A New β-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides
Abstract
:1. Introduction
2. Results
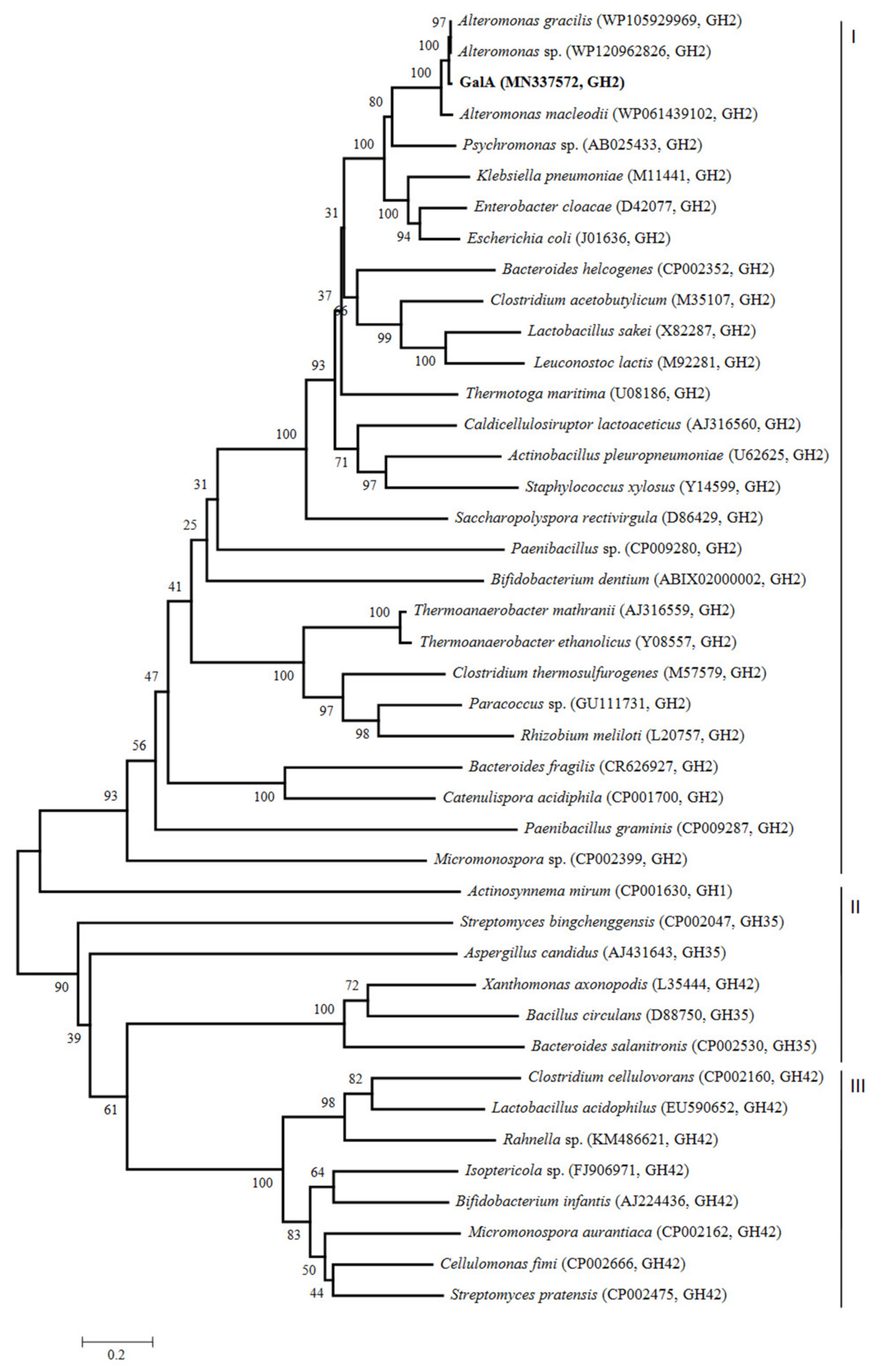
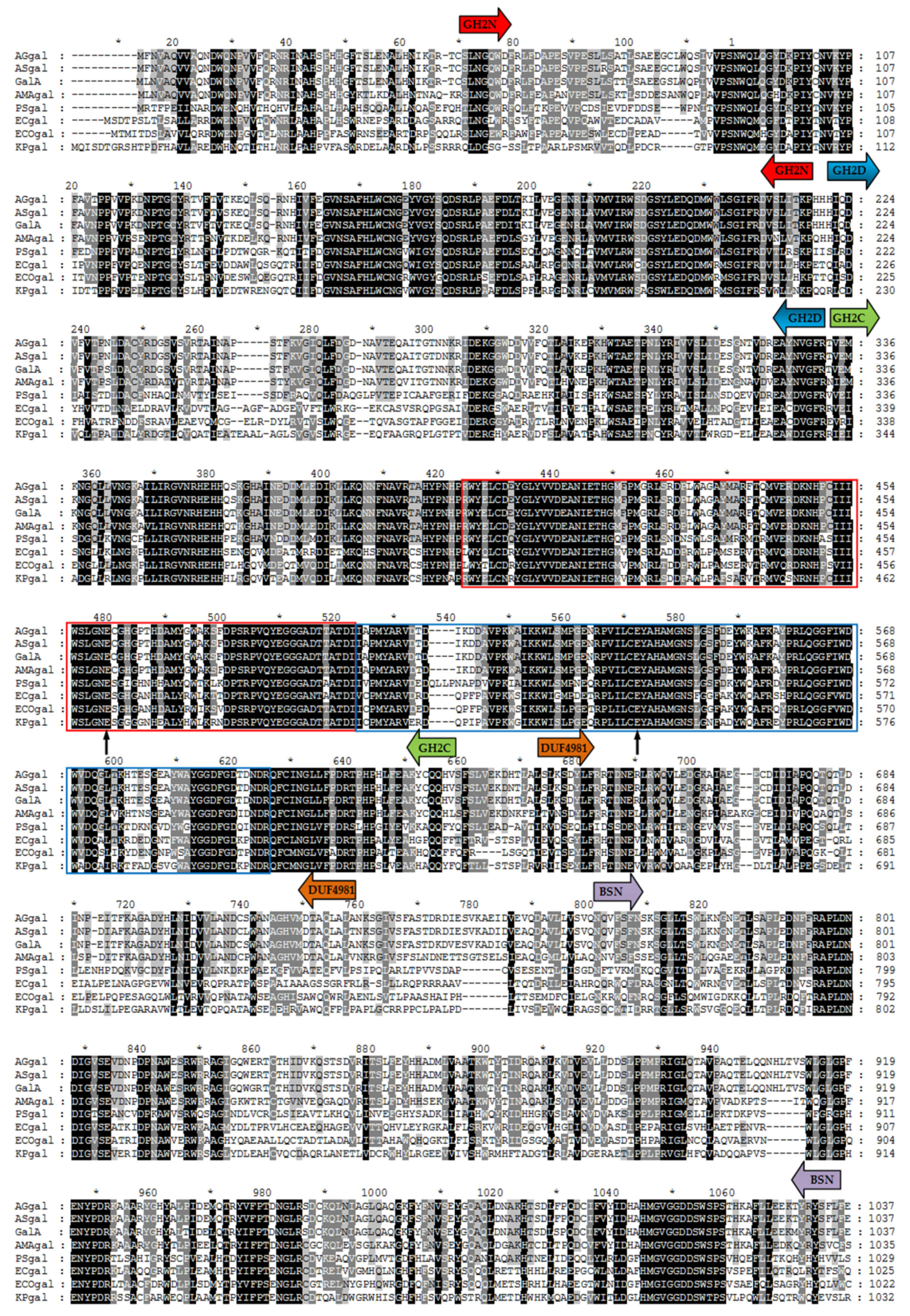
2.1. Cloning and Sequence Analysis of GalA
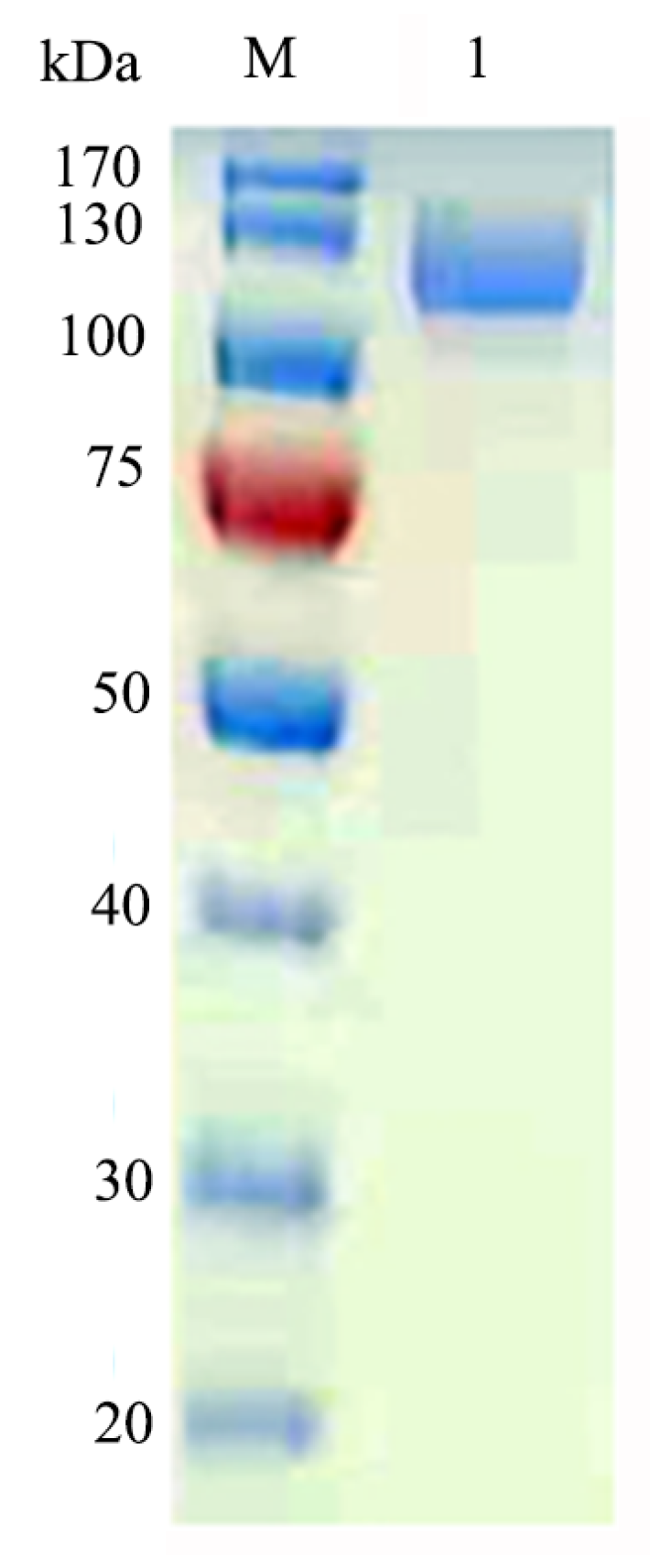
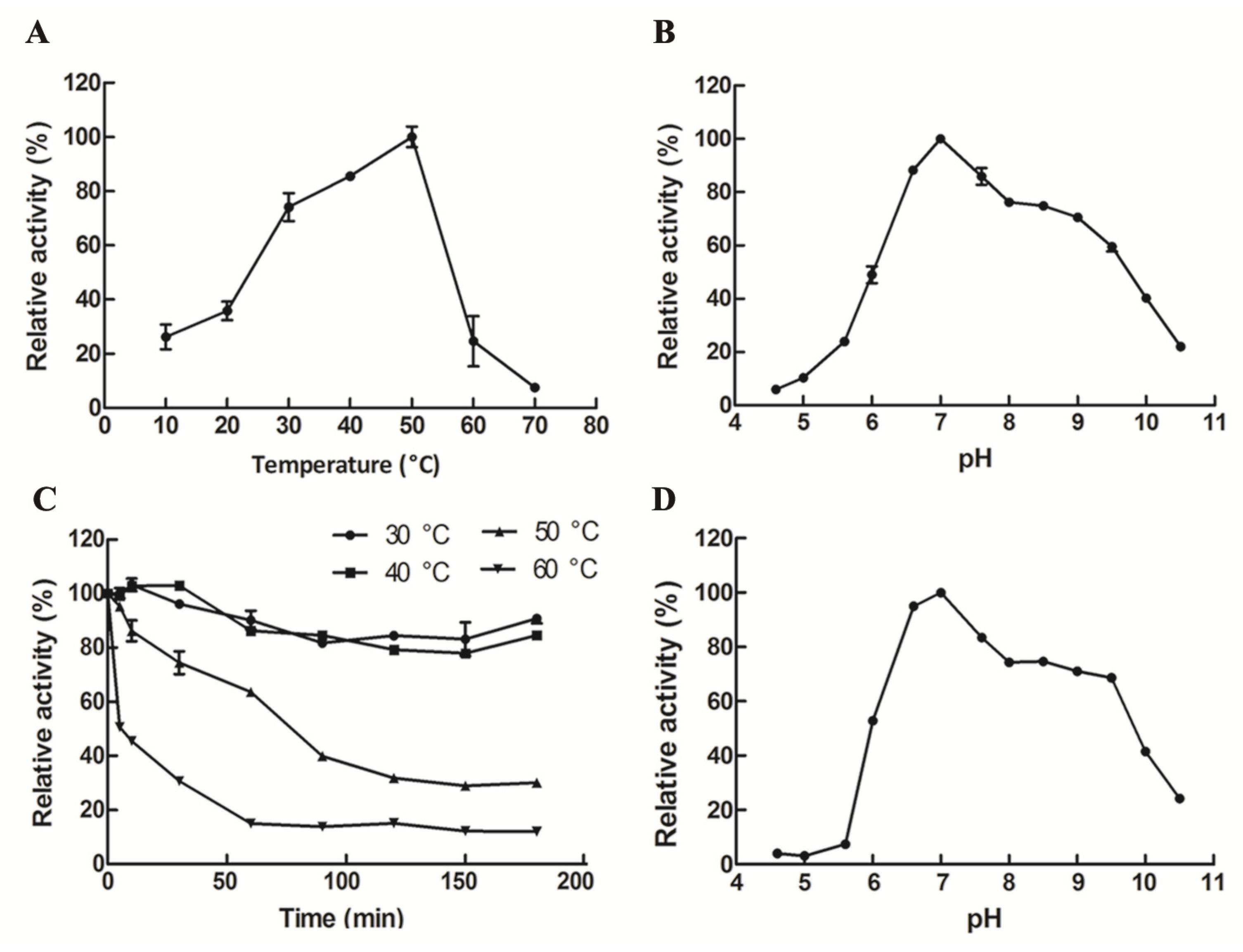
2.2. Expression, Purification, and Characterization of GalA
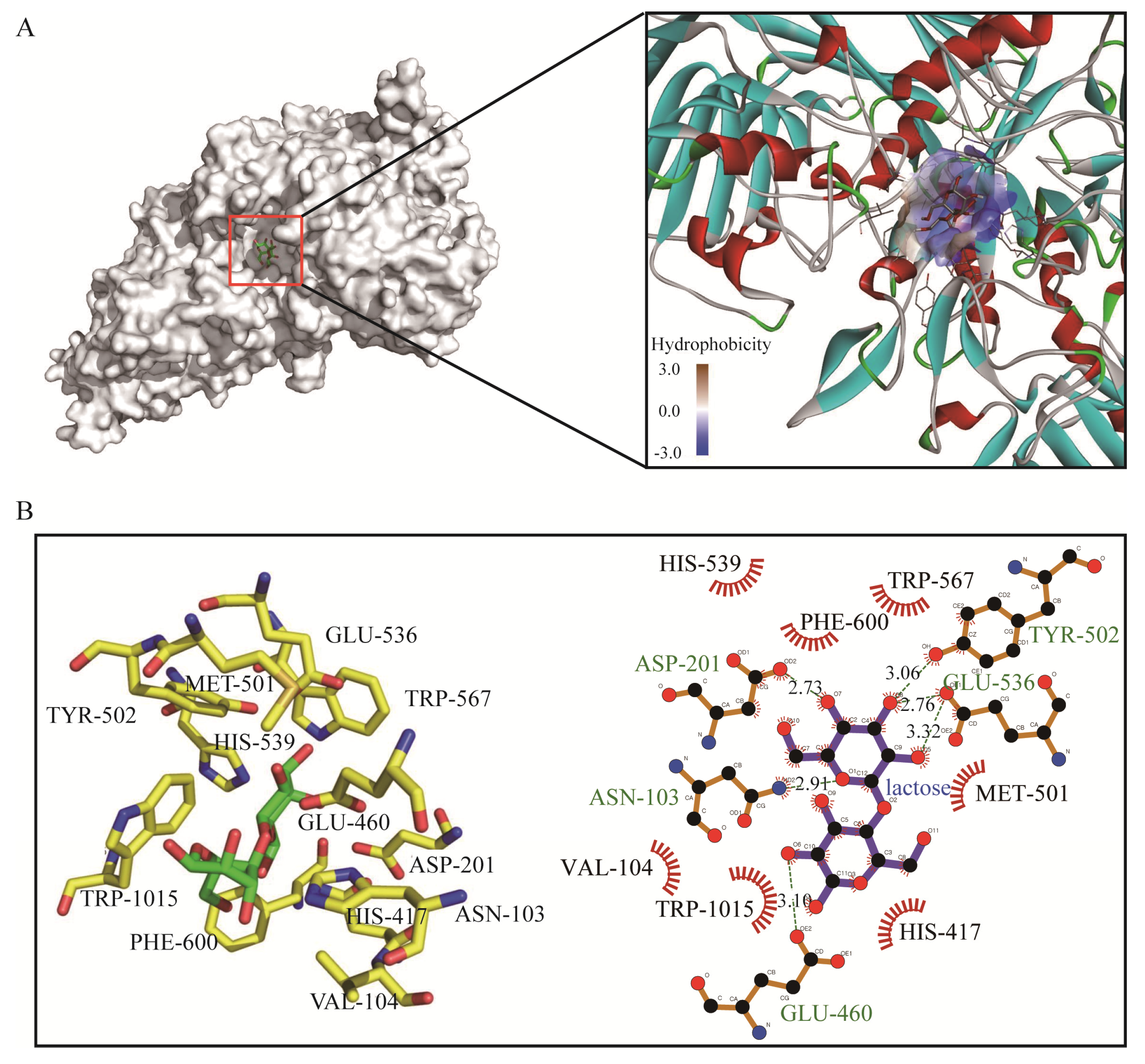
2.3. Docking Analysis of GalA
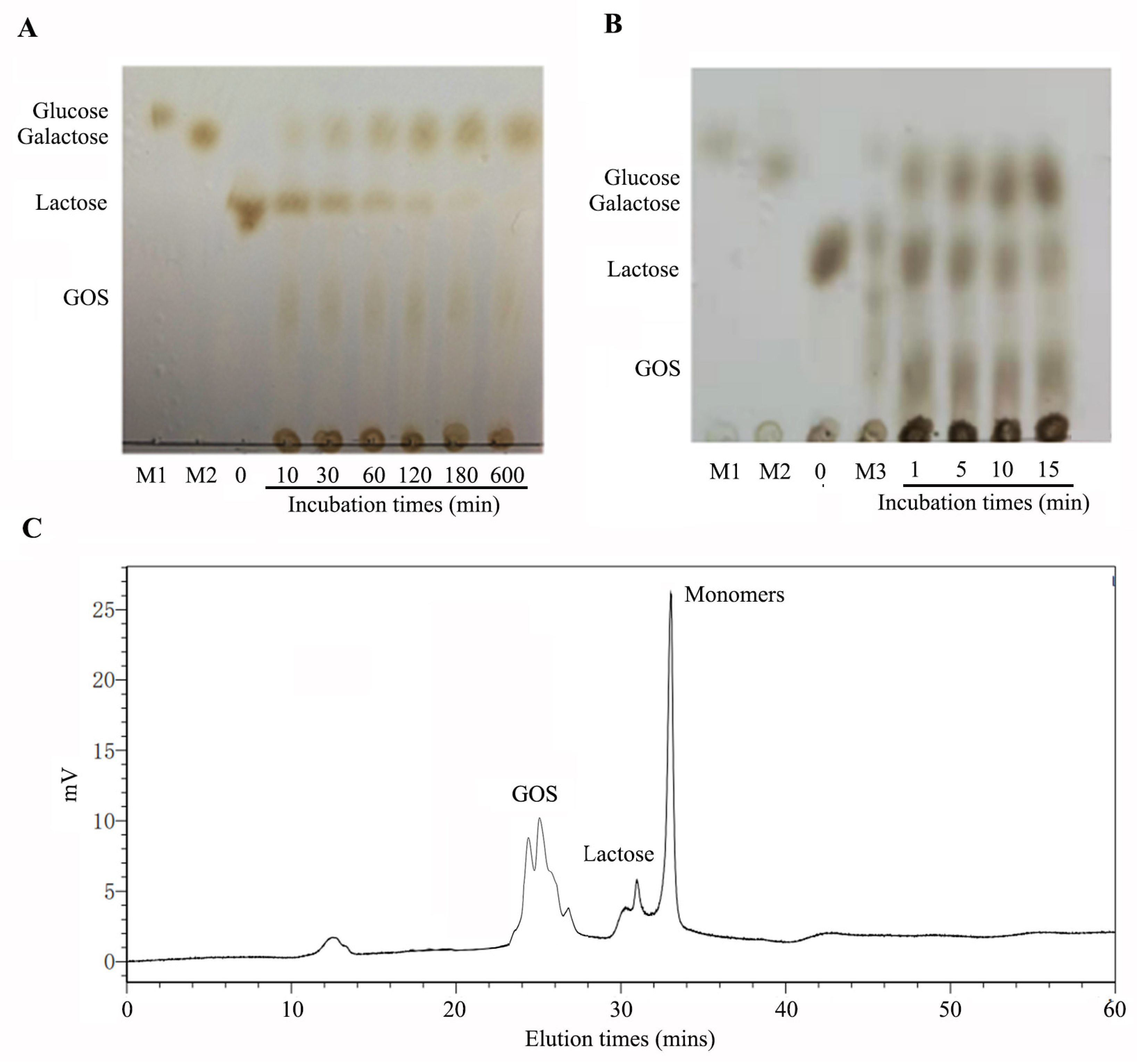
2.4. Hydrolysis of Lactose in Milk
3. Material and Methods
3.1. Bacterial Strains and Cell Culture Conditions
3.2. Sequence Analysis
3.3. Overexpression and Purification of Recombinant GalA
3.4. Characterization of Recombinant GalA
3.5. Docking Analysis
3.6. Activity Assay of β-Galactosidase
3.7. Analysis of the Hydrolysis of Lactose and Galactooligosaccharides Production
3.8. Nucleotide Sequence Accession Numbers
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Saqib, S.; Akram, A.; Halim, S.A.; Tassaduq, R. Sources of β-galactosidase and its applications in food industry. 3 Biotech 2017, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Temuujin, U.; Chi, W.J.; Park, J.S.; Chang, Y.K.; Song, J.Y.; Hong, S.K. Identification and characterization of a novel β-galactosidase from Victivallis vadensis ATCC BAA-548, an anaetobic fecal bacterium. J. Microbiol. 2012, 50, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Splechtna, B.; Nguyen, T.H.; Steinböck, M.; Kulbe, K.D.; Lorenz, W.; Haltrich, D. Production of prebiotic galacto-oligosaccharides from lactose using β-galactosidases from Lactobacillus reuteri. J. Agric. Food Chem. 2006, 54, 4999–5006. [Google Scholar] [CrossRef] [PubMed]
