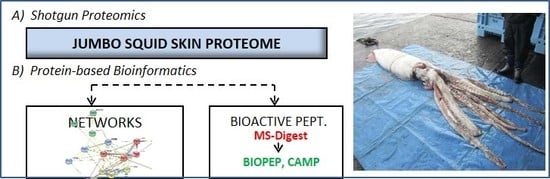
Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics
Abstract
1. Introduction
2. Results and Discussion
2.1. Jumbo Squid (Dosidicus gigas) Skin Proteome
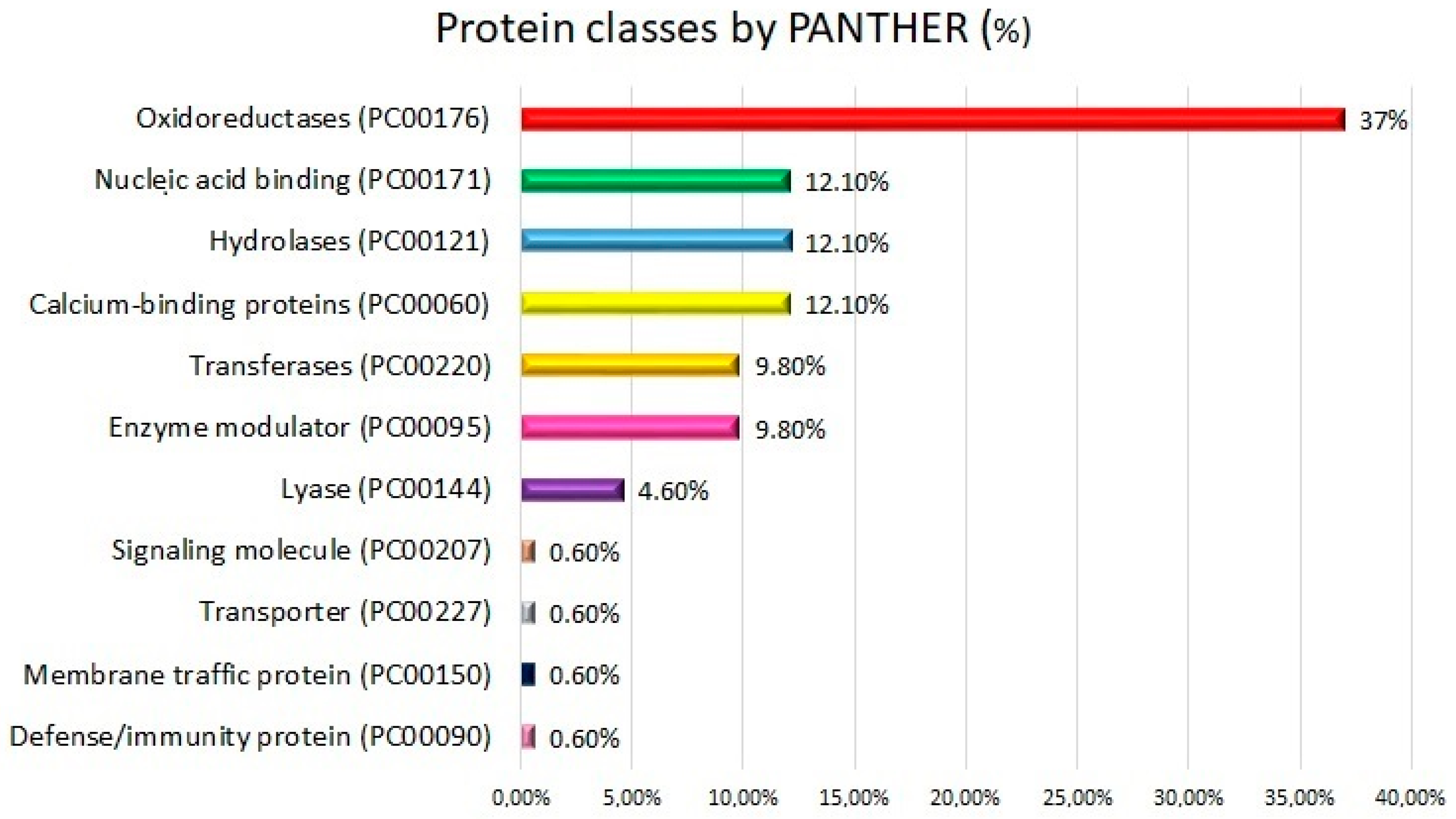
2.2. Functional Analysis: Gene Ontologies and Pathways Analysis
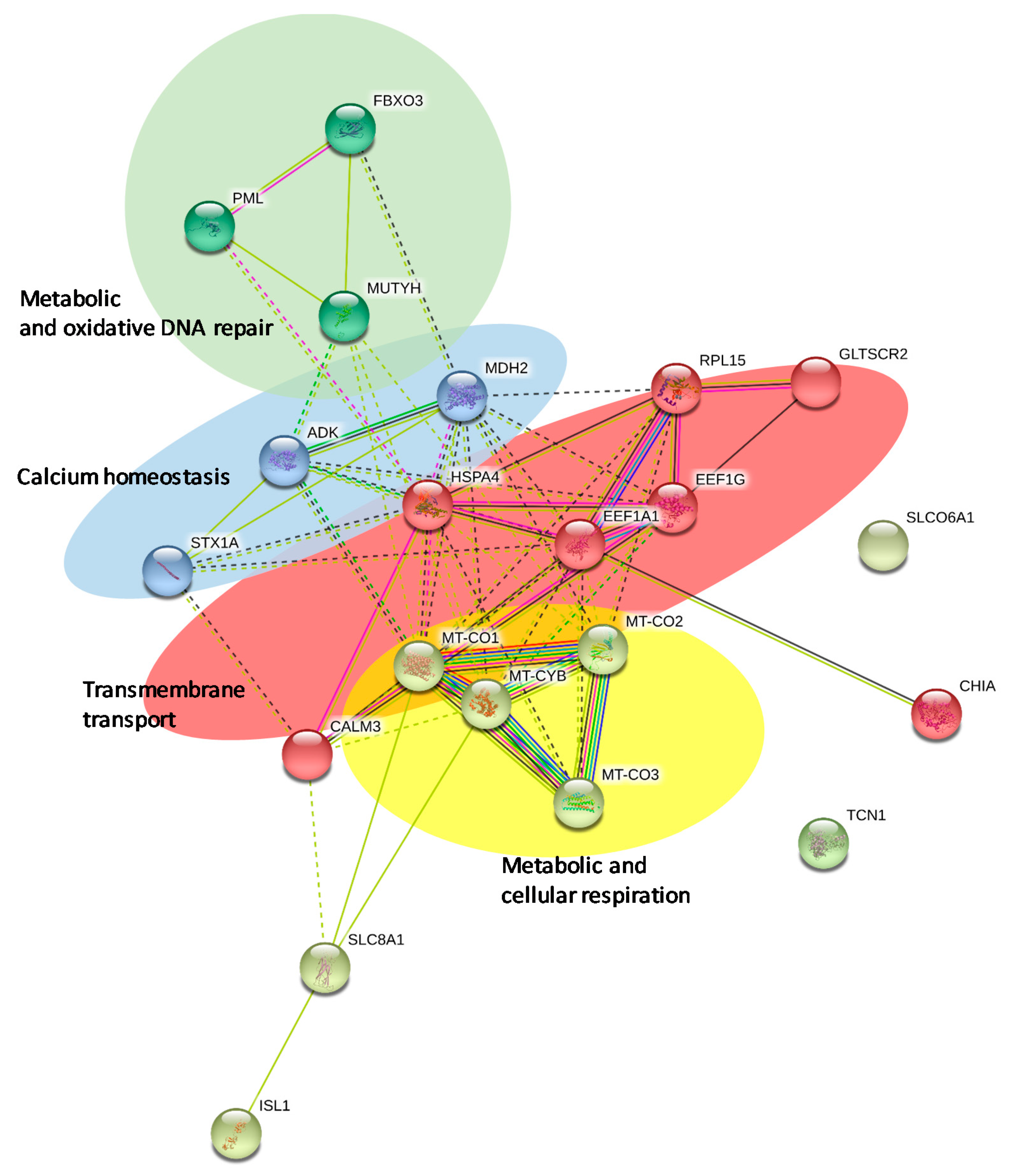
2.3. Network Analysis
2.4. Putative Bioactive Peptides
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Jumbo Squids
3.3. Skin Protein Samples
3.4. SDS-Polyacrylamide Gel Electrophoresis
3.5. In-Solution Protein Digestion with Trypsin
3.6. Shotgun LC-MS/MS Analysis
3.7. Processing of the Mass Spectrometry Data
3.8. Functional Gene Ontologies and Pathways Analysis
3.9. Network Analysis
3.10. Bioactive Peptides Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rustad, T.; Storrø, I.; Slizyte, R. Possibilities for the utilization of marine by-products. Int. J. Food Sci. Technol. 2011, 46, 2001–2014. [Google Scholar] [CrossRef]
- Blanco, M.; Vázquez, J.A.; Pérez-Martín, R.I.; Sotelo, C.G. Hydrolysates of fish skin collagen: An opportunity for valorizing fish industry byproducts. Mar. Drugs 2017, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Regulation (EU) No 1380/2013 of the European Parliament and the Council of 11 December 2013 on the Common Fisheries Policy, Amending Council Regulations (EC) No 1954/2003 and (EC) No 1224/2009 and Repealing Council Regulations (EC) No 2371/2002 and (EC) No 639/2004 and Council Decision 2004/585/EC; European Commission: Brussels, Belgium, 2013. [Google Scholar]
- Carrera, M.; Cañas, B.; Gallardo, J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteomics 2013, 78, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, J.; Anil, S.; Kim, S.K.; Shim, M.S. Marine fish proteins and peptides for cosmeceuticals: A review. Mar. Drugs 2017, 15, 143. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Blanco, M.; Correa, B.; Pérez-Martín, R.I.; Sotelo, C.G. Effect of fish collagen hydrolysates on type I collagen mRNA levels of human dermal fibroblast culture. Mar. Drugs 2018, 16, 144. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Meduíña, A.; Durán, A.I.; Nogueira, M.; Fernández-Compás, A.; Pérez-Martín, R.I.; Rodríguez-Amado, I. Production of valuable compounds and bioactive metabolites from by-products of fish discards using chemical processing, enzymatic hydrolysis, and bacterial fermentation. Mar. Drugs 2019, 17, 139. [Google Scholar] [CrossRef]
- Food and Agriculture Organisation of the United Nations. Global Production Statistics-Fisheries and Aquaculture. Available online: http://www.fao.org/fishery/statistics/global-aqua (accessed on 27 May 2017).
- Ezquerra-Brauer, J.M.; Aubourg, S. Recent trends for the employment of jumbo squid (Dosidicus gigas) by products as a source of bioactive compounds with nutritional, functional and preservative applications: A review. Int. J. Food Sci. Technol. 2019, 54, 987–998. [Google Scholar] [CrossRef]
- Mäthger, L.M.; Denton, E.J.; Marshall, N.J.; Hanlon, R.T. Mechanisms and behavioral functions of structural coloration in cephalopods. J. R. Soc. Interface. 2009, 6, S149–S163. [Google Scholar] [CrossRef]
- Deravi, L.F.; Magyar, A.P.; Sheehy, S.P.; Bell, G.R.; Mäthger, L.M.; Senft, S.L.; Wardill, T.J.; Lane, W.S.; Kuzirian, A.M.; Hanlon, R.T.; et al. The structure-function relationships of a natural nanoscale photonic device in cuttlefish chromatophores. J. R. Soc. Interface 2014, 11, 20130942. [Google Scholar] [CrossRef]
- Aubourg, S.P.; Torres-Arreola, W.; Trigo, M.; Ezquerra-Brauer, J.M. Partial characterization of jumbo squid skin pigment extract and its antioxidant potential in a marine oil system. Eur. J. Lipid Sci. Technol. 2016, 118, 1293–1304. [Google Scholar] [CrossRef]
- Mosquera, M.; Giménez, B.; Montero, P.; Gómez-Guillén, M.C. Incorporation of liposomes containing squid tunic ACE-inhibitory peptides into fish gelatin. J. Sci. Food Agric. 2016, 96, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, S.; Jamili, S.; Ghavam Mostafavi, P.; Rezaie, S.; Khorramizadeh, M. Assessment of the inhibitory effects of ficin-hydrolyzed gelatin derived from squid (Uroteuthis duvauceli) on breast cancer cell lines and animal model. Iran. J. Allergy Asthma Immunol. 2018, 17, 436–452. [Google Scholar] [CrossRef] [PubMed]
- Mendis, E.; Rajapakse, N.; Byun, H.G.; Kim, S.K. Investigation of jumbo squid (Dosidicus gigas) skin gelatin peptides for their in vitro antioxidant effects. Life Sci. 2005, 77, 2166–2178. [Google Scholar] [CrossRef] [PubMed]
- Chan-Higuera, J.E.; Santacruz-Ortega, H.D.C.; Carbonell-Barrachina, A.A.; Burgos-Hernández, A.; Robles-Sánchez, R.M.; Cruz-Ramírez, S.G.; Ezquerra-Brauer, J.M. Xanthommatin is behind the antioxidant activity of the skin of Dosidicus gigas. Molecules 2019, 24, 3420. [Google Scholar] [CrossRef] [PubMed]
- Amado, I.R.; Vázquez, J.A.; Gónzález, P.; Esteban-Fernández, D.; Carrera, M.; Piñeiro, C. Identification of the major ACE-inhibitory peptides produced by enzymatic hydrolysis of a protein concentrate from cuttlefish wastewater. Mar. Drugs 2014, 12, 1390–1405. [Google Scholar] [CrossRef]
- Kumar, P.; Kannan, M.; ArunPrasanna, V.; Vaseeharan, B.; Vijavakumar, S. Proteomic analysis of crude squid ink isolated from Sepia esculenta for their antimicrobial, antibiofilm and cytotoxic properties. Microb. Pathog. 2018, 116, 345–350. [Google Scholar] [CrossRef]
