Abstract
The molecular mechanism that contributes to nitrogen source dependent omega-3 polyunsaturated fatty acid (n-3 PUFA) synthesis in marine oleaginous protists Thraustochytriidae sp., was explored in this study. The fatty acid (FA) synthesis was significantly influenced by the supplement of various levels of sodium nitrate (SN) (1–50 mM) or urea (1–50 mM). Compared with SN (50 mM) cultivation, cells from urea (50 mM) cultivation accumulated 1.16-fold more n-3 PUFAs (49.49% docosahexaenoic acid (DHA) (w/w, of total FAs) and 5.28% docosapentaenoic acid (DPA) (w/w, of total FAs)). Strikingly higher quantities of short chain FAs (<18 carbons) (52.22-fold of that in urea cultivation) were produced from SN cultivation. Ten candidate reference genes (RGs) were screened by using four statistical methods (geNorm, NormFinder, Bestkeeper and RefFinder). MFT (Mitochondrial folate transporter) and NUC (Nucleolin) were determined as the stable RGs to normalize the RT-qPCR (real-time quantitative polymerase chain reaction) data of essential genes related to n-3 PUFAs-synthesis. Our results elucidated that the gene transcripts of delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, enoyl-CoA hydratase, fatty acid elongase 3, long-chain fatty acid acyl-CoA ligase, and acetyl-CoA carboxylase were up-regulated under urea cultivation, contributing to the extension and unsaturated bond formation. These findings indicated that regulation of the specific genes through nitrogen source could greatly stimulate n-3 PUFA production in Thraustochytriidae sp.
1. Introduction
Fatty acids (FAs) are essential compounds that play significant roles in cellular structure, energy storage, physiological metabolism as well as genetic regulation [1]. Omega-3 polyunsaturated fatty acids (n-3 PUFAs) [2] have various health benefits such as healthy brain and eye development [3,4], cardiovascular disease prevention [5,6], anti-inflammatory [7], and anti-cancer effects [8]. Currently, marine oily fish is the main source of n-3 PUFAs, which has shown obvious drawbacks, including accumulated contaminants and unstable wild fish stocks, hence alternative sources are required to meet the increasing global market demand [9,10]. Thraustochytrids, heterotrophic eukaryotic marine protists, have attracted increasing attention for their significant lipid production capacities. Being considered as a sustainable alternative source of n-3 PUFAs, Thraustochytrids have advantages of high lipids contents, fast growth rate and easily large-scale fermentation [11]. For instance, maximum lipid yield (% dry weight basis) achieved were 40.5% and 49.4%, respectively, in Schizochytrium sp. S31 (ATCC) and Schizochytrium sp. DT3 [12]. Different fermentation strategies have been employed to improve lipid yield and FA profile, such as optimization in salinity, pH, dissolved oxygen, temperature, carbon source and nitrogen source [13,14]. Several desaturases and elongases participated in FAS pathway and synthase involved in PKS pathway have been identified [15].
Nitrogen is an indispensable nutrient affecting microalgal growth and lipid production [16,17,18]. Previous research has shown that nitrogen starvation can effectively increase lipid content in Thraustochytrids. Aurantiochytrium sp. strain T66, which accumulated the highest lipid content of 63% (w/w) of dry cell weight under the condition of nitrogen limitation combined with O2 limitation [19]. Ren, Sun, Zhuang, Qu, Ji and Huang [14] reported that in the fermentation of Schizochytrium sp. ABC101, limited nitrogen feeding increased lipid yield by 1.5-fold. With sodium nitrate addition, Aurantiochytrium sp. KRS101 cultivated in orange peel extract medium achieved 50.23% total fatty acid (TFA, % DCW) and 28.51% docosahexaenoic acid (DHA) [20]. When using urea as a nitrogen source, Aurantiochytrium sp. SD116 accumulated 71.09% TFA and 34.79% DHA [21]. However, the molecular mechanisms underlying the effect of nitrogen source on lipids production in remains unclear.
The reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) is a widely used technique for the determination of gene [22]. Reference genes (RGs) are of great importance to normalize data and assure accurate results of RT-qPCR. For each experimental condition, it is essential to validate the expression stability of RGs to avoid false results and/or erroneous interpretations [23]. To date, research about RGs in Thraustochytrids is limited. 18S rRNA was used as an internal RG of Aurantiochytrium sp. under low temperature conditions [24]. Chen et al. [25] also used 18S rRNA as an internal standard in Schizochytrium sp. S056 when glycerol is used as a carbon source in fermentation. ACT (actin) was set as a RG to validate genes involved in biosynthetic pathways of docosahexaenoic acid (DHA) and the ketocarotenoid astaxanthin in Aurantiochytrium sp. SK4 [26]. However, previous publications have indicated that housekeeping genes are not universally suitable RGs for various microorganisms and experimental conditions [27,28,29]. Currently the suitable RGs in Thraustochytriidae sp. PKU#Mn16 under nitrogen stress is lacking.
In this study, we investigated the effects of different nitrogen sources (e.g., sodium nitrate or urea) on the lipid production in Thraustochytriidae sp. PKU#Mn16. To further explore the molecular mechanism that contributes to nitrogen source dependent n-3 PUFAs synthesis in Thraustochytriidae sp. PKU#Mn16, several crucial genes (delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, enoyl-CoA hydratase, fatty acid elongase 3, long-chain fatty acid acyl-CoA ligase, acetyl-CoA carboxylase and nitrate reductase) identified in our previous transcriptome studies were quantitated by using RT-qPCR. Due to the lack of available RGs for abovementioned experimental conditions, novel candidate genes were selected and evaluated to obtain the most suitable RGs for gene expression in Thraustochytrids. This study provides a better understanding of underlying mechanisms of the biosynthesis of n-3 PUFAs in Thraustochytrids cultivated under different nitrogen sources.
2. Results
2.1. Effects of Sodium Nitrate and Urea on the Growth of Thraustochytriidae sp. PKU#Mn16
To investigate the effects of different nitrogen sources on the growth of Thraustochytriidae sp. PKU#Mn16, inorganic nitrogen source sodium nitrate (SN) and organic nitrogen source urea were chosen as the sole nitrogen source, respectively. Seven concentrations of N in the medium (1, 5, 10, 15, 20, 30, and 50 mM) were tested for Thraustochytriidae sp. PKU#Mn16 flask cultivations. As shown in Figure 1a, the growth trends of Thraustochytriidae sp. PKU#Mn16 were generally similar to each other for all the tested SN concentrations. Cell growth was relatively slow within the first 20 h, then entered the logarithmic phase with the time to peak biomass of 68 h, after which cell growth started to decline. At 68 h, the highest peak biomass (dry cell weight (DCW), 1.01 g/L) was observed in 30 mM SN cultivation, followed by 20 mM (1.00 g/L), 5 mM (0.99 g/L), 15 mM (0.95 g/L), 10 mM (0.93 g/L), 50 mM (0.86 g/L), and the lowest peak biomass occurred in 1 mM cultivation (0.63 g/L). Comparing to the sodium nitrate cultivation, the growth trends varied in different concentration of urea (Figure 1b). The highest peak biomass occurred at 68 h in cultivation (0.93 g/L) supplemented with 50 mM urea, followed by 30 mM (0.71 g/L). In cultivation with 15 mM and 20 mM urea, the time to peak biomass (at 0.44 g/L and 0.53 g/L) was 92 h, while in those with 5 mM and 10 mM urea the time to peak biomass (at 0.23 g/L and 0.33 g/L) was 116 h. The differential impact of SN and urea on biomass was also reported in previously published literatures as follows [20,30].