- Park, A.R.; Oh, D.K. Effects of galactose and glucose on the hydrolysis reaction of a thermostable β-galactosidase from Caldicellulosiruptor saccharolyticus. Appl. Microbiol. Biotechnol. 2010, 85, 1427–1435. [Google Scholar] [CrossRef]
- Halbmayr, E.; Mathiesen, G.; Nguyen, T.H.; Maischberger, T.; Peterbauer, C.K.; Eijsink, V.G.H.; Haltrich, D. High-level expression of recombinant β-galactosidases in Lactobacillus plantarum and Lactobacillus sakei using a sakacin P-based expression system. J. Agric. Food Chem. 2008, 56, 4710–4719. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Splechtna, B.; Steinböck, M.; Kneifel, W.; Lettner, P.H.; Kulbe, K.D.; Haltrich, D. Purification and characterization of two novel β-galactosidases from Lactobacillus reuteri. J. Agric. Food Chem. 2006, 54, 4989–4998. [Google Scholar] [CrossRef]
- Møller, P.L.; Jørgensen, F.; Hansen, O.C.; Madsen, S.M.; Stougaard, P. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: Molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 2001, 67, 2276–2283. [Google Scholar]
- Kazemi, S.; Khayati, G.; Faezi-Ghasemi, M. β-galactosidase production by Aspergillus niger ATCC 9142 using inexpensive substrates in solid-state fermentation: Optimization by orthogonal arrays design. Iran. Biomed. J. 2016, 20, 287–294. [Google Scholar]
- Nagy, Z.; Kiss, T.; Szentirmai, A.; Biro, S. β-galactosidase of Penicillium chrysogenum: Production, purification, and characterization of the enzyme. Protein Expr. Purif. 2001, 21, 24–29. [Google Scholar] [CrossRef]
- Zhou, D.Z.K.; Chen, X.D. Effects of temperature and pH on the catalytic activity of the immobilized β-galactosidase from Kluyveromyces lactis. Biochem. Eng. J. 2001, 9, 33–40. [Google Scholar] [CrossRef]
