Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5
Abstract
1. Introduction
2. Results
2.1. Cloning and Sequence Analysis of TsAly7B
2.2. Expression and Purification of Recombinant TsAly7B and Three Truncated Mutants
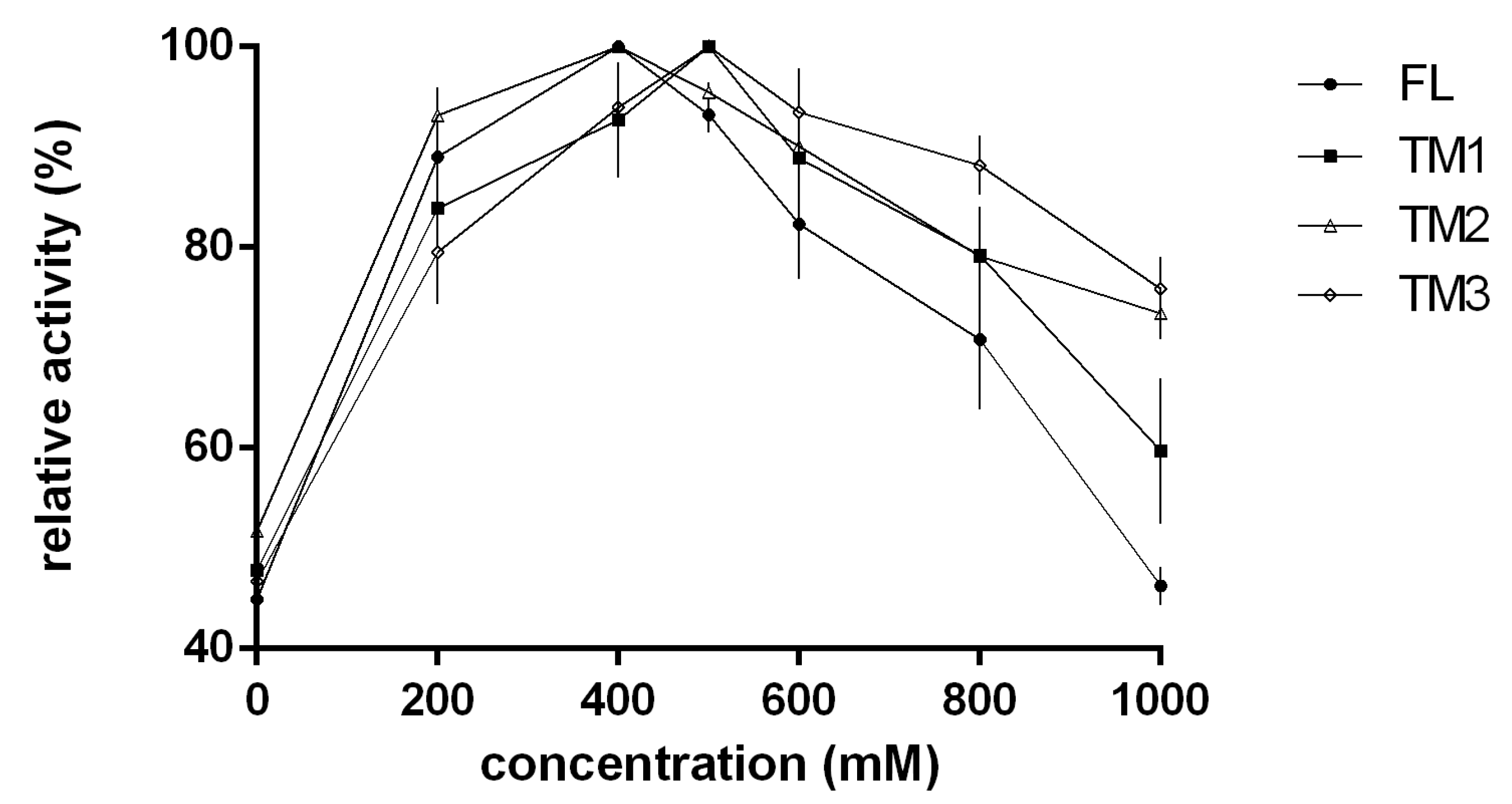
2.3. Biochemical Characterization of TsAly7B and Three Truncated Mutants
2.4. Analysis of the Substrate Specificity and Final Reaction Products
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Chemicals
4.2. Cloning and Sequence Analysis of TsAly7B
4.3. Expression of TsAly7B and Three Truncated Mutants
4.4. Alginate Lyase Activity Assay
4.5. Biochemical Characterization of Recombinant TsAly7B and Three Truncated Mutants
4.6. End Products Analyzed by TsAly7B and Three Truncated Mutants
Author Contributions
Funding
Conflicts of Interest
References
- Panikkar, R.; Brasch, D.J. Composition and block structure of alginates from New Zealand brown seaweeds. Carbohydr. Res. 1996, 293, 119–132. [Google Scholar] [CrossRef]
- Falkeborg, M.; Cheong, L.Z.; Gianfico, C.; Sztukiel, K.M.; Kristensen, K.; Glasius, M.; Xu, X.; Guo, Z. Alginate oligosaccharides: Enzymatic preparation and antioxidant property evaluation. Food Chem. 2014, 164, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Steinbüchel, A.; Rhee, S.K. Polysaccharides and Polyamides in the Food Industry: Properties, Production, and Patents; Wiley-VCH Verlag GmbH & CO. KGaA: Weinheim, Germany, 2005. [Google Scholar]
- Sawabe, T.; Takahashi, H.; Ezura, Y.; Gacesa, P. Cloning, sequence analysis and expression of Pseudoalteromonas elyakovii IAM 14594 gene (alyPEEC) encoding the extracellular alginate lyase. Carbohydr. Res. 2001, 335, 11–21. [Google Scholar] [CrossRef]
- Tusi, S.K.; Khalaj, L.; Ashabi, G.; Kiaei, M.; Khodagholi, F. Alginate oligosaccharide protects against endoplasmic reticulum- and mitochondrial-mediated apoptotic cell death and oxidative stress. Biomaterials 2011, 32, 5438–5458. [Google Scholar] [CrossRef]
- Wang, P.; Jiang, X.; Jiang, Y.; Hu, X.; Mou, H.; Li, M.; Guan, H. In vitro antioxidative activities of three marine oligosaccharides. Nat. Prod. Res. 2007, 21, 646–654. [Google Scholar] [CrossRef]