- Ezquerra-Brauer, J.M.; Miranda, J.M.; Cepeda, A.; Barros-Velázquez, J.; Aubourg, S.P. Effect of jumbo squid (Dosidicus gigas) skin extract on the microbial activity in chilled mackerel (Scomber scombrus). LWT-Food Sci. Technol. 2016, 72, 134–140. [Google Scholar] [CrossRef]
- Ezquerra-Brauer, J.M.; Miranda, J.M.; Chan-Higuera, J.E.; Barros-Velázquez, J.; Aubourg, S.P. New icing media for quality enhancement of chilled hake (Merluccius merlucius) using a jumbo squid (Dosidicus gigas) skin extract. J. Sci. Agric. 2017, 97, 3412–3419. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Proteomics for the assessment of quality and safety of fishery products. Food Res. Int. 2013, 54, 972–979. [Google Scholar] [CrossRef]
- Stryiński, R.; Mateos, J.; Pascual, S.; González, A.F.; Gallardo, J.M.; Łopieńska-Biernat, E.; Medina, I.; Carrera, M. Proteome profiling of L3 and L4 Anisakis simplex development stages by TMT-based quantitative proteomics. J. Proteomics 2019, 201, 1–11. [Google Scholar] [CrossRef]
- Gallardo, J.M.; Carrera, M.; Ortea, I. Proteomics in food science. In Foodomics: Advanced Mass Spectrometry in Modern Food Science and Nutrition; Cifuentes, A., Ed.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2013; pp. 125–165. [Google Scholar]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Advanced proteomics and systems biology applied to study food allergy. Curr. Opin. Food Sci. 2018, 22, 9–16. [Google Scholar] [CrossRef]
- Carrera, M.; González-Fernández, A.; Magadán, S.; Mateos, J.; Pedrós, L.; Medina, I.; Gallardo, J.M. Molecular characterization of B-cell epitopes for the major fish allergen, parvalbumin, by shotgun proteomics, protein-based bioinformatics and IgE-reactive approaches. J. Proteomics 2019, 200, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; McCormack, A.L.; Yates, J.R., III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 4, D1087–D1093. [Google Scholar] [CrossRef]
- Iwaniak, A.; Dziuba, J.; Niklewicz, M. The BIOPEP database—A tool for the in silico method of classification of food proteins as the source of peptides with antihypertensive activity. Acta Aliment. Hung. 2005, 34, 417–425. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, Q.; Rui, W.; Lu, M.; Jing, X.; Shang, T.; Tang, J. BioPD: A web-based information center for bioactive peptides. Regul. Pept. 2004, 120, 1–3. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, C.; Chen, H.; Xue, J.; Guo, X.; Liang, M.; Chen, M. BioPepDB: An integrated data platform for food-derived bioactive peptides. Int. J. Food Sci. Nutr. 2018, 69, 963–968. [Google Scholar] [CrossRef]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef] [PubMed]
- Rong, M.; Zhou, B.; Zhou, R.; Liao, Q.; Zeng, Y.; Xu, S.; Liu, Z. PPIP: Automated software for identification of bioactive endogenous peptides. J. Proteome Res. 2019, 18, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Beltran, J.A.; Tellez Ibarra, R.; Guillen-Ramirez, H.A.; Brizuela, C.A. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: Towards an organized collection enabling visual network analysis. Bioinformatics 2019, 35, 4739–4747. [Google Scholar] [CrossRef]
- Wang, J.; Yin, T.; Xiao, X.; He, D.; Xue, Z.; Jiang, X.; Wang, Y. StraPep: A structure database of bioactive peptides. Database (Oxford) 2018. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, Y.; Yoshioka, T.; Kato, S.; Konno, K. Color development of squid skin as affected by oxygen concentrations. J. Food Sci. 2009, 74, S142–S146. [Google Scholar] [CrossRef] [PubMed]
- Celio, M.R.; Heizmann, C.W. Calcium-binding protein parvalbumin is associated with fast contracting muscle fibres. Nature 1982, 297, 504–506. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.J.; Cavallaro, S.; Yi, C.L.; McPhie, D.; Schreurs, B.G.; Gusey, P.A.; Favit, A.; Zohar, O.; Kim, J.; Beushausen, S. Calexcitin: A signaling protein that binds calcium and GTP, inhibits potassium channels, and enhances membrane excitability. Proc. Natl. Acad. Sci. USA 1996, 93, 13808–13813. [Google Scholar] [CrossRef]
- Keil, B. Specificity of proteolysis, 1st ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 1992. [Google Scholar]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Giménez, B.; Gómez-Estaca, J.; Alemán, A.; Gómez-Guillén, M.C.; Montero, P. Physico-chemical and film forming properties of giant squid (Dosidicus gigas) gelatin. Food Hydrocoll. 2009, 23, 585–592. [Google Scholar] [CrossRef]
- Cai, J.; Li, Y.; Zhang, Y.; Tong, Q.; Wang, F.; Su, X. Protective effects of collagen extracted from Dosidicus gigas skin on MC3T3-E1 cell induced by H2O2. J. Chin. Inst. Food Sci. Technol. 2015, 15, 6–12. [Google Scholar]
- Cai, J.; Li, Y.; Quan, J.; Lin, J.; Zhang, Y.; Wang, F.; Su, X. Effect of collagen peptide extracted from Dosidicus gigas skin on proliferation, differentiation and calcification of MC3T3-E1 cell induced by Cd. J. Chin. Inst. Food Sci. Technol. 2015, 15, 18–24. [Google Scholar]
- Liu, S.; Aweya, J.J.; Zheng, L.; Wang, F.; Zheng, Z.; Zhong, M.; Lun, J.; Zhang, Y. A Litopenaeus vannamei hemocyanin-derived antimicrobial peptide (peptide B11) attenuates cancer cells’ proliferation. Molecules 2018, 23, 3202. [Google Scholar] [CrossRef] [PubMed]
- Atala, A. This month in investigative urology. J. Urol. 2006, 176, 2335–2336. [Google Scholar] [CrossRef]