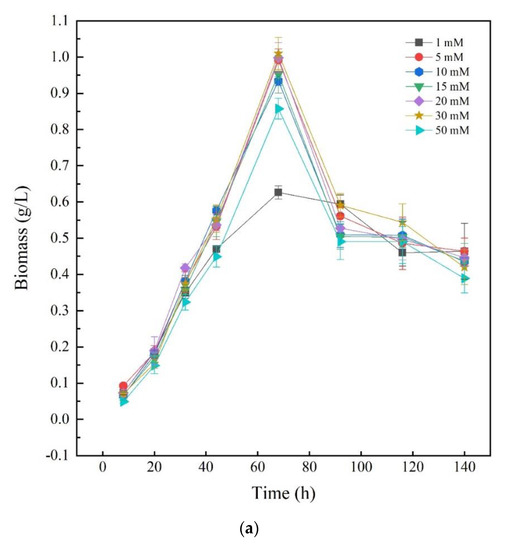
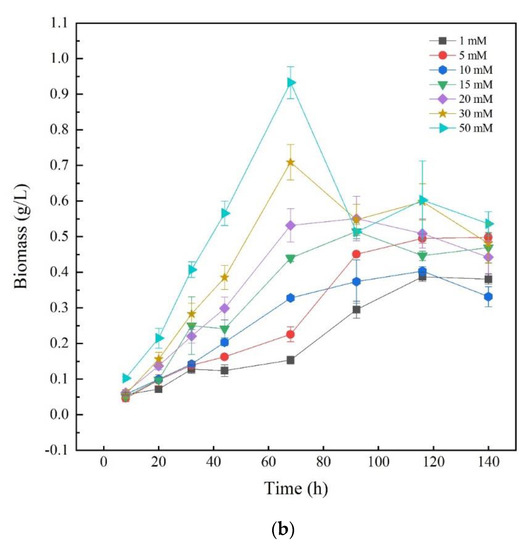
Figure 1.
Growth of Thraustochytriidae sp. PKU#Mn16 cultivated with sodium nitrate (a) or urea (b) as nitrogen sources. All tests were performed in biological triplicate. Values were means ± standard deviation.
In the large-scale cultivation of filamentous cyanobacteria Anabaena sp. PCC 7120, sodium nitrate showed advantages over other nitrogen sources with 65% more growth [30]. Park et al. reported that Thraustochytrid Aurantiochytrium sp. KRS101 showed better biomass when cultivated in orange peel extract supplemented with sodium nitrate as a nitrogen source than with urea [20]. Since similar peak biomass values were obtained at 68 h in two cultivations supplemented with 50 mM SN or urea, 50 mM was chosen for further study to investigate how different nitrogen sources regulate the FA synthesis.
2.2. Effects of Sodium Nitrate and Urea on Fatty Acid Contents and Composition in Thraustochytriidae sp. PKU#Mn16
To examine the effects of sodium nitrate and urea on fatty acid contents and composition for Thraustochytriidae sp. PKU#Mn16, 500 mg cells (dry cell weight, DCW) that cultivated in 50 mM sodium nitrate or 50 mM urea for 68 h were collected for lipids extraction. As shown in Figure 2, the total lipids extracted from SN cultivation accounted for 66.27% of DCW, which is 21.93% more than from urea cultivation (44.34% of DCW). The total lipids extracted mainly comprised six types of fatty acids, namely tetradecanoic acid (TDA, C14:0), hexadecanoic acid (HAD, C16:0), octadecanoic acid (ODA, C18:0), eicosapentaenoic acid (EPA, C20:5), docosapentaenoic acid (DPA, C22:5), and docosahexaenoic acid (DHA, C22:6) (Figure 3). The three most abundant fatty acids under sodium nitrate cultivation were HAD, DHA and DPA, whereas those under urea cultivation were DHA, ODA and DPA. It is noticeable that cells grown in the presence of urea tended to produce more n-3 PUFAs. For cells supplemented with urea, DHA was detected as the predominant composition of total fatty acids (TFAs, (49.49%)), which was 1.11-fold of that from SN cultivation (44.51%). Besides, the percentage of DPA was 5.82%, about two times of that in SN incubation (2.99%). EPA accounted for 0.37% of TFAs, also slightly higher than that derived from SN cultivation. Interestingly, a significantly high proportion (40.83%) of ODA was also observed from urea cultivation, while there was just 0.60% in their counterparts. The ratio of TDA was similar between these two kinds of samples, at 1.21% and 0.96% respectively. For cells from SN cultivation, the largest proportion of TFAs was HDA, at 48.92%, while HDA was barely detected in cells from urea cultivation. Overall, using sodium nitrate as nitrogen source benefits the production of TFAs, while urea as nitrogen source showed more advantage for n-3 PUFA accumulation.
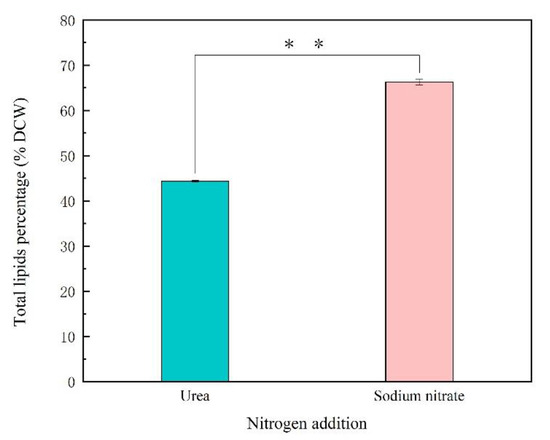
Figure 2.
The total lipids content of Thraustochytriidae sp. PKU#Mn16 in 50 mM urea or 50 mM sodium nitrate cultivation. DCW represents dry cell weight. All tests were performed in biological triplicate. Values were means ± standard deviation. Statistically significant differences (p-value < 0.05 *) were tested by Student’s t-test, p * * <0.01.
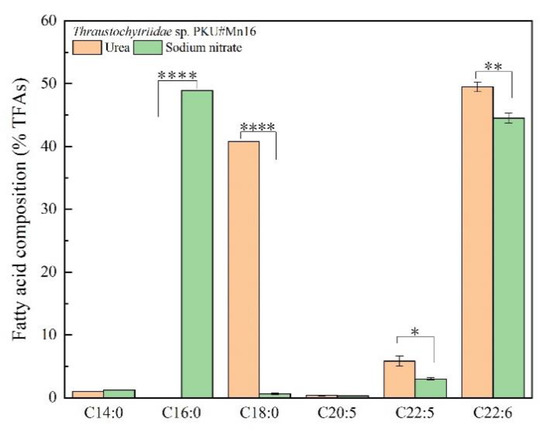
Figure 3.