- Ray, I.; Gupta, M.N. Lactose hydrolysis by lactozym TM immobilized on cellulose beads in batch and fluidized bed modes. Process Biochem. 2003, 39, 325–332. [Google Scholar] [CrossRef]
- Seddigh, S.; Darabi, M. Comprehensive analysis of beta-galactosidase protein in plants based on Arabidopsis thaliana. Turk. J. Biol. 2014, 38, 140–150. [Google Scholar] [CrossRef]
- Priebe, M.G.; Zhong, Y.; Huang, C.; Harmsen, H.J.; Raangs, G.C.; Antoine, J.M.; Welling, G.W.; Vonk, R.J. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose intolerant subjects. J. Appl. Microbiol. 2002, 104, 595–604. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef]
- Juajun, O.; Nguyen, T.H.; Maischberger, T.; Iqbal, S.; Haltrich, D.; Yamabhai, M. Cloning, purification, and characterization of β-galactosidase from Bacillus licheniformis DSM 13. Appl. Microbiol. Biotechnol. 2011, 89, 645–654. [Google Scholar] [CrossRef]
- Huber, R.E.; Kurz, G.; Wallenfels, K. A quantitation of the factors which affect the hydrolase and trangalactosylase activities of β-galactosidase (E. coli) on lactose. Biochemistry 1976, 15, 1994–2001. [Google Scholar] [CrossRef]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef]
- Lukito, W.; Malik, S.G.; Surono, I.S.; Wahlqvist, M.L. From ‘lactose intolerance’ to ‘lactose nutrition’. Asian Pac. J. Chin. Nutr. 2015, 24, S1–S8. [Google Scholar]
- Felicilda-Reynaldo, R.F.; Kenneally, M. Digestive enzyme replacement therapy: Pancreatic enzymes and lactase. Medsurg. Nurs. 2016, 25, 182–185. [Google Scholar]
- Heyman, M.B. Committee on Nutrition. Lactose intolerance in infants, children, and adolescents. Pediatrics 2006, 118, 1279–1286. [Google Scholar] [CrossRef]
