Antibacterial Cyclic Tripeptides from Antarctica-Sponge-Derived Fungus Aspergillus insulicola HDN151418
Abstract
1. Introduction
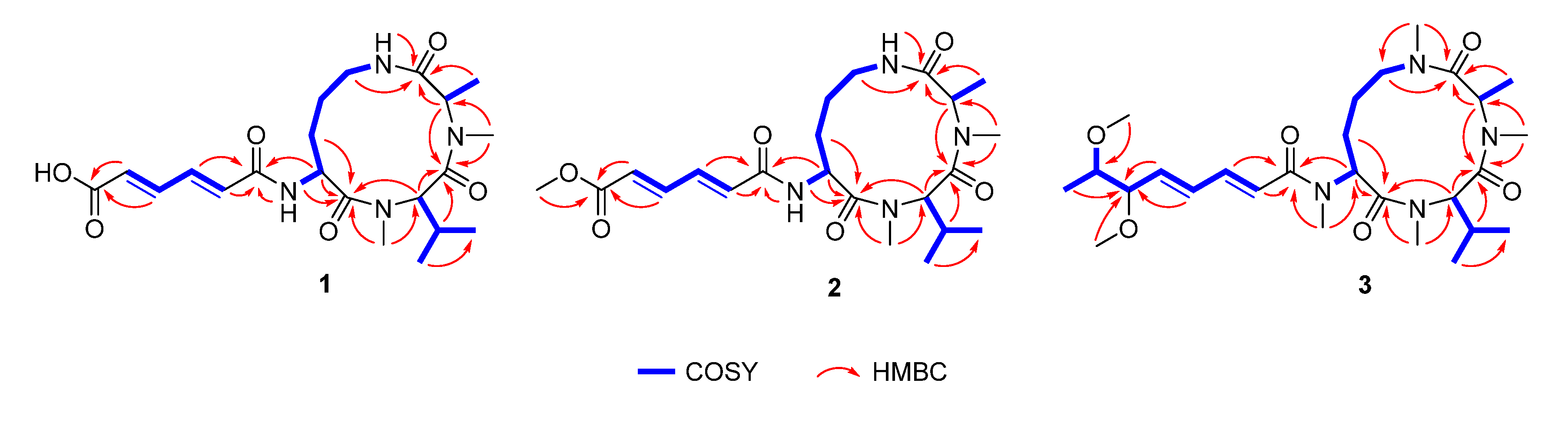
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material and Fermentation
3.3. Isolation and Purification of the Compounds
3.4. Absolute Configuration Assignments of Sclerotiotides M–N (1–2).
3.5. Assay of Cytotoxicity Inhibitory Activity
3.6. Assay of Antimicrobial Activity
3.7. Preparation of MPA Esters Derived from 4 (4g and 4h)
3.8. Chemical Transformation of 4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Velle, A.; Cebollada, A.; Macias, R.; Iglesias, M.; Gil-Moles, M.; Sanz Miguel, P.J. From imidazole toward imidazolium salts and N-heterocyclic carbene ligands: Electronic and geometrical redistribution. ACS Omega 2017, 2, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liao, H.; Liu, L.Y.; Sun, F.; Chen, H.F.; Jiao, W.H.; Zhu, H.R.; Yang, F.; Huang, G.; Zeng, D.Q.; et al. Phakefustatins A-C: Kynurenine-bearing cycloheptapeptides as RXRalpha modulators from the marine sponge Phakellia Fusca. Org. Lett. 2020, 22, 6703–6708. [Google Scholar] [CrossRef]
- Xu, W.J.; Liao, X.J.; Xu, S.H.; Diao, J.Z.; Pan, S.S. Isolation, structure determination, and synthesis of galaxamide, a rare cytotoxic cyclic pentapeptide from a marine algae Galaxaura filamentosa. Org. Lett. 2010, 40, 4569–4572. [Google Scholar] [CrossRef]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an antibiotic lipophilic cyclopeptide from the icelandic thermophilic bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, Z.; Wang, Y.; Hong, K.; Liu, P.; Zhu, W. Cyclic tripeptides from the halotolerant fungus Aspergillus sclerotiorum PT06-1. J. Nat. Prod. 2010, 73, 1133–1137. [Google Scholar] [CrossRef]
- Motohashi, K.; Inaba, S.; Takagi, M.; Shin-ya, K. JBIR-15, a new aspochracin derivative, isolated from a sponge-derived fungus, Aspergillus sclerotiorum Huber Sp080903f04. Biosci. Biotechnol. Biochem. 2009, 73, 1898–1900. [Google Scholar] [CrossRef][Green Version]
- Liu, J.; Gu, B.; Yang, L.; Yang, F.; Lin, H. New Anti-inflammatory cyclopeptides from a sponge-derived fungus Aspergillus violaceofuscus. Front. Chem. 2018, 6, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Myokei, R.; Sakurai, A.; Chang, C.F.; Kodaira, Y.; Tamura, S. Structure of aspochracin, an insecticidal metabolite of Aspergillus ochraceus. Tetrahedron Lett. 1969, 10, 695–698. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Wu, C.; Kurtan, T.; Mandi, A.; Liu, Y.; Gu, Q.; Zhu, T.; Guo, P.; Li, D. Penipyridones A-F, pyridone alkaloids from Penicillium funiculosum. J. Nat. Prod. 2016, 79, 1783–1790. [Google Scholar] [CrossRef]