Fatty acid profile of Thraustochytriidae sp. PKU#Mn16 in 50 mM urea or 50 mM sodium nitrate cultivation. The name of fatty acids was shown in total carbon numbers: C14:0 = tetradecanoic acid (TDA), C16:0 = hexadecanoic acid (HDA), C18:0 = octadecanoic acid (ODA), C20:5 = eicosapentaenoic acid (EPA), C22:5 = docosapentaenoic acid (DPA), C22:6 = docosahexaenoic acid (DHA). All tests were performed in biological triplicate. Data were shown as weigh percentage of TFAs. Values were means ± standard deviation. Statistically significant differences (p-value < 0.5 *) were tested by Student’s t-test, p * <0.05, p **<0.01, p **** <0.0001.
2.3. Screen of the Reference Genes for RT-qPCR
To explore the molecular mechanism of how nitrogen sources affect lipids production in Thraustochytriidae sp. PKU#Mn16, the expression levels of key genes involved in the fatty acid metabolism pathway and nitrate metabolism pathway were quantified by RT-qPCR. However, due to the lack of available RGs for RT-qPCR normalization, it is necessary to screen novel RGs for reliable quantification. Ten putative RGs (CHIA, RPPK, NUC, MFT, CAMK1, HSF, VPS, SAC, RBATP and HTATP) were selected from the RNA-seq dataset (Table 1). Cycle threshold (Ct) value, the number of cycles that the fluorescence signal reaches the set threshold level in every reaction, is used to determine the expression levels of each gene. The higher the Ct value, the lower gene expression level, and vice versa. The mean Ct values of ten candidate RGs ranged from 24 to 35. The RBATP displayed the maximal Ct value of 24, while HTATP showed the minimal Ct value of 35. The Ct values of the other RGs mainly fell between 27–34. To identify optimal RGs for Thraustochytriidae sp. PKU#Mn16 under sodium nitrate or urea cultivation, four statistical methods (geNorm, NormFinder, BestKeeper and RefFinder) were employed to evaluate and rank the stability of their expression level (Table 2).

Table 1.
Candidate reference genes and primer sets for RT-qPCR (real-time quantitative polymerase chain reaction).

Table 2.
Ranking of the expression stability of 10 candidate reference genes (ranking order: Better-Good-Average).
GeNorm analysis showed that among ten selected RGs, VPS had the lowest expression stability (M) value of 0.127, which was regarded as the best RG under the experimental condition when sodium nitrate and urea used as nitrogen sources. On the contrary, HTATP displayed the biggest M value of 0.94 and considered as the least stable RG in the same experimental treatment. Besides, the pairwise variation V2/3 value was 0.112 (Figure 4), which suggested that the optimal number of RG for accurate normalization was two. According to the stability value measured by NormFinder, MFT, CHIA and NUC were the best three stably expressed RGs, with stability values of 0.238, 0.315 and 0.351 respectively. The least stable RG determined with NormFinder was the same as by geNorm—HTATP, with a stability value of 1.168. The result of BestKeeper analysis showed that the most stable RG was HTATP, followed by SAC, RPPK and CHIA. VPS appeared to have the least stable expression in this algorithm. RefFinder integrates four computational programs (geNorm, NormFinder, BestKeeper and delta Ct) to compare and rank the candidate RGs. The comprehensive ranking recommended by RefFinder from the highest to the lowest stability was as follow: MFT > NUC > CHIA > HSF > VPS > RPPK > CAMK > HTATP > SAC > RBATP (Table 2). According to the comprehensive ranking of expression stability, MFT and NUC were the best two RGs and used to normalize the expression of target genes in Thraustochytriidae sp. PKU#Mn16 under the experimental condition of sodium nitrate and urea as the respective nitrogen source.
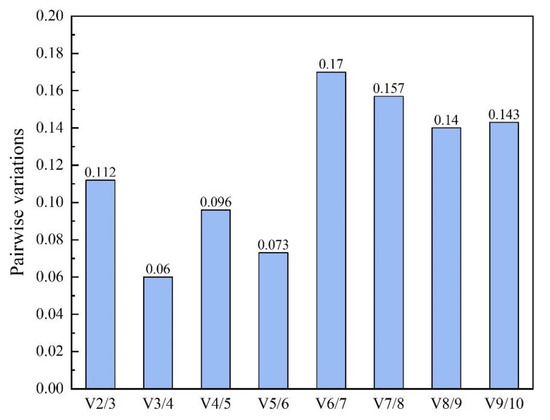
Figure 4.
Determination of the optimal number of reference genes for normalization. Pairwise variation (V) of the candidate reference genes calculated by geNorm. To obtain reliable normalization, Vn/Vn + 1 should be smaller than a threshold of 0.15, otherwise, another reference gene (n + 1) should be added.
2.4. Effect of Various Nitrogen Sources on the Expression of the Key FA Synthesis Genes of Thraustochytriidae sp. PKU#Mn16
Six genes that play important roles in fatty acid biosynthesis and metabolism were chosen to explore the mechanism on how the different nitrogen source influence the production of the fatty acids. Similar expression profiles were obtained with using either MFT or NUC as a reference gene (Figure 5), which confirmed that the methods to quantify gene expression were reliable in the present study.
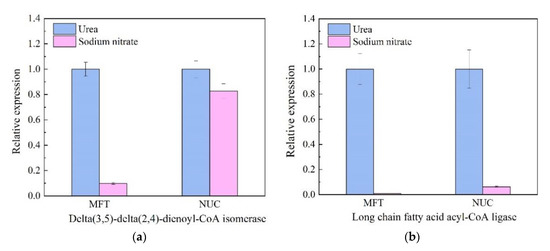
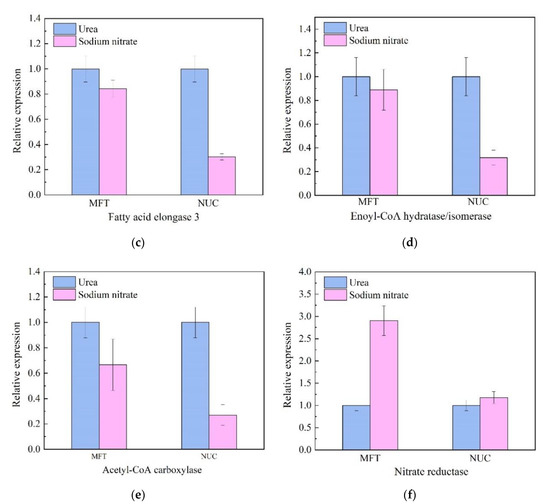
Figure 5.
Relative expression levels of key genes involved in nitrogen metabolism and fatty acid biosynthesis using the best stable reference genes (MFT and NUC). Two best stable reference genes, MFT (mitochondrial folate transporter) and NUC (nucleolin) were selected from Thraustochytriidae sp. PKU#Mn16 transcriptome data and identified by four statistical methods (geNorm, NormFinder, BestKeeper and RefFinder) for stability. Relative quantification of six lipids related genes (delta(3,5)-delta(2,4)-dienoyl-CoA isomerase (a), long-chain fatty acid acyl-CoA ligase (b), fatty acid elongase 3 (c), enoyl-CoA hydratase/isomerase (d), Acetyl-CoA carboxylase (e) and nitrate reductase (f)) were normalized by reference genes MFT and NUC. The average Ct value was calculated from three biological and technical replicates and used for relative quantification of genes expression using 2−ΔΔCT method. Values were means ± standard deviation.