- McFadden, D.W.; Riggs, D.R.; Jackson, B.J.; Vona-Davis, L. Keyhole limpet hemocyanin, a novel immune stimulant with promising anticancer activity in Barrett’s esophageal adenocarcinoma. Am. J. Surg. 2003, 186, 552–555. [Google Scholar] [CrossRef]
- Dipolo, R. Ca pump driven by ATP in squid axons. Nature 1978, 274, 390–392. [Google Scholar] [CrossRef]
- Cheung, H.S.; Wang, F.L.; Ondetti, M.; Sabo, E.; Cushman, D. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme: Importance of the COOH-terminal dipeptide sequences. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar]
- Kasamatsu, C.; Kimura, S.; Kagawa, M.; Hatae, K. Identification of high molecular weight proteins in squid muscle by western blotting analysis and postmortem rheological changes. Biosci. Biotechnol. Biochem. 2004, 68, 1119–1124. [Google Scholar] [CrossRef]
- Chan-Higuera, J.E.; Carbonell-Barrachina, A.A.; Cárdenas-López, J.L.; Kačániová, M.; Burgos-Hernández, A.; Ezquerra-Brauer, J.M. Jumbo squid (Dosidicus gigas) skin pigments: Chemical analysis and evaluation of antimicrobial and antimutagenic potential. J. Microbiol. Biotech. Food Sci. 2019, 9, 349–353. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
| N | Accession | Description | Gene | Uni. Pep. | PSM | Cov. (%) |
|---|---|---|---|---|---|---|
| 1 | A0A1Y1DCG9 | Paramyosin OS = Dosidicus gigas | DgPm | 17 | 46 | 22 |
| 2 | A0A2Z5EQ31 | Symplectin/biotinidase-like protein OS = Dosidicus gigas | sympp | 1 | 2 | 3 |
| 3 | A0A0P0UX03 | Hemocyanin subunit 1 OS = Todarodes pacificus | Tphcy | 116 | 3007 | 38 |
| 4 | A0A077B1P8 | Hemocyanin subunit 2 OS = Euprymna scolopes | HCY2 | 10 | 1608 | 24 |
| 5 | A0A077B6R8 | Hemocyanin subunit 1 OS = Euprymna scolopes | HCY1 | 13 | 1437 | 19 |
| 6 | T2F8L5 | Hemocyanin OS = Sepiella maindroni | HCY1 | 8 | 1544 | 18 |
| 7 | W6CNR9 | Hemocyanin subunit 3 OS = Sepia officinalis | HCY3 | 10 | 1035 | 13 |
| 8 | A0A1Q2SJF4 | Hemocyanin-like protein OS = Uroteuthis edulis | hc | 8 | 746 | 14 |
| 9 | F1ADJ4 | Myosin heavy chain OS = Todarodes pacificus | MYH | 16 | 456 | 15 |
| 10 | I0JGT9 | Actin I OS = Sepia officinalis | ACTI | 11 | 202 | 53 |
| 11 | G4V4Y8 | Myosin heavy chain isoform C OS = Doryteuthis pealeii | MYH | 3 | 411 | 12 |
| 12 | A4D0I0 | Hemocyanin subunit 1 OS = Todarodes pacificus | Tphcy | 6 | 174 | 50 |
| 13 | A0A0P0UX01 | Hemocyanin subunit2 OS = Todarodes pacificus | Tphcy | 4 | 171 | 51 |
| 14 | A0A0L8G4B4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000685mg | 27 | 53 | 13 |
| 15 | V6A729 | Myosin heavy chain isoform A OS = Octopus bimaculoides | MYH | 2 | 348 | 8 |
| 16 | Q2V0V2 | Tropomyosin OS = Todarodes pacificus | tp-tm | 27 | 127 | 46 |
| 17 | A0A0L8GFI1 | Spectrin beta chain OS = Octopus bimaculoides | OCBIM_22034275mg | 24 | 72 | 12 |
| 18 | I7H9I6 | Haemocyanin OS = Nautilus pompilius | hc | 1 | 532 | 5 |
| 19 | A0A075IT96 | Heat shock protein 70 OS = Sepiella maindroni | HSP70 | 3 | 59 | 23 |
| 20 | A0A0L8HMH4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011261mg | 12 | 35 | 3 |
| 21 | E7CLR5 | Hemocyanin (Fragment) OS = Spirula spirula | HCY1 | 1 | 315 | 12 |
| 22 | A0A0L8IA52 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026555mg | 1 | 49 | 18 |
| 23 | A0A0L8GPG8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030693mg | 11 | 59 | 17 |
| 24 | A0A0L8FFZ3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22022789mg | 2 | 394 | 30 |
| 25 | A0A0L8H027 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024964mg | 8 | 48 | 5 |
| 26 | A0A0L8G0V9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003270mg | 6 | 32 | 16 |
| 27 | Q06270 | Intermediate filament protein OS = Nototodarus sloanii | OCBIM_22025455mg | 9 | 38 | 18 |
| 28 | Q76EJ2 | Cathepsin D OS = Todarodes pacificus | tpaD | 9 | 49 | 22 |
| 29 | P08052 | Myosin regulatory light chain LC-2, mantle muscle OS = Todarodes pacificus | MYL | 8 | 23 | 50 |
| 30 | A0A0L8HC80 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017953mg | 8 | 16 | 5 |
| 31 | A0A0L8G3E9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22001601mg | 1 | 31 | 11 |
| 32 | P30842 | Omega-crystallin OS = Nototodarus sloanii | N/A | 5 | 22 | 9 |
| 33 | Q68LN1 | Filamin OS = Euprymna scolopes | OCBIM_22031719mg | 4 | 20 | 34 |
| 34 | A0A0L8FU30 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007941mg | 1 | 12 | 33 |
| 35 | A0A0L8I9I4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028792mg | 1 | 18 | 22 |
| 36 | A0A0L8FNC4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013362mg | 5 | 12 | 4 |
| 37 | Q6E216 | Tropomysin-like protein OS = Todarodes pacificus | ATRP | 5 | 9 | 26 |
| 38 | A0A0L8HDP4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22016840mg | 3 | 27 | 5 |
| 39 | A0A0L8FVD0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007411mg | 4 | 15 | 27 |
| 40 | A0A0L8GWE3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026600mg | 3 | 13 | 12 |
| 41 | A0A0L8HKK9 | Fructose-bisphosphate aldolase OS = Octopus bimaculoides | OCBIM_22013272mg | 3 | 21 | 7 |
| 42 | A0A0L8FP56 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013360mg | 1 | 16 | 3 |
| 43 | G1CW44 | Triosephosphate isomerase OS = Enteroctopus dofleini | OCBIM_22037419mg | 1 | 27 | 11 |
| 44 | G1CW45 | Triosephosphate isomerase OS = Euprymna scolopes | OCBIM_22037419mg | 1 | 8 | 19 |
| 45 | A0A0L8GN79 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030767mg | 2 | 11 | 9 |
| 46 | A0A0L8FZT7 | Protein disulfide-isomerase OS = Octopus bimaculoides | OCBIM_22003356mg | 3 | 17 | 8 |