- Silanikove, N.; Leitner, G.; Merin, U. The interrelationships between lactose intolerance and the modern dairy industry: Global perspectives in evolutional and historical backgrounds. Nutrients 2015, 7, 7312–7331. [Google Scholar] [CrossRef] [PubMed]
- Domingues, L.; Lima, N.; Teixeira, J.A. Aspergillus niger β-galactosidase production by yeast in a continuous high cell density reactor. Process Biochem. 2005, 40, 1151–1154. [Google Scholar] [CrossRef]
- Caputi, V.; Giron, M.C. Microbiome-Gut-Brain Axis and Toll-Like Receptors in Parkinson’s Disease. Int. J. Mol. Sci. 2018, 19, 1689. [Google Scholar] [CrossRef] [PubMed]
- Dembinski, A.; Warzecha, Z.; Ceranowicz, P.; Dembinski, M.; Cieszkowski, J.; Gosiewski, T. Synergic Interaction of Rifaximin and Mutaflor (Escherichia coli Nissle 1917) in the Treatment of Acetic Acid-Induced Colitis in Rats. Gastroenterol. Res. Pract. 2016, 2016, 3126280. [Google Scholar] [CrossRef]
- Miskiewicz, A.; Ceranowicz, P. The Use of Liquids Ionic Fluids as Pharmaceutically Active Substances Helpful in Combating Nosocomial Infections Induced by Klebsiella pneumoniae New Delhi Strain, Acinetobacter baumannii and Enterococcus species. Int. J. Mol. Sci. 2018, 19, 2779. [Google Scholar] [CrossRef]
- Ding, H.; Zhou, L.; Zeng, Q.; Yu, Y.; Chen, B. Heterologous expression of a thermostable β-1,3-galactosidase and its potential in synthesis of galactooligosaccharides. Mar. Drugs 2018, 16, 415. [Google Scholar] [CrossRef]
- Ji, E.S.; Park, N.H.; Oh, D.K. Galacto-oligosaccharide production by a thermostable recombinant β-galactosidase from Thermotoga maritima. World J. Microbiol. Biotechnol. 2005, 21, 759–764. [Google Scholar] [CrossRef]