Nitrate reductase gene (NR) encoding enzymes that catalyzed the first step in nitrogen assimilation pathway [31] was down-regulated in urea cultivation, as the normalized NR transcripts against MFT and NUC were 34% and 85% of those from SN cultivation respectively. However, five genes (delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, enoyl-CoA hydratase, fatty acid elongase 3, long-chain fatty acid acyl-CoA ligase and acetyl-CoA carboxylase) involved in lipids metabolism were all up-regulated in urea cultivation. Delta(3,5)-delta(2,4)-dienoyl-CoA isomerase gene (DCI) played a significant role in the beta-oxidation of PUFA [13]. It was up-regulated 10.27-fold when using MFT as an RG, while up-regulated 1.20-fold using NUC. Long chain fatty acid acyl-CoA ligase gene (LCFAAC) participated in the fatty acid biosynthesis pathway [32]. The relative expression level of LCFAAC varied significantly between urea cultivation and SN cultivation, with the prominent up-regulation by urea observed (113.82-fold (MFT as RG) and 15.95-fold (NUC as RG), respectively). Fatty acid elongase 3 gene (FAE3) was an indispensable gene in fatty acid elongation [33]. When normalized against MFT, FAE3 was slightly up-regulated (1.22-fold). Enoyl-CoA hydratase gene (ECH) encoded enzyme which catalyzed a critical step of fatty acid metabolism [34]. It was up-regulated by 3.14-fold with NUC normalization. In addition, upregulation by urea was observed for acetyl-CoA carboxylase gene (ACC) (1.50-fold with MFT as RG), which encoded a key regulator in fatty acid biosynthesis. As shown in Figure 3, cells grown in urea medium accumulated more DHA, DPA, EPA and ODA than those in sodium nitrate. These five genes played significant roles in the biosynthesis of these accumulated lipids and were presumably responsible for producing significant quantities of n-3 PUFAs in Thraustochytriidae sp. PKU#Mn16.
3. Discussion
3.1. Essential Genes Involved in FAS and PKS Pathways of Fatty Acid Production
The biosynthesis of fatty acids in microorganism is commonly achieved by a cyclic reaction pathway by stepwise, iterative elongation with acetyl-CoA and malonyl-CoA as precursors. Fatty acid synthase (FAS) pathway mainly utilizes the large multifunctional enzyme--fatty acid synthase to catalyzes the fatty acids biosynthesis [35]. FAS harbors four catalytic domains for a respective reaction as follow: condensation, ketoacyl reduction, hydroxyacyl dehydration, and enoyl reduction [36]. In FAS pathway, saturated acid C16:0 (HDA) was formed after the cyclic FAS-catalyzed reactions using acetyl-CoA and malonyl-CoA as precursors. C16:0 was converted to C18:0 (ODA) by elongase. Then C18:0 was catalyzed by a series of desaturases (delta 9-desaturase, delta 12-desaturase, delta 15-desaturase delta 6-desaturase, delta 5-desaturase and delta 4-desaturase) and elongases to finally synthesize n-3 PUFAs, such as C20:5 (EPA), C22:5 (DPA) and C22:6 (DHA) [37,38].
Cells grown in sodium nitrate medium produced more C16:0, while those grown in urea medium produced less C16:0 but enormous C18:0. In addition, higher contents of C20:5 (EPA), C22:5 (DPA) and C22:6 (DHA) were also observed in cell cultivated in urea medium. Fatty acid elongase 3 (FAE 3) was detected in Thraustochytriidae sp. PKU#Mn16 and was up-regulated under urea cultivation. This indicates that under urea cultivation, a higher level of FAE gene expression was achieved, leading to more conversion of C16:0 to C18:0 and subsequently higher n-3 PUFA production by FAEs. It was reported that overexpressing the endogenous fatty acid elongase genes in Thalassiosira pseudonana led to the improvement of LC-PUFA production, with a 4.5-fold increase in DHA levels [39]. Hamilton et al. also found that overexpression of delta-5 elongase in diatom Phaeodactylum tricornutum led to an 8.0-fold increase in DHA [40]. Acetyl-CoA carboxylase (ACC) catalyzes the rate-limiting step of the FAS pathway, converting acetyl-CoA to malonyl-CoA [41,42]. In this study, the gene expression level of ACC was also up-regulated under urea cultivation, which might correspond to the higher PUFAs accumulation. It was reported that the overexpression of ACC significantly increased the conversion of acetyl-CoA to malonyl-CoA in Aspergillus terreus [43]. The ACC containing mutant S1157A resulted in a 3-fold increase in both polyketide and fatty acid production in yeast [44]. The Enoyl-CoA hydratase (ECH) is a homohexamer which belongs to a low sequence similarity family of CoA-binding proteins that share a hydratase/isomerase sequence motif [45]. It participated in the fatty acid metabolism process which generated acyl-CoA molecules [46]. The abundant expression of ECH in Thraustochytriidae sp. PKU#Mn16 under urea cultivation might translate to the generation of adequate precursors for downstream PUFA production. Long-chain fatty acid acyl-CoA ligase (LCFAAC) is an important enzyme that converts free fatty acids into fatty acyl-CoA esters, which are crucial intermediates for complex lipid biosynthesis [47]. The high expression level of LCFAAC in cells under urea cultivation might also lead to the high PUFAs production in Thraustochytriidae sp. PKU#Mn16.
The polyketide synthase (PKS) pathway synthesized lipids through repetitive cycles of four principal reactions: condensation by ketoacyl synthase (KS), ketoreduction by ketoreductase (KR), dehydration by dehydratase (DH), and enoyl reduction by enoyl reductase (ER). An isomerase was in charge of the conformation of the fatty acid chain [48]. However, this pathway was reported to be lack of complete enzymes in Thraustochytrids [49]. In our genome sequencing annotation of Thraustochytrium sp. SZU445 [13], KS, KR and ER was detected, but DH and isomerase were not identified. However, delta(3,5)-delta(2,4)-dienoyl-CoA isomerase (DCI) that catalyzed the isomerization of 3-cis-octenoyl-CoA to 2-trans-octenoyl-CoA was detected, and it was regarded as the isoenzyme of DH and isomerase in PKS pathway, which lead to the transpose of the unsaturated carbon bonds to the correct sites. In the urea cultivation, a higher level of DCI gene expression was observed, probably contributing to the formation of the carbon-carbon double bonds in DHA and EPA. Rai et al. found that overexpression of human peroxisomal enoyl-CoA delta isomerase2 HsPECI2 changed the polar lipid content of tobacco. The production of phosphatidylcholine, phosphatidylserine and digalactosyldiacylglycerol were moderately upregulated [50].
3.2. Effects of Nitrate Reductase on Nitrogen Assimilation and Lipids Production
Nitrogen is one of the essential components in living organisms due to its participation in the construction of biomolecules such as proteins and nucleic acids [51]. Previous studies have shown that Thraustochytrids harbor the ability to utilized nitrate and a variety of nitrogen-containing compounds as nitrogen sources (Table 3). The preferable and non-preferable nitrogen source for Thraustochytrids is species specific. However, most of the previous study mainly focused on the fermentation optimization by using various nitrogen sources, the molecular mechanism that contributes to the nitrogen source dependent PUFA production was seldom explored.

Table 3.
Effects of different nitrogen sources on lipids production of Thraustochytrids.