- Liu, S.; Liu, G.; Yi, Y. Novel vanadyl complexes of alginate saccharides: Synthesis, characterization, and biological activities. Carbohydr. Polym. 2015, 121, 86–91. [Google Scholar] [CrossRef]
- Burana-Osot, J.; Hosoyama, S.; Nagamoto, Y.; Suzuki, S.; Linhardt, R.J.; Toida, T. Photolytic depolymerization of alginate. Carbohydr. Res. 2009, 344, 2023–2027. [Google Scholar] [CrossRef]
- Uno, T.; Hattori, M.T.; Yoshida, T. Oral administration of alginic acid oligosaccharide suppresses IgE production and inhibits the induction of oral tolerance. Biosci. Biotechnol. Biochem. 2006, 70, 3054–3057. [Google Scholar] [CrossRef]
- Tomoda, Y.; Umemura, K.; Adachi, T. Promotion of barley root elongation under hypoxic conditions by alginate lyase-lysate (A.L.L.). Biosci. Biotechnol. Biochem. 1994, 58, 202–203. [Google Scholar] [CrossRef]
- Xu, X.; Iwamoto, Y.; Kitamura, Y.; Oda, T.; Muramatsu, T. Root growth-promoting activity of unsaturated oligomeric uronates from alginate on carrot and rice plants. Biosci. Biotechnol. Biochem. 2003, 67, 2022–2025. [Google Scholar] [CrossRef]
- Duan, G.; Han, F.; Yu, W. Cloning, sequence analysis, and expression of gene alyPI encoding an alginate lyase from marine bacterium Pseudoalteromonas sp. CY24. Can. J. Microbiol. 2009, 55, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Bao, M.; Wu, Y.; Yu, W.; Han, F. Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, and alters its substrate preference and product distribution. FEMS Microbiol. Lett. 2015, 362, fnv054. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Carvalho, V.; Domingues, L.; Gama, F.M. Recombinant CBM-fusion technology—Applications overview. Biotechnol. Adv. 2015, 33, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Tomme, P.; Driver, D.P.; Amandoron, E.A.; Miller, R.C., Jr.; Antony, R.; Warren, J.; Kilburn, D.G. Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. J. Bacteriol. 1995, 177, 4356–4363. [Google Scholar] [CrossRef] [PubMed]
- Freelove, A.C.; Bolam, D.N.; White, P.; Hazlewood, G.P.; Gilbert, H.J. A novel carbohydrate-binding protein is a component of the plant cell wall-degrading complex of Piromyces equi. J. Biol. Chem. 2001, 276, 43010–43017. [Google Scholar] [CrossRef] [PubMed]