- Talens-Perales, D.; Górska, A.; Huson, D.H.; Polaina, J.; Marín-Navarro, J. Analysis of domain architecture and phylogenetics of family 2 glycoside hydrolases (GH2). PLoS ONE 2016, 11, e0168035. [Google Scholar] [CrossRef]
- Crittenden, R.G.; Playne, M.J. Production, properties and applications of food-grade oligosaccharides. Trends Food Sci. Technol. 1996, 7, 353–361. [Google Scholar] [CrossRef]
- Warmerdam, A.; Paudel, E.; Jia, W.; Boom, R.M.; Janssen, A.E. Characterization of beta-galactosidase isoforms from Bacillus circulans and their contribution to GOS production. Appl. Biochem. Biotechnol. 2013, 170, 340–358. [Google Scholar] [CrossRef]
- Yanahira, S.; Kobayashi, T.; Suguri, T.; Nakakoshi, M.; Miura, S.; Ishikawa, H.; Nakajima, I. Formation of oligosaccharides from lactose by Bacillus circulans beta-galactosidase. Biosci. Biotechnol. Biochem. 1995, 59, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, F.; Hansen, O.C.; Stougaard, P. High-efficiency synthesis of oligosaccharides with a truncated beta-galactosidase from Bifidobacterium bifidum. Appl. Microbiol. Biotechnol. 2001, 57, 647–652. [Google Scholar] [PubMed]
- Song, J.; Imanaka, H.; Imamura, K.; Minoda, M.; Katase, T.; Hoshi, Y.; Yamaguchi, S.; Nakanishi, K. Cloning and expression of a beta-galactosidase gene of Bacillus circulans. Biosci. Biotechnol. Biochem. 2011, 75, 1194–1197. [Google Scholar] [CrossRef] [PubMed]
- Karan, R.; Capes, M.D.; DasSarma, P.; DasSarma, S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Haloruburm lacusprofundi. BMC Biotechnol. 2013, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Shumway, M.V.; Sheridan, P.P. Site-directed mutagenesis of a family 42 β-galactosidase from an Antarctic bacterium. Int. J. Biochem. Mol. Biol. 2012, 3, 209–218. [Google Scholar] [PubMed]