In the nitrate assimilation pathway, nitrate reductase (NR) plays an important role and can be found in bacteria, microalgae, fungi and plants [51,54,55]. NR is a cytosolic enzyme that catalyzes the very first step in nitrogen assimilation, that is, to convert nitrate (NO3−) to nitrite (NO2−). The nitrite is then catalyzed by nitrite reductase to ammonium (NH4+) [56]. It has been demonstrated that the activities of NR are high in nitrate cultivated cells and low when ammonium used as a nitrogen source to cultivate cells [57]. Urea is an economical friendly organic nitrogen source. In the urea metabolism, urease, one of the extracellular enzymes of Thraustochytrids [58], catalyzes the conversion of urea to carbon dioxide and ammonia. Interestingly, in the present study the gene expression of NR was down-regulated in Thraustochytriidae sp. PKU#Mn16 under urea cultivation compared to that under sodium nitrate cultivation, which was consistent with NR activities. In the study of Benhima et al., sodium tungstate was used to inhibit NR in microalgae Dunaliella tertiolecta, resulting in a 50% increase of neutral lipids. Besides, fatty acid methyl esters composition showed a slight variation of polyunsaturation and elongation [59]. McCarthy et al. found that the knockout (KO) of NR gene impacted the lipids metabolism in model pennate diatom Phaeodactylum tricornutum. The concentration of triacylglycerol was increased in NR-KO cells, consistent with up-regulated key genes of triacylglycerol biosynthesis [56]. Consistent with published works, our results showed sodium nitrate induced a relatively high expression level of NR, and urea down-regulated NR expression resulting in more long chain PUFAs.
4. Materials and Methods
4.1. Microorganism Cultivation
Thraustochytriidae sp. PKU#Mn16 was preserved in China General Microbiological Culture Center (CGMCC) under the accession nos. 8095. Modified artificial seawater [60] (0.05 g/L KH2PO4, 0.6 g/L KCl, 2.44 g/L MgSO4, 0.3 g/L CaCl2.2H2O, 1 g/L Tris-HCl (pH 8.0), 10 mL/L PI metal, 3 mL/L chelated iron solution, 18 g/L NaCl and 20 g/L glucose dissolved in deionized water) was used to prepare nitrogen addition medium. 2 M sodium nitrate solution and urea solution were prepared and added into the modified artificial seawater respectively to make a series of concentrations of nitrogen medium: 1 mM, 5 mM, 10 mM, 15 mM, 20 mM, 30 mM and 50 mM. Cultures were then incubated on an orbital shaker (LYZ-123CD, Longyue Company, Shanghai, China) at 23 °C, 200 rpm for 140 h. Three parallel cell samples were collected from each medium at regular intervals to analyze biomass and generate growth curve. Cells were washed three times with deionized water and centrifugated (Z366K, HERMLE, Wehingen, Germany) at 25 °C, 7871× g for 6 min, then cell pellets were lyophilized in a freeze dryer (Triad 2.51, Labconco, Kansas City, MO, USA) to calculate dry cell weight (DCW).
4.2. Lipids Extraction and Fatty Acid Composition Analysis
Lipids of three biological replicates were extracted by the modified Bligh and Dyer procedure [61]. 500 mg freeze-dried cells were extracted with a solvent mixture of chloroform/methanol (1:2, v/v) in a Soxhlet extractor at 70 °C for 96 h. Crude total lipids were gained after the solvent mixture was evaporated. Fatty acid methyl esters (FAMEs) were prepared by a direct acid-catalyzed transesterification [62,63]. Crude lipids were added to 4 mL 4% sulfuric acid in methanol and incubated at 70 °C for 1 h. Subsequently, FAMEs were extracted in 2 mL n-hexane and 2 mL deionized water. After the upper organic layer was transferred to a new tube and dried with nitrogen stream, 1 mL dichloromethane was added and the whole solution was transferred to a chromatography bottle, ready for gas chromatography-mass spectrometry (GC-MS, 7890-5975 Agilent, Santa Clara, CA, USA) analysis.
In the GC-MS analysis, GC column used for the determination of FAMEs is HP-5MS (19091S-433, 30.0 m × 250 μm, I.D. × 0.25 μm film thickness, Agilent J&W) with a maximum temperature of 350 °C. An aliquot of 1 μL of each sample was injected into the GC column. The inlet temperature of GC was set to 250 °C. High purity Helium was used as the carrier gas. Constant pressure mode was used, and the split ratio was 10:1. The temperature program was set as follows: first, the temperature is raised from 60 °C at a rate of 25 °C/min to 180 °C, then the temperature was raised to 240 °C at a rate of 3 °C/min, holding at 240 °C for 1 min, and then raised to 250 °C at a rate of 5 °C/min. The GC-MS transfer line temperature was set to 250 °C, and the full scan mode was employed for the GC-MS detection. A 37-component fatty acid standard mixture was used to validate the temperature program that separated sequentially the peaks of 37 fatty acids. Nonadecanoic acid was used as an internal standard and the content of the fatty acids was quantified by comparing the peak areas of that of the internal standards. Three biological replicates were examined.
4.3. RNA Extraction and cDNA Synthesis
Cells were collected (7871× g for 6 min) after cultivation for 68 h. Cell pellets were then washed three times with distilled water and subsequently frozen at −80 °C for 2 h. The frozen cell pellets were ground in liquid nitrogen to fine powder then 100 mg powder was transferred into a clean 1.5 mL centrifuge tube. Subsequently 1 mL TRIzol (Life Technologies, Carlsbad, CA, USA) reagent was added into the tube and set in room temperature (RT) for 30 min, after that 0.2 mL chloroform was added and the mixture was vortexed for 30 s, rest in RT for 5 min then centrifuged at 17,709× g for 15 min. The upper layer was transferred to a new centrifuge tube and was added 0.5 mL isopropanol and placed in RT for 10 min. Total RNA precipitate was obtained after centrifugation at 17,709× g for 15 min at 4 °C, and then it was washed twice with 75% ethanol and dissolved with DNase/RNase free water. The integrity of RNA was checked by 1.5% (w/v) agarose gel electrophoresis. The quality and concentration of RNA were determined using a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Total RNA with 260/280 nm ratio of 1.9–2.2 and 260/230 nm ratio greater than 1.9 was used to synthesize cDNA with PrimerScriptTM RT reagent Kit (Takara, Japan) according to the manufacturer’s guidelines. The cDNA was stored at −20 °C for later use.
4.4. Selection of Candidate Reference Genes from RNA-Seq Data
Candidate reference genes (RGs) were selected from the RNA-seq dataset of Thraustochytriidae sp. PKU#Mn16 under nitrogen addition cultivation [15]. The coefficient of variation (CV) of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) was calculated across all the treatments and genes with lower CV values were considered stably expressed. Candidate RGs were chosen under the following requirements that the CV of FPKM was less than 10% and annotation of genes was adequate.
4.5. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
The primers used for real-time quantitative PCR (RT-qPCR) were designed according to transcriptomic sequence data using Primer Premier 5 software. Primers were synthesized with the following parameters: Primer length of 18–25 bp, product lengths of 80–200 bp, Tm values of 50–65 °C and GC content of 45–55%. RT-qPCR reactions were performed in the QuantStudio 6 Flex Real-Time PCR System (ABI, Vernon, CA, USA). The SYBR TaqTM ExPremix (Tli RNaseH Plus) Kit (Takara, Shiga, Japan) was used with the following cycling conditions: initial denaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The programs SDS v2.4 (ABI, Vernon, CA, USA) were used for monitoring the qPCR reactions and for analyzing primer efficiencies, R2 of efficiencies and gene expression level. To confirm primer specificity, only melting curves of amplicons presenting single peaks were selected for further analysis. Relative gene expression was quantified using the 2−△△CT method [64]. Three biological and three technical replicates were performed in all RT-qPCR analysis.