- Zhou, H.; Li, L.; Wang, W.; Che, Q.; Li, D.; Gu, Q.; Zhu, T. Chrodrimanins I and J from the antarctic moss-derived fungus Penicillium funiculosum GWT2-24. J. Nat. Prod. 2015, 78, 1442–1445. [Google Scholar] [CrossRef]
- Shah, M.; Sun, C.; Sun, Z.; Zhang, G.; Che, Q.; Gu, Q.; Zhu, T.; Li, D. Antibacterial polyketides from antarctica sponge-derived fungus Penicillium sp. HDN151272. Mar. Drugs 2020, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Lin, A.; Gu, Q.; Zhu, T.; Li, D. Four new chloro-eremophilane sesquiterpenes from an Antarctic deep-sea derived fungus, Penicillium sp. PR19N-1. Mar. Drugs 2013, 11, 1399–1408. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591. [Google Scholar] [CrossRef]
- Sun, C.; Ge, X.; Mudassir, S.; Zhou, L.; Yu, G.; Che, Q.; Zhang, G.; Peng, J.; Gu, Q.; Zhu, T.; et al. New glutamine-containing azaphilone alkaloids from deep-sea-derived fungus Chaetomium globosum HDN151398. Mar. Drugs 2019, 17, 253. [Google Scholar] [CrossRef]
- Latypov, S.K.; Seco, J.M.; Quinoa, E.; Riguera, R. MTPA vs MPA in the determination of the absolute configuration of chiral alcohols by 1H NMR. J. Org. Chem. 1996, 61, 8569–8577. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- Yu, G.; Sun, Z.; Peng, J.; Zhu, M.; Che, Q.; Zhang, G.; Zhu, T.; Gu, Q.; Li, D. Secondary metabolites produced by combined culture of Penicillium crustosum and a Xylaria sp. J. Nat. Prod. 2019, 82, 2013–2017. [Google Scholar] [CrossRef]




| No. | 1 a | 2 b | 3 b |
|---|---|---|---|
| 2 | 4.50, q (7.1) | 4.52, q (7.1) | 4.65, q (6.4) |
| 3 | 1.37, d (7.1) | 1.39, d (7.1) | 1.20, d (6.4) |
| NAla-CH3 | 2.83, s | 2.85, s | 2.63, s |
| 5 | 4.97, d (10.4) | 4.99, d (10.3) | 4.94, d (10.6) |
| 6 | 2.21, m | 2.22, m | 2.28, m |
| 7 | 0.63, d (6.7) | 0.64, d (6.7) | 0.74, d (6.7) |
| 8 | 0.79, d (6.4) | 0.80, d (6.3) | 0.81, d (6.2) |
| NVal-CH3 | 2.83, s | 2.85, s | 2.91, s |
| 10 | 4.71, m | 4.73, m | 5.33, t (5.5) |
| NOrn (α)H/ CH3 | 8.39, d (7.6) | 8.46, d (7.5) | 3.08, s |
| 11 | 1.61. m | 1.71, m | 1.70, m |
| 1.96, m | 1.96, m | 1.95, m | |
| 12 | 1.46, m | 1.47, m | 1.48, m |
| 1.67, m | 1.63, m | 1.66, m | |
| 13 | 2.86, m | 2.89, m | 2.97, m |
| 3.01, m | 3.01, m | 3.12, m | |
| NOrn (ω)H/CH3 | 7.47, t (6.0) | 7.51, t (5.7) | 2.83, s |
| 2′ | 6.55, d (14.3) | 6.60, d (14.6) | 6.64, d (15.0) |
| 3′ | 7.12, ov. | 7.17, ov. | 7.11, dd (15.0, 11.2) |
| 4′ | 7.16, ov. | 7.26, ov. | 6.46, dd (15.5, 11.2) |
| 5′ | 6.22, d (14.4) | 6.36, d (14.8) | 6.01, dd (15.5, 7.1) |
| 6′ | 3.71, dd (7.1, 3.9) | ||
| 7′ | 3.69, s | 3.34, m | |
| 8′ | 1.02, d (6.4) | ||