- Hildebrandt, P.; Wanarska, M.; Kur, J. A new cold-adapted beta-D-galactosidase from the Antarctic Arthrobacter sp. 32c—Gene cloning, overexpression, purification and properties. BMC Microbiol. 2009, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Tang, X.; Gai, Y.; Mehmood, M.; Xiao, X.; Wang, F. Molecular characterization of cold-inducible beta-galactosidase from Arthrobacter sp. ON14 isolated from Antarctica. J. Microbiol. Biotechnol. 2011, 21, 236–242. [Google Scholar]
- Lee, D.H.; Kang, S.G.; Suh, S.G.; Byun, J.K. Purification and characterization of a beta-galactosidase from peach (Prunus persica). Mol. Cells 2003, 15, 68–74. [Google Scholar]
- Husain, Q. Beta galactosidases and their potential applications: A review. Crit. Rev. Biotechnol. 2010, 30, 41–62. [Google Scholar] [CrossRef]
- Chen, W.; Chen, H.; Xia, Y.; Zhao, J.; Tian, F.; Zhang, H. Production, purification, and characterization of a potential thermostable galactosidase for milk lactose hydrolysis from Bacillus stearothermophilus. J. Dairy Sci. 2008, 91, 1751–1758. [Google Scholar] [CrossRef]
- Hoyoux, A.; Jennes, I.; Dubois, P.; Genicot, S.; Dubail, F.; Francois, J.M.; Baise, E.; Feller, G.; Gerday, C. Cold-adapted beta-galactosidase from the antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 2001, 67, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Maischberger, T.; Leitner, E.; Nitisinprasert, S.; Juajun, O.; Yamabhai, M.; Nguyen, T.H.; Haltrich, D. Beta-galactosidase from Lactobacillus