4.6. Assessment of Gene Expression Stability
Four statistical programs, geNorm, NormFinder, BestKeeper and RefFinder, were used to assess the stability of the RGs across all experimental conditions. The geNorm program calculates the gene expression stability (M) value for each gene. Genes with an M value less than 1.5 are considered to be stably expressed, the lower M value the more stable expression. In addition, this program also calculates a pairwise variation (Vn/n + 1) value between genes. The value of “n” is the optimal number of RGs when the pairwise value of variation (Vn/n + 1) is below a cut-off value of 0.15. The NormFinder program evaluates the stability of candidate RGs by an assessment of within and between-group variations. A lower variation value indicates a better reference gene. The BestKeeper program examines the coefficient of variation (CV) and standard deviation (SD) based on the raw Ct values of all candidate RGs. Any gene with an SD value lower than 1 was considered as a gene with stable expression. The most stable RG recommended by BestKeeper program is the one with both the lowest CV and SD values. The Delta Ct approach compares the difference in Ct values of RG pairwise and ranks the candidate RGs using the variability of averaged SD. RefFinder (https://www.heartcure.com.au/reffinder/) integrates the four algorithms (geNorm, NormFind, BestKeeper and delta Ct) methods. The stability of candidate RGs was comprehensively validated and ranked by calculating the geometric mean of their weights for the overall final ranking.
4.7. Validation of Reference Genes
Six genes (delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, enoyl-CoA hydratase, fatty acid elongase 3, long chain fatty acid acyl-CoA ligase, acetyl-CoA carboxylase and nitrate reductase ) which was annotated in RNA-seq of Thraustochytriidae sp. PKU#Mn16 as responsible genes for lipid metabolism were used to verify the stability of selected RGs. The top two best RGs were used to normalize the expression of target genes in the present study. The RT-qPCR thermocycling protocol was the same as described above.
4.8. Statistical Analysis
Statistical analysis of the experimental data was performed by using Students’s t-test in the GraphPad Prism 8.0.1 software, and the statistical difference amongst groups was determined by the p value at p *< 0.05, p * * < 0.01, p * ** < 0.001, p * ** * < 0.0001 [65].
5. Conclusions
In the present study, sodium nitrate as a nitrogen source is beneficial to the short-chain fatty acids’ accumulation in Thraustochytriidae sp. PKU#Mn16, while urea as nitrogen source has more advantage for n-3 PUFA biosynthesis. Out of ten reference genes, two genes (MFT and NUC) showed higher expression stability and were chosen for RT-qPCR normalization. Five essential genes related to the long-chain fatty acid synthesis encompassing (delta(3,5)-delta(2,4)-dienoyl-CoA isomerase, enoyl-CoA hydratase, fatty acid elongase 3, long-chain fatty acid acyl-CoA ligase and acetyl-CoA carboxylase) were up-regulated in the cultivation using urea as the nitrogen source, while nitrate reductase was up-regulated in sodium nitrate cultivation. These findings shed light on the underlying molecular mechanisms of n-3 PUFA production in Thraustochytrids and revealed the key genes which could be exploited in the improvement of n-3 PUFA production.
Author Contributions
S.L. and X.Y. conceived the study and designed the experiments. S.L. performed the experimental work. S.L., X.Y. and Y.L. analyzed the experimental data. S.L. and X.Y. drafted the manuscript. S.L., X.Y. and Y.L. contributed to the manuscript revision. Z.H. supervised the overall work. Y.L. and X.Y. supervised the student. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by [National Key Research and Development Project] grant number [2018YFA0902500]. The [Natural Science Foundation of Guangdong Province] grant number [2018A030313139], [Joint R&D Project of Shenzhen-Hong Kong Innovation] grant number [SGLH20180622152010394], [Natural Science Foundation of Shenzhen] grant number [KQJSCX20180328093806045], [Shenzhen Grant Plan for Science &Technology] grant number [CKCY2016042710211071] and [Shenzhen University Start-up Research Funding] grant number [827-000192, 2016100]. Siting Li is a recipient of the AUT-Shenzhen University Ph.D. Tuition Fees Award from the School of Science.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Keum, Y.-S. Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance—A review. Life Sci. 2018, 203, 255–267. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.S.; Sampson, E.L. Fatty acid and fatty aldehyde composition of the major brain lipids in normal human gray matter, white matter, and myelin. J. Lipid Res. 1965, 6, 545–551. [Google Scholar] [PubMed]
- Guesnet, P.; Alessandri, J.-M. Docosahexaenoic acid (DHA) and the developing central nervous system (CNS) —Implications for dietary recommendations. Biochimie 2011, 93, 7–12. [Google Scholar] [CrossRef]
- Bernasconi, A.A.; Wiest, M.M.; Lavie, C.J.; Milani, R.V.; Laukkanen, J.A. Effect of Omega-3 Dosage on Cardiovascular Outcomes: An Updated Meta-Analysis and Meta-Regression of Interventional Trials. Mayo Clin. Proc. 2020. [Google Scholar] [CrossRef]
- Abdelhamid, A.S.; Brown, T.J.; Brainard, J.S.; Biswas, P.; Thorpe, G.C.; Moore, H.J.; Deane, K.H.; AlAbdulghafoor, F.K.; Summerbell, C.D.; Worthington, H.V.; et al. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018, 11, Cd003177. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 45, 1105–1115. [Google Scholar] [CrossRef]
- Gillet, L.; Roger, S.; Bougnoux, P.; Le Guennec, J.-Y.; Besson, P. Beneficial effects of omega-3 long-chain fatty acids in breast cancer and cardiovascular diseases: Voltage-gated sodium channels as a common feature? Biochimie 2011, 93, 4–6. [Google Scholar] [CrossRef]
- Ward, O.P.; Singh, A. Omega-3/6 fatty acids: Alternative sources of production. Process Biochem. 2005, 40, 3627–3652. [Google Scholar] [CrossRef]
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Schenk, P.M. Towards sustainable sources for omega-3 fatty acids production. Curr. Opin. Biotechnol. 2014, 26, 14–18. [Google Scholar] [CrossRef]
- Yokochi, T.; Honda, D.; Higashihara, T.; Nakahara, T. Optimization of docosahexaenoic acid production by Schizochytrium limacinum SR21. Appl. Microbiol. Biotechnol. 1998, 49, 72–76. [Google Scholar] [CrossRef]