| 47 | A0A0L8H0K3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024969mg | 3 | 8 | 7 |
| 48 | A0A0L8GNQ0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030666mg | 2 | 5 | 10 |
| 49 | A0A0L8IA72 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025549mg | 4 | 8 | 7 |
| 50 | A0A0L8IAK7 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025100mg | 5 | 9 | 1 |
| 51 | A0A0L8HDG9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017348mg | 3 | 4 | 20 |
| 52 | Q86DP6 | Malate dehydrogenase (Fragment) OS = Sepia officinalis | Mdh | 3 | 7 | 11 |
| 53 | P05945 | Myosin catalytic light chain LC-1, mantle muscle OS = Todarodes pacificus | MYL | 2 | 6 | 19 |
| 54 | A0A0L8GQL2 | Tubulin beta chain OS = Octopus bimaculoides | OCBIM_22029847mg | 3 | 8 | 8 |
| 55 | A0A0L8HMP5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011994mg | 3 | 10 | 16 |
| 56 | A0A0L8IAD9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025091mg | 3 | 3 | 6 |
| 57 | A0A0L8FJA0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017780mg | 2 | 6 | 3 |
| 58 | A0A0L8G425 | Adenosylhomocysteinase OS = Octopus bimaculoides | OCBIM_22000532mg | 3 | 6 | 7 |
| 59 | A0A0L8FXP2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004658mg | 3 | 3 | 5 |
| 60 | A0A0L8I198 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22039192mg | 3 | 8 | 19 |
| 61 | A0A0L8I871 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028797mg | 2 | 7 | 18 |
| 62 | A0A2S1FRU3 | Elongation factor 1-alpha OS = Callistoctopus minor | EEF1A1 | 4 | 6 | 7 |
| 63 | A0A0L8FFD9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22023810mg | 2 | 3 | 2 |
| 64 | A0A0L8I874 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028979mg | 2 | 6 | 16 |
| 65 | A0A0L8FK19 | Tubulin alpha chain OS = Octopus bimaculoides | OCBIM_22016917mg | 2 | 5 | 3 |
| 66 | A0A0K0WTY3 | Arginine kinase OS = Sepia pharaonis | AK | 4 | 7 | 7 |
| 67 | A0A0L8GXA0 | Glucosamine-6-phosphate isomerase OS = Octopus bimaculoides | OCBIM_22026276mg | 1 | 3 | 9 |
| 68 | F8V2T7 | Sodium/potassium-transporting ATPase subunit alpha OS = Bathypolypus arcticus | OCBIM_22028074mg | 2 | 4 | 2 |
| 69 | A0A0L8H4W4 | Proteasome subunit alpha type OS = Octopus bimaculoides | OCBIM_22022293mg | 2 | 3 | 10 |
| 70 | A0A0L8GSZ5 | Histone H4 OS = Octopus bimaculoides | OCBIM_22029078mg | 2 | 5 | 10 |
| 71 | A0A0L8GDJ1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22035502mg | 2 | 4 | 6 |
| 72 | A0A159BRC2 | ColAa OS = Sepia pharaonis | N/A | 2 | 6 | 1 |
| 73 | A0A0L8FIB5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020215mg | 1 | 2 | 5 |
| 74 | A0A0L8G4U5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000359mg | 2 | 4 | 6 |
| 75 | Q9NL93 | G protein a subunit o class OS = Octopus vulgaris | OvGao | 2 | 5 | 6 |
| 76 | A0A0L8IG11 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004528mg | 2 | 8 | 11 |
| 77 | A0A0L8GG89 | Proteasome subunit alpha OS = Octopus bimaculoides | OCBIM_22033871mg | 2 | 3 | 9 |
| 78 | A0A0L8H716 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020867mg | 2 | 12 | 11 |
| 79 | A0A0S1U346 | Triosephosphate isomerase OS = Amphioctopus fangsiao | OCBIM_22037419mg | 1 | 3 | 18 |
| 80 | A0A0L8H4E7 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22022663mg | 2 | 4 | 6 |
| 81 | A0A0L8I919 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22027793mg | 1 | 7 | 5 |
| 82 | A0A0L8HN83 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22010679mg | 1 | 1 | 3 |
| 83 | A0A0L8ICB5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22019476mg | 2 | 4 | 4 |
| 84 | A0A0L8FMD3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22014986mg | 1 | 2 | 4 |
| 85 | A0A0L8H0E1 | Sorting nexin OS = Octopus bimaculoides | OCBIM_22024936mg | 1 | 5 | 3 |
| 86 | A0A0L8IA39 | Tubulin alpha chain OS = Octopus bimaculoides | OCBIM_22026381mg | 1 | 2 | 3 |
| 87 | A0A0L8IG73 | Malic enzyme OS = Octopus bimaculoides | OCBIM_22004207mg | 1 | 1 | 3 |
| 88 | A0A0L8H635 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22021483mg | 1 | 2 | 8 |
| 89 | A0A0L8GYT6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026168mg | 1 | 3 | 10 |
| 90 | A0A0L8GFD5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22034343mg | 1 | 2 | 3 |
| 91 | A0A0L8HKN4 | Ornithine aminotransferase OS = Octopus bimaculoides | OCBIM_22012517mg | 1 | 4 | 3 |
| 92 | A0A0L8G0I6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003454mg | 2 | 2 | 4 |
| 93 | A0A0L8HE61 | AP complex subunit beta OS = Octopus bimaculoides | OCBIM_22016805mg | 1 | 1 | 1 |
| 94 | A0A0L8HMS6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011048mg | 1 | 3 | 3 |
| 95 | A0A0L8FWD6 | Calcium-transporting ATPase OS = Octopus bimaculoides | OCBIM_22006279mg | 2 | 6 | 2 |
| 96 | A0A0L8GP54 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030838mg | 1 | 2 | 7 |
| 97 | A0A0L8G9P1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22037676mg | 1 | 4 | 9 |
| 98 | A0A0L8HTA6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007620mg | 1 | 4 | 6 |
| 99 | A0A0L8IAN9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025097mg | 1 | 1 | 8 |
| 100 | A0A0L8HCU8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22018310mg | 1 | 3 | 2 |
| 101 | A0A0A7NZU2 | Putative chitotriosidase OS = Euprymna scolopes | Chia | 1 | 1 | 4 |
| 102 | A0A0L8G3Z0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000581mg | 1 | 3 | 4 |