pentosus: Purification, characterization and formation of galacto-oligosaccharides. Biotechnol. J. 2010, 5, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Nguyen, T.H.; Nguyen, T.T.; Maischberger, T.; Haltrich, D. beta-Galactosidase from Lactobacillus plantarum WCFS1: Biochemical characterization and formation of prebiotic galacto-oligosaccharides. Carbohydr. Res. 2010, 345, 1408–1416. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.A.; Lee, S.L.; Chou, C. Enzymatic production of galactooligosaccharides by β-galactosidase from Lactobacillus pentosus purification characterization and formation of galacto-oligosaccharides from Bifidobacterium longum BCRC 15708. J. Agric. Food Chem. 2007, 55, 2225–2230. [Google Scholar] [CrossRef] [PubMed]
- Mahdian, S.M.A.; Karimi, E.; Tanipour, M.H.; Parizadeh, S.M.R.; Glayour-Mobarhan, M.; Bazaz, M.M.; Mashkani, B. Expression of a functional cold active β-galactosidase from Planococcus sp.-L4 in Pichia pastoris. Protein Expr. Purif. 2016, 125, 19–25. [Google Scholar] [CrossRef]
- Mahoney, R.R. Enzymes exogenous to milk in dairy, β-d-galactosidase. Encycl. Dairy Sci. 2003, 2, 907–914. [Google Scholar]
- Sun, J.; Yao, C.; Wang, W.; Zhuang, Z.; Liu, J.; Dai, F.; Hao, J. Cloning, expression and characterization of a novel cold-adapted β-galactosidase from the deep-sea bacterium Alteromonas sp. ML52. Mar. Drugs 2018, 16, 469. [Google Scholar] [CrossRef]
| Reagent Added | Concentration (mM) | Relative Activity (%) |
|---|---|---|
| None | - | 100.0 ± 0.0 |
| NaCl | 10 | 54.9 ± 2.3 |
| SDS | 1 | 17.9 ± 3.7 |
| EDTA | 1 | 26.2 ± 3.9 |
| Al2(SO4)3 | 1 | 21.7 ± 3.0 |
| KCI | 1 | 67.5 ± 0.9 |
| CuSO4 | 1 | 8.1 ± 1.0 |