- Byreddy, A.R.; Barrow, C.J.; Puri, M. Bead milling for lipid recovery from thraustochytrid cells and selective hydrolysis of Schizochytrium DT3 oil using lipase. Bioresour. Technol. 2016, 200, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, S.; Liu, L.; Li, S.; Luo, Y.; Lv, C.; Wang, B.; Cheng, C.H.; Chen, H.; Yang, X.J.M.D. Genome Sequencing and Analysis of Thraustochytriidae sp. SZU445 Provides Novel Insights into the Polyunsaturated Fatty Acid Biosynthesis Pathway. Mar. Drugs 2020, 18, 118. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.-J.; Sun, L.-N.; Zhuang, X.-Y.; Qu, L.; Ji, X.-J.; Huang, H. Regulation of docosahexaenoic acid production by Schizochytrium sp.: Effect of nitrogen addition. Bioprocess Biosyst. Eng. 2014, 37, 865–872. [Google Scholar] [CrossRef]
- Yang, X.; Li, S.; Li, S.; Liu, L.; Hu, Z. De Novo Transcriptome Analysis of Polyunsaturated Fatty Acid Metabolism in Marine Protist Thraustochytriidae sp. PKU#Mn16. J. Am. Oil Chem. Soc. 2019, 97, 35–48. [Google Scholar] [CrossRef]
- Shen, X.F.; Chu, F.F.; Lam, P.K.; Zeng, R.J. Biosynthesis of high yield fatty acids from Chlorella vulgaris NIES-227 under nitrogen starvation stress during heterotrophic cultivation. Water Res 2015, 81, 294–300. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Nitrogen starvation-induced cellular crosstalk of ROS-scavenging antioxidants and phytohormone enhanced the biofuel potential of green microalga Acutodesmus dimorphus. Biotechnol. Biofuels 2017, 10, 60. [Google Scholar] [CrossRef]
- Chen, B.; Wan, C.; Mehmood, M.A.; Chang, J.-S.; Bai, F.; Zhao, X. Manipulating environmental stresses and stress tolerance of microalgae for enhanced production of lipids and value-added products–A review. Bioresour. Technol. 2017, 244, 1198–1206. [Google Scholar] [CrossRef]
- Jakobsen, A.N.; Aasen, I.M.; Josefsen, K.D.; Strom, A.R. Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp. strain T66: Effects of N and P starvation and O2 limitation. Appl. Microbiol. Biotechnol. 2008, 80, 297–306. [Google Scholar] [CrossRef]
- Park, W.-K.; Moon, M.; Shin, S.-E.; Cho, J.M.; Suh, W.I.; Chang, Y.K.; Lee, B. Economical DHA (Docosahexaenoic acid) production from Aurantiochytrium sp. KRS101 using orange peel extract and low cost nitrogen sources. Algal Res. 2018, 29, 71–79. [Google Scholar] [CrossRef]
- Gao, M.; Song, X.; Feng, Y.; Li, W.; Cui, Q. Isolation and characterization of Aurantiochytrium species: High docosahexaenoic acid (DHA) production by the newly isolated microalga, Aurantiochytrium sp. SD116. J. Oleo Sci. 2013, 62, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Paiva-Cavalcanti, M.; Regis-da-Silva, C.G.; Gomes, Y.M. Comparison of real-time PCR and conventional PCR for detection of Leishmania (Leishmania) infantum infection: A mini-review. J. Venom. Anim. Toxins Incl. Trop. Dis. 2010, 16, 537–542. [Google Scholar] [CrossRef]
- Bin, S.; Pu, X.; Shu, B.; Kang, C.; Luo, S.; Tang, Y.; Wu, Z.; Lin, J. Selection of Reference Genes for Optimal Normalization of Quantitative Real-Time Polymerase Chain Reaction Results for Diaphorina citri Adults. J. Econ. Entomol. 2018, 355. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Tan, Y.; Cui, G.; Feng, Y.; Cui, Q.; Song, X. Transcriptome and gene expression analysis of DHA producer Aurantiochytrium under low temperature conditions. Sci. Rep. 2015, 5, 14446. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, P.-p.; Zhang, M.; Zhu, Y.-m.; Wang, X.-p.; Luo, X.-a.; Bao, Z.-d.; Yu, L.-j. Transcriptome analysis reveals that up-regulation of the fatty acid synthase gene promotes the accumulation of docosahexaenoic acid in Schizochytrium sp. S056 when glycerol is used. Algal Res. 2016, 15, 83–92. [Google Scholar] [CrossRef]
- Ye, J.; Liu, M.; He, M.; Ye, Y.; Huang, J. Illustrating and Enhancing the Biosynthesis of Astaxanthin and Docosahexaenoic Acid in Aurantiochytrium sp. SK4. Mar Drugs 2019, 17. [Google Scholar] [CrossRef]
- Sagri, E.; Koskinioti, P.; Gregoriou, M.E.; Tsoumani, K.T.; Bassiakos, Y.C.; Mathiopoulos, K.D. Housekeeping in Tephritid insects: The best gene choice for expression analyses in the medfly and the olive fly. Sci. Rep. 2017, 7, 45634. [Google Scholar] [CrossRef]
- Martins, P.K.; Mafra, V.; de Souza, W.R.; Ribeiro, A.P.; Vinecky, F.; Basso, M.F.; da Cunha, B.A.; Kobayashi, A.K.; Molinari, H.B. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 2016, 6, 28348. [Google Scholar] [CrossRef]
- Pinto, F.; Pacheco, C.C.; Ferreira, D.; Moradas-Ferreira, P.; Tamagnini, P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE 2012, 7, e34983. [Google Scholar] [CrossRef]
- Johnson, T.J.; Jahandideh, A.; Isaac, I.C.; Baldwin, E.L.; Muthukumarappan, K.; Zhou, R.; Gibbons, W.R. Determining the optimal nitrogen source for large-scale cultivation of filamentous cyanobacteria. J. Appl. Phycol. 2016, 29, 1–13. [Google Scholar] [CrossRef]
- Arora, V.; Ghosh, M.K.; Singh, P.; Gangopadhyay, G. Light regulation of Nitrate reductase gene expression and enzyme activity in the leaves of mulberry. Indian J. Biochem. Biophys. 2018, 55, 62–66. [Google Scholar]
- Murphy, M.G.; Spence, M.W. Long-chain fatty acid:CoA ligase in rat brain in vitro: A comparison of activities with oleic and cis-vaccenic acids. J. Neurochem. 1980, 34, 367–373. [Google Scholar] [CrossRef]
- Inagaki, K.; Aki, T.; Fukuda, Y.; Kawamoto, S.; Shigeta, S.; Ono, K.; Suzuki, O. Identification and expression of a rat fatty acid elongase involved in the biosynthesis of C18 fatty acids. Biosci. Biotechnol. Biochem. 2002, 66, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, G.; Liu, H.-w. Enoyl-CoA hydratase. Bioorg. Med. Chem. 2003, 11, 9–20. [Google Scholar] [CrossRef]
- Maier, T.; Leibundgut, M.; Boehringer, D.; Ban, N. Structure and function of eukaryotic fatty acid synthases. Q. Rev. Biophys. 2010, 43, 373–422. [Google Scholar] [CrossRef] [PubMed]
- Schonauer, M.S.; Kastaniotis, A.J.; Hiltunen, J.K.; Dieckmann, C.L. Intersection of RNA Processing and the Type II Fatty Acid Synthesis Pathway in Yeast Mitochondria. Mol. Cell. Biol. 2008, 28, 6646. [Google Scholar] [CrossRef]