| 103 | A0A0L8I836 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028993mg | 1 | 3 | 3 |
| 104 | A0A0L8IDP3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22014847mg | 1 | 1 | 4 |
| 105 | A0A0L8FZ08 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004461mg | 1 | 1 | 1 |
| 106 | A0A0L8GZM9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025211mg | 1 | 4 | 2 |
| 107 | A0A193PD55 | Chitinase OS = Todarodes pacificus | TpChi | 1 | 2 | 2 |
| 108 | Q8IS80 | 60S acidic ribosomal protein OS = Euprymna scolopes | OCBIM_22035130mg | 1 | 3 | 19 |
| 109 | A0A0L8FQ90 | Serine/threonine-protein phosphatase OS = Octopus bimaculoides | OCBIM_22011907mg | 1 | 1 | 4 |
| 110 | A0A0L8FIY8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22018177mg | 1 | 3 | 13 |
| 111 | A0A0L8I107 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22039276mg | 1 | 2 | 4 |
| 112 | A0A0L8G4M6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000216mg | 1 | 2 | 0 |
| 113 | A0A0L8GLC5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22031874mg | 1 | 3 | 8 |
| 114 | A0A0L8HDX1 | Superoxide dismutase OS = Octopus bimaculoides | OCBIM_22016770mg | 1 | 2 | 6 |
| 115 | A0A0L8HU31 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22005978mg | 1 | 2 | 3 |
| 116 | Q8SWQ7 | Non-muscle myosin II heavy chain OS = Doryteuthis pealeii | MYH | 1 | 1 | 1 |
| 117 | B8Q2 × 2 | G alpha q subunit OS = Euprymna scolopes | COI | 1 | 1 | 5 |
| 118 | A0A0L8G1S2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22001882mg | 1 | 1 | 3 |
| 119 | A0A0L8HAV5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22019117mg | 1 | 1 | 7 |
| 120 | A0A0L8IDX1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013485mg | 1 | 4 | 4 |
| 121 | A0A0L8GRX5 | Histone H2B OS = Octopus bimaculoides | OCBIM_22029075mg | 1 | 1 | 6 |
| 122 | A0A0L8FS75 | Proteasome subunit alpha type OS = Octopus bimaculoides | OCBIM_22010113mg | 1 | 2 | 4 |
| 123 | A0A0L8FRK2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22010655mg | 1 | 2 | 6 |
| 124 | A0A0L8GZX1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025682mg | 1 | 5 | 1 |
| 125 | A0A0L8G456 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000796mg | 1 | 1 | 6 |
| 126 | A0A0L8FF63 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024380mg | 1 | 1 | 10 |
| 127 | A0A0L8H8U9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020735mg | 1 | 1 | 5 |
| 128 | A0A0L8I5N4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22033390mg | 1 | 2 | 3 |
| 129 | A0A0L8I398 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22037157mg | 1 | 1 | 11 |
| 130 | A0A0L8GP93 | Nicotinamide-nucleotide adenylyltransferase OS = Octopus bimaculoides | OCBIM_22030204mg | 1 | 1 | 6 |
| 131 | A0A0L8IIH3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025740mg | 1 | 3 | 0 |
| 132 | A0A0L8GZD4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025455mg | 1 | 1 | 1 |
| 133 | A0A0L8HQW9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22008430mg | 1 | 4 | 2 |
| 134 | A0A0L8G2Z7 | Small ubiquitin-related modifier OS = Octopus bimaculoides | OCBIM_22001102mg | 1 | 1 | 11 |
| 135 | A0A0L8G8L3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22038063mg | 1 | 2 | 2 |
| 136 | O46345 | S-syntaxin OS = Doryteuthis pealeii | STX1 | 1 | 1 | 3 |
| 137 | A0A0L8GDD2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22036000mg | 1 | 1 | 2 |
| 138 | C4N147 | Sodium/calcium exchanger regulatory protein 1 OS = Doryteuthis pealeii | SLC8A1 | 1 | 4 | 7 |
| 139 | A0A0L8FJE4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017696mg | 1 | 2 | 2 |
| 140 | A0A0L8I067 | Kinesin-like protein OS = Octopus bimaculoides | OCBIM_22000619mg | 1 | 1 | 1 |
| 141 | A0A0L8FYB6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22005155mg | 1 | 1 | 1 |
| 142 | A0A0L8GUV0 | Serine/threonine-protein phosphatase OS = Octopus bimaculoides | OCBIM_22027338mg | 1 | 1 | 2 |
| 143 | A0A0L8GJ12 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22032700mg | 1 | 2 | 1 |
| 144 | A0A0L8GLG2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22032112mg | 1 | 1 | 1 |
| 145 | A0A0L8GY97 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026356mg | 1 | 2 | 2 |
| 146 | Q27Q56 | Hemocyanin subunit 2 OS = Sepia officinalis | HCY2 | 1 | 961 | 7 |
| 147 | A0A161HPY5 | Actin OS = Crassostrea brasiliana | ACTI | 3 | 96 | 38 |
| 148 | D2YZ90 | Beta actin OS = Idiosepius paradoxus | ACTI | 2 | 95 | 37 |
| 149 | K1QFR9 | Spectrin beta chain OS = Crassostrea gigas | CGI_10013845 | 1 | 34 | 4 |
| 150 | C1KC83 | Heat shock cognate protein 70 OS = Haliotis diversicolor | HSP70 | 1 | 37 | 16 |
| 151 | A0A2C9K1T4 | Uncharacterized protein OS = Biomphalaria glabrata | 106078167 | 1 | 68 | 13 |
| 152 | A0A0B7B7H2 | Uncharacterized protein OS = Arion vulgaris | ORF162822 | 1 | 40 | 11 |
| 153 | A0A2T7NLR4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_17913 | 1 | 18 | 5 |
| 154 | K1RH58 | Alpha-actinin, sarcomeric OS = Crassostrea gigas | CGI_10003110 | 1 | 43 | 10 |
| 155 | A0A2P1H676 | Heat shock protein 70 OS = Diplodon chilensis | HSP70 | 1 | 36 | 12 |
| 156 | K1PMY9 | Calmodulin OS = Crassostrea gigas | CGI_10006482 | 1 | 22 | 13 |
| 157 | A0A2T7NGU8 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_18553 | 5 | 12 | 29 |
| 158 | Q564J1 | Haemocyanin OS = Aplysia californica | hc | 2 | 927 | 2 |
| 159 | A0A2T7NV41 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_15545 | 4 | 18 | 25 |