| 9′ | 3.23, s | ||
| 10′ | 3.25, s | ||
| COOH | 12.5, brs |
| No. | 1 a | 2 b | 3 c |
|---|---|---|---|
| 1 | 171.1 | 171.2 | 168.5 |
| 2 | 54.9 | 55.0 | 52.9 |
| 3 | 16.7 | 16.7 | 17.2 |
| NAla-CH3 | 30.1 | 30.1 | 28.7 |
| 4 | 169.5 | 169.6 | 168.2 |
| 5 | 58.0 | 58.1 | 57.6 |
| 6 | 26.8 | 26.9 | 26.3 |
| 7 | 18.1 | 18.2 | 18.1 |
| 8 | 20.2 | 20.3 | 19.9 |
| NVal-CH3 | 30.2 | 30.3 | 29.7 |
| 9 | 171.9 | 172.0 | 171.8 |
| 10 | 50.1 | 50.2 | 52.7 |
| NOrn (α)-CH3 | 31.7 | ||
| 11 | 28.4 | 28.4 | 23.0 |
| 12 | 23.1 | 23.2 | 24.5 |
| 13 | 39.5 | 39.5 | 47.3 |
| NOrn (ω)-CH3 | 33.6 | ||
| 1′ | 163.8 | 163.8 | 166.6 |
| 2′ | 132.3 | 133.0 | 122.3 |
| 3′ | 141.8 | 142.5 | 141.7 |
| 4′ | 136.7 | 136.7 | 131.5 |
| 5′ | 128.2 | 126.8 | 138.8 |
| 6′ | 167.5 | 166.7 | 83.9 |
| 7′ | 52.1 | 78.9 | |
| 8′ | 15.6 | ||
| 9′ | 57.1 | ||
| 10′ | 56.8 |
| No. | B. cereus | P. species | M. phlei | E. tarda | B. subtilis | MRCNS | MRSA | V. parahemolyticus |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.13 | 3.13 | 3.13 | 1.56 | 6.25 | 12.5 | 25.0 | 3.13 |
| 2 | 6.25 | 6.25 | 12.5 | 1.56 | 12.5 | 25.0 | 25.0 | 6.25 |
| 3 | >50.0 | >50.0 | >50.0 | 25.0 | >50.0 | >50.0 | >50.0 | 25.0 |
| 4 | 25.0 | 25.0 | >50.0 | 25.0 | >50.0 | >50.0 | >50.0 | 25.0 |
| 5 | 25.0 | 25.0 | >50.0 | 25.0 | >50.0 | >50.0 | >50.0 | 25.0 |
| 6 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 |
| CIP a | 0.780 | 0.195 | 0.780 | 0.0125 | 0.195 | 25.0 | 25.0 | 0.390 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Zhang, Z.; Ren, Z.; Yu, L.; Zhou, H.; Han, Y.; Shah, M.; Che, Q.; Zhang, G.; Li, D.; et al. Antibacterial Cyclic Tripeptides from Antarctica-Sponge-Derived Fungus Aspergillus insulicola HDN151418. Mar. Drugs 2020, 18, 532. https://doi.org/10.3390/md18110532
Sun C, Zhang Z, Ren Z, Yu L, Zhou H, Han Y, Shah M, Che Q, Zhang G, Li D, et al. Antibacterial Cyclic Tripeptides from Antarctica-Sponge-Derived Fungus Aspergillus insulicola HDN151418. Marine Drugs. 2020; 18(11):532. https://doi.org/10.3390/md18110532
Chicago/Turabian StyleSun, Chunxiao, Ziping Zhang, Zilin Ren, Liu Yu, Huan Zhou, Yaxin Han, Mudassir Shah, Qian Che, Guojian Zhang, Dehai Li, and et al. 2020. "Antibacterial Cyclic Tripeptides from Antarctica-Sponge-Derived Fungus Aspergillus insulicola HDN151418" Marine Drugs 18, no. 11: 532. https://doi.org/10.3390/md18110532
APA StyleSun, C., Zhang, Z., Ren, Z., Yu, L., Zhou, H., Han, Y., Shah, M., Che, Q., Zhang, G., Li, D., & Zhu, T. (2020). Antibacterial Cyclic Tripeptides from Antarctica-Sponge-Derived Fungus Aspergillus insulicola HDN151418. Marine Drugs, 18(11), 532. https://doi.org/10.3390/md18110532