- Ohara, J.; Sakaguchi, K.; Okita, Y.; Okino, N.; Ito, M.J.M.b. Two fatty acid elongases possessing C18-Δ6/C18-Δ9/C20-Δ5 or C16-Δ9 elongase activity in Thraustochytrium sp. ATCC 26185. Mar. Biotechnol. 2013, 15, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Heggeset, T.M.B.; Ertesvag, H.; Liu, B.; Ellingsen, T.E.; Vadstein, O.; Aasen, I.M. Lipid and DHA-production in Aurantiochytrium sp.—Responses to nitrogen starvation and oxygen limitation revealed by analyses of production kinetics and global transcriptomes. Sci. Rep. 2019, 9, 19470. [Google Scholar] [CrossRef]
- Cook, O.; Hildebrand, M. Enhancing LC-PUFA production in Thalassiosira pseudonana by overexpressing the endogenous fatty acid elongase genes. J. Appl. Phycol. 2015, 28, 897–905. [Google Scholar] [CrossRef]
- Hamilton, M.L.; Haslam, R.P.; Napier, J.A.; Sayanova, O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 2014, 22, 3–9. [Google Scholar] [CrossRef]
- Khozin-Goldberg, I.; Cohen, Z. Unraveling algal lipid metabolism: Recent advances in gene identification. Biochimie 2011, 93, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hunkeler, M.; Hagmann, A.; Stuttfeld, E.; Chami, M.; Guri, Y.; Stahlberg, H.; Maier, T. Structural basis for regulation of human acetyl-CoA carboxylase. Nature 2018, 558, 470–474. [Google Scholar] [CrossRef]
- Hasan, H.; Abd Rahim, M.H.; Campbell, L.; Carter, D.; Abbas, A.; Montoya, A. Overexpression of acetyl-CoA carboxylase in Aspergillus terreus to increase lovastatin production. New Biotechnol. 2018, 44, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Da Silva, N.A. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J. Biotechnol. 2014, 187, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.K.; Mathieu, M.; Zeelen, J.P.; Hiltunen, J.K.; Wierenga, R.K. Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: A spiral fold defines the CoA-binding pocket. EMBO J. 1996, 15, 5135–5145. [Google Scholar] [CrossRef] [PubMed]
- Goepfert, S.; Hiltunen, J.K.; Poirier, Y. Identification and functional characterization of a monofunctional peroxisomal enoyl-CoA hydratase 2 that participates in the degradation of even cis-unsaturated fatty acids in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 35894–35903. [Google Scholar] [CrossRef]
- Cao, Y.; Traer, E.; Zimmerman, G.A.; McIntyre, T.M.; Prescott, S.M. Cloning, expression, and chromosomal localization of human long-chain fatty acid-CoA ligase 4 (FACL4). Genomics 1998, 49, 327–330. [Google Scholar] [CrossRef]
- Ratledge, C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef]
- Morabito, C.; Bournaud, C.; Maes, C.; Schuler, M.; Aiese Cigliano, R.; Dellero, Y.; Marechal, E.; Amato, A.; Rebeille, F. The lipid metabolism in thraustochytrids. Prog. Lipid Res. 2019, 76, 101007. [Google Scholar] [CrossRef]
- Rai, V.; Sarkar, S.; Satpati, S.; Dey, N. Overexpression of human peroxisomal enoyl-CoA delta isomerase2 HsPECI2, an ortholog of bamboo expressed during gregarious flowering alters salinity stress responses and polar lipid content in tobacco. Funct. Plant Biol. 2016, 43, 232–243. [Google Scholar] [CrossRef]
- González, P.J.; Correia, C.; Moura, I.; Brondino, C.D.; Moura, J.J.G. Bacterial nitrate reductases: Molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 2006, 100, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.D.; Anh, H.T.; Thu, N.T. Study on biological characteristics of heterotrophic marine microalga-Schizochytrium Mangrove PQ6 isolated from Phu Quoc island, Kien Giang Province, Vietnam. J. Phycol. 2011, 47, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Auma, K.; Hamid, A.A.; Yusoff, W.M.W. Effect of nitrogen sources on biomass, lipid and docosahexanoic acid production by Aurantiochytrum sp. SW1. In Proceedings of AIP Conference Proceedings; AIP Publishing: Melville, NY, USA, 1940; p. 020065. [Google Scholar]
- Crawford, N.M.; Arst, H.N., Jr. The molecular genetics of nitrate assimilation in fungi and plants. Annu. Rev. Genet. 1993, 27, 115–146. [Google Scholar] [CrossRef]
- Sanz-Luque, E.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 2015, 6, 899. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.K.; Smith, S.R.; McCrow, J.P.; Tan, M.; Zheng, H.; Beeri, K.; Roth, R.; Lichtle, C.; Goodenough, U.; Bowler, C.P.; et al. Nitrate Reductase Knockout Uncouples Nitrate Transport from Nitrate Assimilation and Drives Repartitioning of Carbon Flux in a Model Pennate Diatom. Plant Cell 2017, 29, 2047–2070. [Google Scholar] [CrossRef] [PubMed]
- Syrett, P.; Leftley, J. Nitrate and urea assimilation by algae. Perspect. Exp. Biol. 2016, 2, 221–234. [Google Scholar]
- Taoka, Y.; Nagano, N.; Okita, Y.; Izumida, H.; Sugimoto, S.; Hayashi, M. Extracellular enzymes produced by marine eukaryotes, thraustochytrids. Biosci. Biotechnol. Biochem. 2009, 73, 180–182. [Google Scholar] [CrossRef]
- Benhima, R.; El Arroussi, H.; Kadmiri, I.M.; El Mernissi, N.; Wahby, I.; Bennis, I.; Smouni, A.; Bendaou, N. Nitrate Reductase Inhibition Induces Lipid Enhancement of Dunaliella Tertiolecta for Biodiesel Production. Sci. World J. 2018, 2018, 6834725. [Google Scholar] [CrossRef]
- Sun, L.; Ren, L.; Zhuang, X.; Ji, X.; Yan, J.; Huang, H. Differential effects of nutrient limitations on biochemical constituents and docosahexaenoic acid production of Schizochytrium sp. Bioresour. Technol. 2014, 159, 199–206. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Liu, Y.; Singh, P.; Sun, Y.; Luan, S.; Wang, G. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl. Microbiol. Biotechnol. 2014, 98, 3241–3255. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhang, J.; Zhao, J.; Gao, Z.; Zhang, C.; Chen, M. Regulation of lipid accumulation in Schizochytrium sp. ATCC 20888 in response to different nitrogen sources. Eur. J. Lipid Sci. Technol. 2017, 119. [Google Scholar] [CrossRef]
- Carvalho, D.M.; de Sa, P.H.; Castro, T.L.; Carvalho, R.D.; Pinto, A.; Gil, D.J.; Bagano, P.; Bastos, B.; Costa, L.F.; Meyer, R.; et al. Reference genes for RT-qPCR studies in Corynebacterium pseudotuberculosis identified through analysis of RNA-seq data. Antonie Van Leeuwenhoek 2014, 106, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Irvine, N.A.; Ruyter, B.; Ostbye, T.K.; Sonesson, A.K.; Lillycrop, K.A.; Berge, G.; Burdge, G.C. Dietary Fish Oil Alters DNA Methylation of Genes Involved in Polyunsaturated Fatty Acid Biosynthesis in Muscle and Liver of Atlantic Salmon (Salmo salar). Lipids 2019, 54, 725–739. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).