacher, R. Application of an LC–MS/MS based multi-mycotoxin method for the semi-quantitative determination of mycotoxins occurring in different types of food infected by moulds. Food Chem. 2010, 119, 408–416. [Google Scholar] [CrossRef]
- Abdallah, M.F.; Krska, R.; Sulyok, M. Occurrence of ochratoxins, fumonisin B2, aflatoxins (B1and B2), and other secondary fungal metabolites in dried date palm fruits from Egypt: A Mini-Survey. J. Food Sci. 2018, 83, 559–564. [Google Scholar] [CrossRef]
- Rundberget, T.; Wilkins, A.L. Determination of Penicillium mycotoxins in foods and feeds using liquid chromatography-mass spectrometry. J. Chromatogr. A 2002, 964, 189–197. [Google Scholar] [CrossRef]
- Sulyok, M.; Berthiller, F.; Krska, R.; Schuhmacher, R. Development and validation of a liquid chromatography/tandem mass spectrometric method for the determination of 39 mycotoxins in wheat and maize. Rapid Commun. Mass Spectrom. 2006, 20, 2649–2659. [Google Scholar] [CrossRef]
- El-Sayed, Y.S.; Khalil, R.H. Toxicity, biochemical effects and residue of aflatoxin B1 in marine water-reared sea bass (Dicentrarchus labrax L.). Food Chem. Toxicol. 2009, 47, 1606–1609. [Google Scholar] [CrossRef]
- Guan, S.; He, J.; Young, J.C.; Zhu, H.; Li, X.-Z.; Ji, C.; Zhou, T. Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 2009, 290, 290–295. [Google Scholar] [CrossRef]
- Deng, S.X.; Tian, L.X.; Liu, F.J.; Jin, S.J.; Liang, G.Y.; Yang, H.J.; Du, Z.Y.; Liu, Y.J. Toxic effects and residue of aflatoxin B1 in tilapia (Oreochromis niloticus × O. aureus) during long-term dietary exposure. Aquaculture 2010, 307, 233–240. [Google Scholar] [CrossRef]
- Pietsch, C.; Michel, C.; Kersten, S.; Valenta, H.; Dänicke, S.; Schulz, C.; Kloas, W.; Burkhardt-Holm, P. In vivo effects of deoxynivalenol (DON) on innate immune responses of carp (Cyprinus carpio L.). Food Chem. Toxicol. 2014, 68, 44–52. [Google Scholar] [CrossRef]
- Tola, S.; Bureau, D.P.; Hooft, J.M.; Beamish, F.W.; Sulyok, M.; Krska, R.; Encarnação, P.; Petkam, R. Effects of wheat naturally contaminated with fusarium mycotoxins on growth performance and selected health indices of red tilapia (Oreochromis niloticus × O. mossambicus). Toxins 2015, 7, 1929–1944. [Google Scholar] [CrossRef]
- EFSA. Scientific Opinion on the risks for animal and public health related to the presence of T-2 and HT-2 toxin in food and feed. EFSA J. 2011, 9, 2481. Available online: https://efsa.onlinelibrary.wiley.com/doi/epdf/ (accessed on 16 April 2019). [CrossRef]
- Prosperini, A.; Font, G.; Ruiz, M. Interaction effects of Fusarium enniatins (A, A1, B and B1) combinations on in vitro cytotoxicity of Caco-2 cells. Toxicol. Vitr. 2014, 28, 88–94. [Google Scholar] [CrossRef]
- Juan-García, A.; Manyes, L.; Ruiz, M.-J.; Font, G. Involvement of enniatins-induced cytotoxicity in human HepG2 cells. Toxicol. Lett. 2013, 218, 166–173. [Google Scholar] [CrossRef]
- Au, T.K.; Wallace, S.H.; Leung, P.C. The biology of ophiobolins. Life Sci. 2000, 67, 733–742. [Google Scholar] [CrossRef]
- Tolosa, J.; Font, G.; Mañes, J.; Ferrer, E. Mitigation of enniatins in edible fish tissues by thermal processes and identification of degradation products. Food Chem. Toxicol. 2017, 101, 67–74. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
Sample Availability: Samples are available from the authors in the laboratory of food chemistry in the Preventive Medicine Department, Faculty of Pharmacy, University of Valencia. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).