| FeCI2 | 1 | 77.8 ± 4.2 |
| (NH4)2SO4 | 1 | 93.7 ± 1.7 |
| MnSO4 | 1 | 164.1 ± 1.4 |
| Li2SO4 | 1 | 64.7 ± 2.0 |
| ZnCI2 | 1 | 77.2 ± 2.1 |
| BaCI2 | 1 | 26.9 ± 5.1 |
| CoCI2 | 1 | 69.4 ± 2.2 |
| MgCI2 | 1 | 149.0 ± 3.8 |
| CaCI2 | 1 | 82.9 ± 2.2 |
| FeCI3 | 1 | 142.7 ± 1.0 |
| Substrate | Relative Activity (%) 1 |
|---|---|
| 2-Nitrophenyl-β-d-galactopyranoside (ONPG) | 100 ± 7.9 |
| 4-Nitrophenyl-β-d-galactopyranoside (PNPG) | 14.6 ± 3.1 |
| 4-Nitrophenyl-ɑ-d-galactopyranoside | 0 |
| 4-Nitrophenyl-β-d-glucopyranoside | 0 |
| 4-Nitrophenyl-β-d-xylopyranoside | 0 |
| Parameters | Value |
|---|---|
| X coordinate of the center | 81.551 |
| Y coordinate of the center | 130.691 |
| Z coordinate of the center | 103.074 |
| Size in the X dimension | 24 Å |
| Size in the Y dimension | 24 Å |
| Size in the Z dimension | 24 Å |
| Maximum number of binding modes to generate | 9 |
| Maximum energy difference between the best | 4 kcal/mol |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhu, X.; Xing, M. A New β-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides. Mar. Drugs 2019, 17, 599. https://doi.org/10.3390/md17110599
Li S, Zhu X, Xing M. A New β-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides. Marine Drugs. 2019; 17(11):599. https://doi.org/10.3390/md17110599
Chicago/Turabian StyleLi, Shangyong, Xiangjie Zhu, and Mengxin Xing. 2019. "A New β-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides" Marine Drugs 17, no. 11: 599. https://doi.org/10.3390/md17110599
APA StyleLi, S., Zhu, X., & Xing, M. (2019). A New β-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides. Marine Drugs, 17(11), 599. https://doi.org/10.3390/md17110599