- Sim, P.F.; Furusawa, G.; Teh, A.H. Functional and structural studies of a multidomain alginate lyase from Persicobacter sp. CCB-QB2. Sci. Rep. 2017, 7, 13656. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Q.; Zhang, K.; Zhu, Q.; Li, Z.; Liu, Y.; Fitzek, E.; Yohe, T.; Zhao, L.; Li, W.; Liu, T.; et al. Structural and biochemical characterization of a multidomain alginate lyase reveals a novel role of CBM32 in CAZymes. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 1862–1869. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.; Li, S.; Su, H.; Tang, L.; Tan, Y.; Yu, W.; Han, F. Characterization of a new endo-type polysaccharide lyase (PL) family 6 alginate lyase with cold-adapted and metal ions-resisted property. Int. J. Biol. Macromol. 2018, 120, 729–735. [Google Scholar] [CrossRef]
- Yang, A.; Cheng, J.; Liu, M.; Shangguan, Y.; Liu, L. Sandwich fusion of CBM9_2 to enhance xylanase thermostability and activity. Int. J. Biol. Macromol. 2018, 117, 586–591. [Google Scholar] [CrossRef]
- Kim, D.R.; Lim, H.K.; Lee, K.I.; Hwang, I.T. Identification of a novel cellulose-binding domain within the endo-β-1,4-xylanase KRICT PX-3 from Paenibacillus terrae HPL-003. Enzym. Microb. Technol. 2016, 93–94, 166–173. [Google Scholar] [CrossRef]
- Peng, C.; Wang, Q.; Lu, D.; Han, W.; Li, F. A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, K.; Sakka, M.; Kimura, T.; Sakka, K. Essential role of a family-32 carbohydrate-binding module in substrate recognition by Clostridium thermocellum mannanase CtMan5A. FEBS Lett. 2014, 588, 1726–1730. [Google Scholar] [CrossRef] [PubMed]



| Reagents Added | Concentration (mM) | Relative Activity (%) | |||
|---|---|---|---|---|---|
| TsAly7B-FL | TsAly7B-TM1 | TsAly7B-TM2 | TsAly7B-TM3 | ||
| None | - | 100 | 100 | 100 | 100 |
| NH4Cl | 1 | 108.13 ± 3.56 | 102.04 ± 4.39 | 104.4 ± 12.19 | 102.31 ± 2.41 |
| LiCl | 1 | 105.58 ± 9.71 | 99.78 ± 4.37 | 77.14 ± 9.68 | 95.81 ± 9.31 |
| KCl | 1 | 104.28 ± 5.86 | 96.6 ± 3.53 | 107.47 ± 5.3 | 103.05 ± 2.94 |
| MgCl2 | 1 | 100.39 ± 2.37 | 100.7 ± 7.84 | 83.19 ± 3.92 | 100.57 ± 5.58 |
| CaCl2 | 1 | 114.36 ± 6.95 | 89.91 ± 12.71 | 100.82 ± 4.81 | 105.11 ± 10 |
| MnCl2 | 1 | 89.31 ± 7.92 | 93.31 ± 24.01 | 84.89 ± 3.58 | 50.47 ± 3.85 |
| BaCl2 | 1 | 91.29 ± 16.32 | 86.71 ± 10.09 | 50.34 ± 3.6 | 83.76 ± 7.6 |
| EDTA | 1 | 3.29 ± 0.58 | 1.1 ± 0.77 | 4.61 ± 3.59 | 2.26 ± 1.72 |