| 160 | E7DS67 | Actin (Fragment) OS = Gonospira metablata | ACTI | 1 | 38 | 18 |
| 161 | K1RBG6 | Actin-1/3 OS = Crassostrea gigas | CGI_10017112 | 1 | 39 | 8 |
| 162 | P02595 | Calmodulin OS = Patinopecten sp. | CAM | 1 | 16 | 30 |
| 163 | V6A758 | Myosin heavy chain isoform C OS = Sepia officinalis | MYH | 1 | 17 | 16 |
| 164 | A0A0B7BLG3 | Uncharacterized protein OS = Arion vulgaris | ORF192624 | 3 | 23 | 2 |
| 165 | K1PPW8 | Coatomer subunit beta OS = Crassostrea gigas | CGI_10006442 | 2 | 8 | 7 |
| 166 | A0A210R0F2 | Fructose-bisphosphate aldolase OS = Mizuhopecten yessoensis | KP79_PYT16607 | 2 | 8 | 6 |
| 167 | A0A2T7PZW7 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01565 | 1 | 6 | 1 |
| 168 | A0A0B7B4N1 | Uncharacterized protein OS = Arion vulgaris | ORF158201 | 1 | 10 | 4 |
| 169 | A0A210QY92 | Coatomer subunit beta’ OS = Mizuhopecten yessoensis | KP79_PYT21841 | 1 | 5 | 5 |
| 170 | V3ZPS1 | Uncharacterized protein OS = Lottia gigantea | LOTGIDRAFT_222012 | 2 | 9 | 12 |
| 171 | E3VWM3 | Fructose-bisphosphate aldolase OS = Meretrix meretrix | FBA | 1 | 20 | 4 |
| 172 | A0A2T7PSV4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03483 | 2 | 10 | 11 |
| 173 | A0A0B7AZA8 | Uncharacterized protein OS = Arion vulgaris | ORF148015 | 2 | 10 | 19 |
| 174 | K7WKX6 | Fructose-bisphosphate aldolase OS = Haliotis rufescens | FBA | 1 | 3 | 9 |
| 175 | A0A2T7NF32 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_20261 | 1 | 5 | 4 |
| 176 | A0A2T7NMW4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_18325 | 2 | 4 | 5 |
| 177 | K1QZU8 | Calcium-transporting ATPase OS = Crassostrea gigas | CGI_10023684 | 1 | 2 | 1 |
| 178 | A0A0L8IAE8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025089mg | 1 | 2 | 8 |
| 179 | A0A2C9KC89 | Uncharacterized protein OS = Biomphalaria glabrata | 106056965 | 2 | 5 | 3 |
| 180 | A0A210R746 | Ras-related protein Rab-6A OS = Mizuhopecten yessoensis | KP79_PYT20147 | 1 | 9 | 11 |
| 181 | A0A0B6Z4Q3 | Uncharacterized protein OS = Arion vulgaris | ORF48472 | 2 | 12 | 8 |
| 182 | A0A2T7PZP4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01513 | 1 | 4 | 3 |
| 183 | A0A2C9JIZ4 | Uncharacterized protein OS = Biomphalaria glabrata | 106056849 | 1 | 9 | 13 |
| 184 | K1PTH4 | ADP-ribosylation factor OS = Crassostrea gigas | CGI_10020174 | 1 | 2 | 1 |
| 185 | Q6PTL0 | Triosephosphate isomerase OS = Nucula proxima | OCBIM_22037419mg | 1 | 5 | 6 |
| 186 | A0A2C9JZR8 | Uncharacterized protein OS = Biomphalaria glabrata | 106074442 | 1 | 2 | 2 |
| 187 | A0A2C9JIA9 | Uncharacterized protein OS = Biomphalaria glabrata | 106056539 | 1 | 6 | 4 |
| 188 | A0A385NHM7 | Glutathione S-transferase OS = Tegillarca granosa | GST | 1 | 8 | 5 |
| 189 | A0A210QUP5 | Malic enzyme OS = Mizuhopecten yessoensis | KP79_PYT06884 | 1 | 1 | 3 |
| 190 | V3YXF9 | Adenosylhomocysteinase OS = Lottia gigantea | LOTGIDRAFT_184532 | 1 | 2 | 3 |
| 191 | A0A210QGP4 | Chitotriosidase-1 OS = Mizuhopecten yessoensis | KP79_PYT06201 | 1 | 1 | 3 |
| 192 | A0A210QHE1 | Adenosylhomocysteinase OS = Mizuhopecten yessoensis | KP79_PYT14445 | 1 | 4 | 3 |
| 193 | A0A210PIA6 | Ornithine aminotransferase OS = Mizuhopecten yessoensis | KP79_PYT16913 | 1 | 3 | 3 |
| 194 | K1QQB6 | 40S ribosomal protein S14 OS = Crassostrea gigas | CGI_10011151 | 1 | 4 | 9 |
| 195 | A0A2C9KEN8 | Tubulin alpha chain OS = Biomphalaria glabrata | 106069694 | 1 | 2 | 3 |
| 196 | A0A2T7PWT6 | Serine/threonine-protein phosph OS = Pomacea canaliculata | C0Q70_00460 | 1 | 1 | 3 |
| 197 | A0A0B7AJW7 | Fructose-bisphosphate aldolase OS = Arion vulgaris | ORF124546 | 1 | 8 | 4 |
| 198 | A0A2C9L7N6 | Uncharacterized protein OS = Biomphalaria glabrata | 106080319 | 1 | 49 | 4 |
| 199 | A0A210QTZ1 | Peptidyl-prolyl cis-trans OS = Mizuhopecten yessoensis | KP79_PYT00632 | 1 | 2 | 6 |
| 200 | A0A2I7M8C2 | Go protein alpha subunit OS = Argopecten irradians | N/A | 1 | 4 | 3 |
| 201 | K1R2G8 | Titin OS = Crassostrea gigas | CGI_10016808 | 1 | 2 | 0 |
| 202 | K1QVD7 | Neuronal acetylcholine receptor subunit non-alpha-2 OS = Crassostrea gigas | CGI_10016138 | 1 | 2 | 1 |
| 203 | K1Q7G5 | Ficolin-2 OS = Crassostrea gigas | CGI_10026202 | 1 | 2 | 3 |
| 204 | A0A2C9K9W9 | Uncharacterized protein OS = Biomphalaria glabrata | 106068683 | 1 | 1 | 1 |
| 205 | A0A0B6ZP87 | Uncharacterized protein OS = Arion vulgaris | ORF71130 | 1 | 3 | 4 |
| 206 | V4AP92 | Elongation factor 1-alpha OS = Lottia gigantea | LOTGIDRAFT_239271 | 1 | 2 | 2 |
| 207 | A0A2T7PU69 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03920 | 1 | 4 | 4 |
| 208 | V3ZN51 | Staphylococcal nuclease domain-cont. OS = Lottia gigantea | LOTGIDRAFT_235720 | 1 | 3 | 1 |
| 209 | A0A2T7PSF5 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03333 | 1 | 2 | 0 |
| 210 | K1PQD4 | Phosphoglucomutase-1 OS = Crassostrea gigas | CGI_10011818 | 1 | 1 | 2 |
| 211 | A0A0B7BF17 | Uncharacterized protein OS = Arion vulgaris | ORF179770 | 1 | 3 | 2 |
| 212 | A0A2T7Q0W0 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01928 | 1 | 1 | 3 |
| 213 | A0A0L8I692 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22034637mg | 1 | 4 | 19 |
| 214 | K1PQ79 | Copine-3 OS = Crassostrea gigas | CGI_10011897 | 1 | 3 | 1 |
| 215 | K1PWB9 | EH domain-containing protein 1 OS = Crassostrea gigas | CGI_10005813 | 1 | 1 | 4 |
| 216 | A0A2T7Q016 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01636 | 1 | 1 | 2 |