| SDS | 1 | 100.9 ± 3.19 | 89.86 ± 3.14 | 79.15 ± 3.94 | 90.24 ± 7.2 |
| Protein | Specific Activity (U/mg) | Molecular Weight (kDa) | Specific Activity (U/nmol) |
|---|---|---|---|
| TsAly7B-FL | 488.8 ± 12.4 | 64.79 | 31.6 ± 0.8 |
| TsAly7B-TM1 | 869.1 ± 9.1 | 51.35 | 44.6 ± 0.5 |
| TsAly7B-TM2 | 1147.2 ± 19.7 | 52.66 | 60.4 ± 1.0 |
| TsAly7B-TM3 | 238.4 ± 5.6 | 33.78 | 8.0 ± 0.2 |
| Relative Activity (%) | ||||
|---|---|---|---|---|
| Substrate | FL | TM1 | TM2 | TM3 |
| Alginate | 100 | 100 | 100 | 100 |
| PolyM | 110.93 ± 3.23 | 96.61 ± 12.46 | 97.08 ± 2.43 | 107.38 ± 28.33 |
| PolyG | 91.29 ± 4.6 | 81.47 ± 18.13 | 103.21 ± 5.81 | 43.05 ± 11.62 |
| Primers | Sequence (5′ to 3′) | Usage |
|---|---|---|
| Expression of TsAly7B and truncated proteins | ||
| TsAly7B-FL-F | CATGCCATGGGCTTGATTAATAATAAATTAAAAAAATGTG | Expression of TsAly7B-FL |
| TsAly7B-FL-R | CCGCTCGAGAGGGTTATAACCCGTATGTGA | |
| TsAly7B-TM1-F | CATGCCATGGGCATCGATAATGGTACTCATGATG | Expression of TsAly7B-TM1 |
| TsAly7B-TM1-R | CCGCTCGAGAGGGTTATAACCCGTATGTGA | |
| TsAly7B-TM2-1-F | CATGCCATGGGCTTGATTAATAATAAATTAAAAAAATGTG | Expression of TsAly7B-TM2 |
| TsAly7B-TM2-1-R | CGGGATCCGGCATTCACAATGGTTTGCG | |
| TsAly7B-TM2-2-F | CGGGATCCACGAAGATCAATGGCTGTACT | |
| TsAly7B-TM2-2-R | CCGCTCGAGAGGGTTATAACCCGTATGTGA | |
| TsAly7B-TM3-F | CATGCCATGGGCCCGCCTTCTGGTAATTTCG | Expression of TsAly7B-TM3 |
| TsAly7B-TM3-R | CCGCTCGAGAGGGTTATAACCCGTATGTGA | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Tang, L.; Bao, M.; Liu, Z.; Yu, W.; Han, F. Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5. Mar. Drugs 2020, 18, 25. https://doi.org/10.3390/md18010025
Zhang Z, Tang L, Bao M, Liu Z, Yu W, Han F. Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5. Marine Drugs. 2020; 18(1):25. https://doi.org/10.3390/md18010025
Chicago/Turabian StyleZhang, Zhelun, Luyao Tang, Mengmeng Bao, Zhigang Liu, Wengong Yu, and Feng Han. 2020. "Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5" Marine Drugs 18, no. 1: 25. https://doi.org/10.3390/md18010025
APA StyleZhang, Z., Tang, L., Bao, M., Liu, Z., Yu, W., & Han, F. (2020). Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5. Marine Drugs, 18(1), 25. https://doi.org/10.3390/md18010025