| 217 | V4AKV4 | Calcium-transporting ATPase OS = Lottia gigantea | LOTGIDRAFT_208914 | 1 | 3 | 1 |
| 218 | A0A2T7NL99 | Proteasome subunit beta OS = Pomacea canaliculata | C0Q70_17739 | 1 | 2 | 4 |
| 219 | A0A0L8HWW8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003772mg | 1 | 2 | 2 |
| KEGG Pathway | p-Value |
|---|---|
| Metabolic pathways (cysteine and methionine metabolism) | 4.53 × 10−4 |
| Endocytosis/phagosome | 1.05 × 10−2 |
| RNA transport | 2.24 × 10−2 |
| Protein methylation | 3.46 × 10−2 |
| Calcium homeostasis | 1.00 × 10−1 |
| InterPro Motifs | p-Value |
|---|---|
| Small GTP-binding protein domain | 3.1 × 10−4 |
| Heat shock protein 70, conserved site | 8.5 × 10−4 |
| Small GTPase superfamily | 8.6 × 10−4 |
| Proteasome, alpha-subunit, N-terminal domain | 1.3 × 10−3 |
| P-loop containing nucleoside triphosphate hydrolase | 8.3 × 10−3 |
| EF-hand-like domain | 2.9 × 10−2 |
| Ubiquitin | 3.4 × 10−2 |
| Proteins | Peptides | PeptideRanker Score | Anti-Microbial Peptide (AMP) | Discriminant Score for AMP |
|---|---|---|---|---|
| ADP-ribosylation factor OS = Crassostrea gigas | SPSPKQMVSCPVCGL | 0.915222 | Non-AMP | 0.043 |
| Collagen ColAa OS = Sepia pharaonis | PGDPGPVGRTGPMGL | 0.934847 | Non-AMP | 0.003 |
| Collagen ColAa OS = Sepia pharaonis | RGPPGPPGL | 0.912657 | Non-AMP | 0.030 |
| Heat shock protein 70 OS = Sepiella maindroni | GGMPGGMPGGMPGGMPNF | 0.92432 | AMP | 0.504 |
| Hemocyanin OS = Sepiella maindroni | KKPMMPF | 0.932566 | AMP | 0.978 |
| Hemocyanin OS = Sepiella maindroni | PNQPMRPF | 0.920777 | AMP | 0.983 |
| Hemocyanin subunit 1 OS = Todarodes pacificus | NDPMRPF | 0.923312 | AMP | 0.795 |
| Hemocyanin subunit 2 OS = Sepia officinalis | SDPMRPF | 0.938433 | AMP | 0.879 |
| Uncharacterized protein OS = Octopus bimaculoides | CPCMGRF | 0.985441 | AMP | 0.622 |
| Uncharacterized protein OS = Octopus bimaculoides | GGPPGMPPF | 0.973279 | Non-AMP | 0.208 |
| Uncharacterized protein OS = Octopus bimaculoides | GRCVMCNCNKHSSTCDPQTGKCVNCQHNTL | 0.969319 | Non-AMP | 0.238 |
| Uncharacterized protein OS = Octopus bimaculoides | GSCVPCNCNGF | 0.952459 | AMP | 0.745 |
| Uncharacterized protein OS = Octopus bimaculoides | QPPQCCPSKGGSF | 0.943546 | AMP | 0.687 |
| Uncharacterized protein OS = Octopus bimaculoides | GSWGNGNRW | 0.915802 | Non-AMP | 0.403 |
| Uncharacterized protein OS = Octopus bimaculoides | PPPSKRF | 0.911736 | AMP | 0.983 |
| Uncharacterized protein OS = Biomphalaria glabrata | PPPPQPVGGGGGNRW | 0.955862 | Non-AMP | 0.092 |
| Uncharacterized protein OS = Biomphalaria glabrata | SRSPPRPF | 0.904351 | AMP | 0.993 |
| Uncharacterized protein OS = Pomacea canaliculata | HDGDGPRPCCF | 0.93215 | Non-AMP | 0.031 |
| Proteins | Peptides | PeptideRanker Score | Anti-Microbial Peptide (AMP) | Discriminant Score for AMP |
|---|---|---|---|---|
| ADP-ribosylation factor OS = Crassostrea gigas | CPICYDFMHTAMILPECSHTFCSFCIR | 0.902646 | Non-AMP | 0.160 |
| Calcium-transporting ATPase OS = Octopus bimaculoides | FSDDYPGFF | 0.970864 | Non-AMP | 0.006 |
| Calcium-transporting ATPase OS = Crassostrea gigas | FLQFQLTVNCVAVMVAFFGACIINDSPLK | 0.979848 | Non-AMP | 0.281 |
| Calcium-transporting ATPase OS=Lottia gigantea | FADAPFMK | 0.93747 | Non-AMP | 0.014 |
| Calmodulin OS = Crassostrea gigas | GAFFVFDR | 0.915228 | Non-AMP | 0.003 |
| Chitinase OS = Todarodes pacificus | MLAVSLLFLLAIGGVSSAGHR | 0.976725 | AMP | 0.746 |
| Chitotriosidase OS = Euprymna scolopes | MASTFATVFGVLSLCFLGLHLTNGEYK | 0.984749 | Non-AMP | 0.106 |
| Coatomer subunit beta’ OS = Mizuhopecten yessoensis | YCLCLFR | 0.924855 | AMP | 0.579 |
| Collagen ColAa OS = Sepia pharaonis | GPPGIPGLPGPK | 0.93716 | AMP | 0.504 |
| Collagen ColAa OS = Sepia pharaonis | GPPGPPGLK | 0.913133 | Non-AMP | 0.119 |
| Collagen ColAa OS = Sepia pharaonis | AGPPGFPGTPGPK | 0.907398 | AMP | 0.682 |
| Ficolin-2 OS = Crassostrea gigas | DQDNDMYVSDNCGILFPSGWWHR | 0.901865 | Non-AMP | 0.008 |
| Fructose-bisphosphate aldolase OS = Mizuhopecten yessoensis | KPWALTFSFGR | 0.93422 | Non-AMP | 0.123 |
| Hemocyanin OS = Aplysia californica | MVGYLGQALMALLLLALSNAALVR | 0.993669 | Non-AMP | 0.380 |
| Hemocyanin OS = Aplysia californica | FEPNPFFSGK | 0.924588 | Non-AMP | 0.093 |
| Hemocyanin OS = Aplysia californica | VACCLHGMPVFPHWHR | 0.903581 | Non-AMP | 0.106 |
| Hemocyanin OS = Nautilus pompilius | MATHWHSLLLFSLQLLVFTYATSDPTNIR | 0.97599 | Non-AMP | 0.008 |
| Hemocyanin OS = Sepiella maindroni | GSPIGVPYWDWTKPMK | 0.917605 | Non-AMP | 0.027 |
| Hemocyanin-like protein OS = Uroteuthis edulis | TNFFFLALIATVWLGNAETETETSK | 0.90323 | Non-AMP | 0.062 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | VFVGFLLHGFGSSAYATFDICNDAGECR | 0.96087 | Non-AMP | 0.233 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | LNHLPLLCLAVILTLWMSGSNTVNGNLVR | 0.926117 | Non-AMP | 0.287 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | VFAGFLFMGIK | 0.904542 | AMP | 0.865 |
| Hemocyanin subunit 2 OS = Euprymna scolopes | VFAGFWFHGIK | 0.943 | AMP | 0.506 |
| Hemocyanin subunit 2 OS = Sepia officinalis | VFGGFWLHGIK | 0.907156 | AMP | 0.739 |
| Hemocyanin subunit 3 OS = Sepia officinalis | TSFLFLAFVATSWFVYAVTASK | 0.905214 | Non-AMP | 0.136 |
| Malate dehydrogenase OS = Sepia officinalis | DLFNTNASIVANLADACAQYCPK | 0.965037 | Non-AMP | 0.251 |
| Myosin heavy chain isoform A OS = Octopus bimaculoides | YQSGFIYTYSGLFCVAINPYR | 0.956725 | Non-AMP | 0.024 |
| Myosin heavy chain OS = Todarodes pacificus | NWEWWR | 0.951523 | Non-AMP | 0.478 |
| Myosin II heavy chain OS = Doryteuthis pealeii | NWQWWR | 0.973264 | AMP | 0.959 |
| Myosin II heavy chain OS = Doryteuthis pealeii | YYSGLIYTYSGLFCVVVNPYK | 0.939159 | Non-AMP | 0.032 |
| Neuronal acetylcholine receptor subunit non-alpha-2 OS = Crassostrea gigas | LLIDLCLSVLVTTLAIVSLYFYDMSDSR | 0.904075 | Non-AMP | 0.015 |
| Peptidyl-prolyl cis-trans isomerase OS = Mizuhopecten yessoensis | MAGAGIGCVLLFLLPALLSAGK | 0.996478 | Non-AMP | 0.159 |
| Phosphoglucomutase-1 OS = Crassostrea gigas | DGLWAVLAWLSVLANQNCSVEECIK | 0.991266 | AMP | 0.904 |
| Protein disulfide-isomerase OS = Octopus bimaculoides | NVFIEFYAPWCGHCK | 0.907443 | Non-AMP | 0.053 |
| S-syntaxin OS = Doryteuthis pealeii | IAILVCLVILVLVIVSTVGGVFGG | 0.965343 | Non-AMP | 0.000 |
| Titin OS = Crassostrea gigas | DGSWQNLVTVLGCLKPQFVNLQR | 0.974127 | AMP | 0.724 |
| Titin OS = Crassostrea gigas | GYPPPIISWYR | 0.917986 | Non-AMP | 0.074 |
| Tubulin alpha chain OS = Octopus bimaculoides | FVDWCPTGFK | 0.923256 | Non-AMP | 0.010 |
| Uncharacterized protein OS = Arion vulgaris | APDFIFYAPR | 0.921198 | Non-AMP | 0.009 |
| Uncharacterized protein OS = Octopus bimaculoides | FLQFQLTVNVVAVLVAFFGACTINVSI | 0.978717 | AMP | 0.916 |
| Uncharacterized protein OS = Octopus bimaculoides | YYTFFVTIFLFATTLCSTIPKPK | 0.984914 | Non-AMP | 0.012 |
| Uncharacterized protein OS = Octopus bimaculoides | LFPAFGFGAR | 0.94902 | AMP | 0.505 |
| Uncharacterized protein OS = Octopus bimaculoides | ATMLGAQGNIFFASLSCCCLILSCS | 0.999233 | AMP | 0.879 |
| Uncharacterized protein OS = Octopus bimaculoides | SGPFYIFSGGMPR | 0.939205 | Non-AMP | 0.089 |
| Uncharacterized protein OS = Octopus bimaculoides | EFSMMFR | 0.931708 | Non-AMP | 0.001 |
| Uncharacterized protein OS = Octopus bimaculoides | YGSCVPCNCNGFSNDCDPVTGECIDCQR | 0.980617 | Non-AMP | 0.243 |
| Uncharacterized protein OS = Octopus bimaculoides | HNPEGCISCFCMGVTEFCTSTSR | 0.964134 | Non-AMP | 0.083 |
| Uncharacterized protein OS = Octopus bimaculoides | APMVELCECPQGYTGVSCQECSPGYSR | 0.963828 | Non-AMP | 0.012 |
| Uncharacterized protein OS = Octopus bimaculoides | GCGCSAGQFECQNGLCINENK | 0.930153 | AMP | 0.982 |
| Uncharacterized protein OS = Octopus bimaculoides | EECMSCFCFK | 0.918951 | AMP | 0.982 |
| Uncharacterized protein OS = Octopus bimaculoides | NSEYGFACFCPQGFAGYQCDTVGER | 0.906197 | AMP | 0.576 |
| Uncharacterized protein OS = Octopus bimaculoides | MIIYILSLAGVALGVYFLSCVR | 0.995663 | Non-AMP | 0.008 |
| Uncharacterized protein OS = Octopus bimaculoides | MILTIFACLMALDIELNTSNSIQEE | 0.968187 | Non-AMP | 0.026 |
| Uncharacterized protein OS = Octopus bimaculoides | AIGALVDACGPGLCPDWADWAPK | 0.948884 | AMP | 0.774 |
| Uncharacterized protein OS = Octopus bimaculoides | QGDWTCPNPACGNNNFGWR | 0.9572 | Non-AMP | 0.286 |
| Uncharacterized protein OS = Octopus bimaculoides | GGFGGGGGGGGGMGGDR | 0.928063 | Non-AMP | 0.065 |
| Uncharacterized protein OS = Octopus bimaculoides | GFFEDDYDEYGGGYGGGMGFGGLNR | 0.944869 | Non-AMP | 0.143 |
| Uncharacterized protein OS = Octopus bimaculoides | LDDGDACLLDMGTEYCCYASDITCSYPVNGK | 0.968621 | Non-AMP | 0.056 |
| Uncharacterized protein OS = Octopus bimaculoides | MAFYTILNVVTIVLLIIVGQCR | 0.998628 | Non-AMP | 0.031 |
| Uncharacterized protein OS = Octopus bimaculoides | GGSFGFNFR | 0.969779 | Non-AMP | 0.355 |
| Uncharacterized protein OS = Octopus bimaculoides | NSTDVCNCSIYVGLFPCNECTK | 0.994975 | Non-AMP | 0.462 |
| Uncharacterized protein OS = Octopus bimaculoides | PPSPPIYFR | 0.946483 | Non-AMP | 0.226 |
| Uncharacterized protein OS = Octopus bimaculoides | CFLCATGTGTSIEVLALVTIGWCLLHATGTR | 0.96344 | AMP | 0.768 |
| Uncharacterized protein OS = Octopus bimaculoides | FDFFYK | 0.96245 | Non-AMP | 0.032 |
| Uncharacterized protein OS = Octopus bimaculoides | FSPIPFLFCTISGTCNFATR | 0.95134 | AMP | 0.505 |
| Uncharacterized protein OS = Octopus bimaculoides | FWELTECCPHQCLEWLSNLVTR | 0.933791 | Non-AMP | 0.106 |
| Uncharacterized protein OS = Octopus bimaculoides | DAFCSSPNFNSWLK | 0.922125 | Non-AMP | 0.058 |
| Uncharacterized protein OS = Octopus bimaculoides | NGYEEDDALIGLLNLCTAILK | 0.917521 | Non-AMP | 0.479 |
| Uncharacterized protein OS = Octopus bimaculoides | DYFWLVCR | 0.911557 | Non-AMP | 0.001 |
| Uncharacterized protein OS = Biomphalaria glabrata | QGELGDCWLLAAVASLTCNPK | 0.919385 | AMP | 0.783 |
| Uncharacterized protein OS = Biomphalaria glabrata | SPPRPFEWK | 0.905581 | Non-AMP | 0.006 |
| Uncharacterized protein OS = Pomacea canaliculata | SVFNIPPNCFSEMM | 0.908085 | Non-AMP | 0.003 |
| Uncharacterized protein OS = Pomacea canaliculata | SCLMGHGSLFGAGAGSLHLQAIAALK | 0.919795 | Non-AMP | 0.315 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics. Mar. Drugs 2020, 18, 31. https://doi.org/10.3390/md18010031
Carrera M, Ezquerra-Brauer JM, Aubourg SP. Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics. Marine Drugs. 2020; 18(1):31. https://doi.org/10.3390/md18010031
Chicago/Turabian StyleCarrera, Mónica, Josafat Marina Ezquerra-Brauer, and Santiago P. Aubourg. 2020. "Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics" Marine Drugs 18, no. 1: 31. https://doi.org/10.3390/md18010031
APA StyleCarrera, M., Ezquerra-Brauer, J. M., & Aubourg, S. P. (2020). Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics. Marine Drugs, 18(1), 31. https://doi.org/10.3390/md18010031