New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis
Abstract
1. Introduction
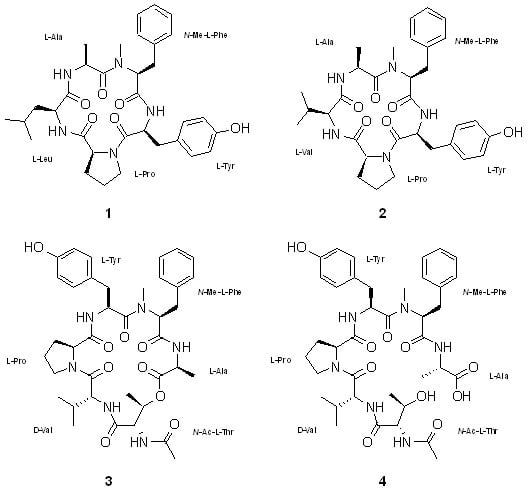
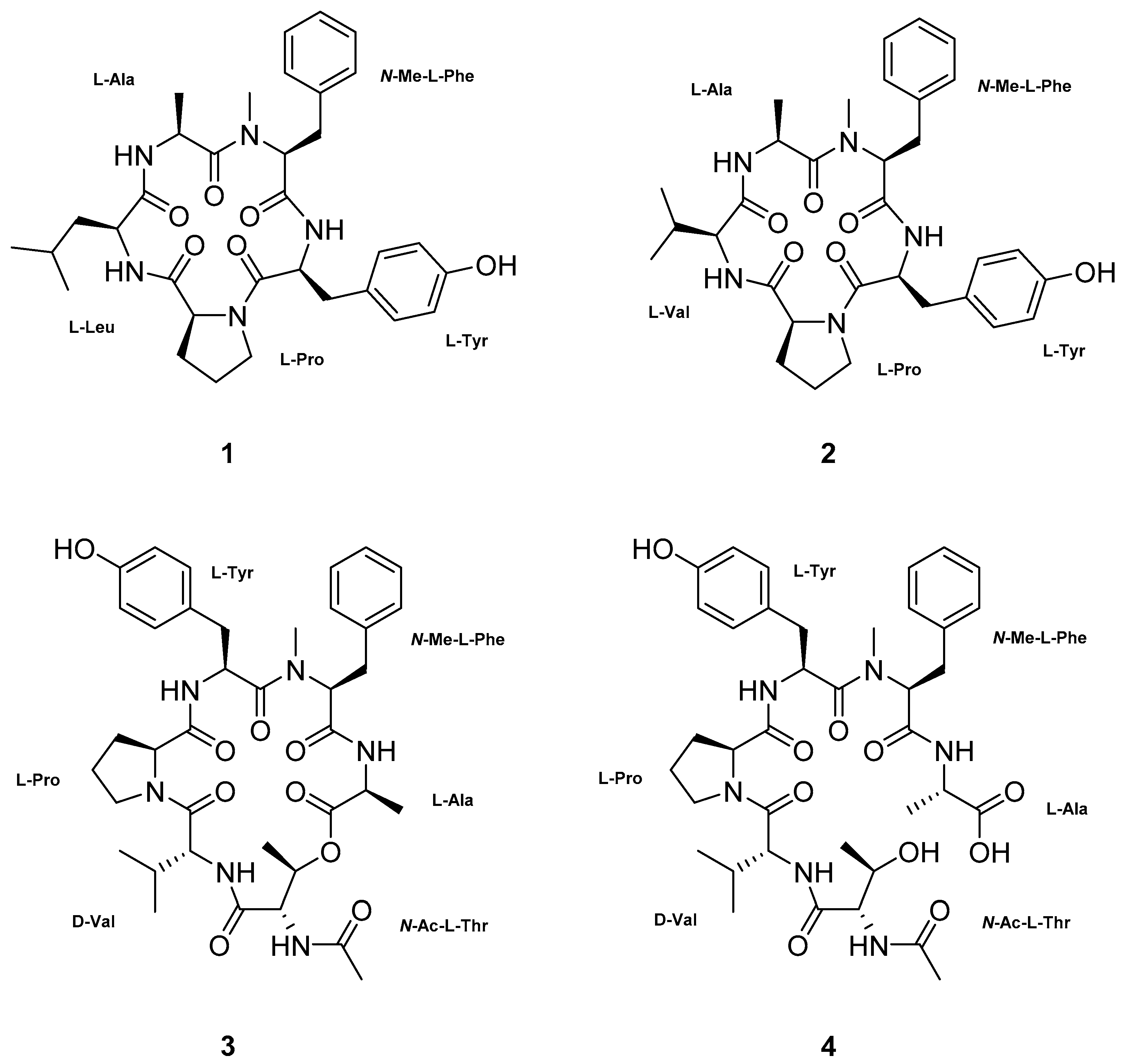
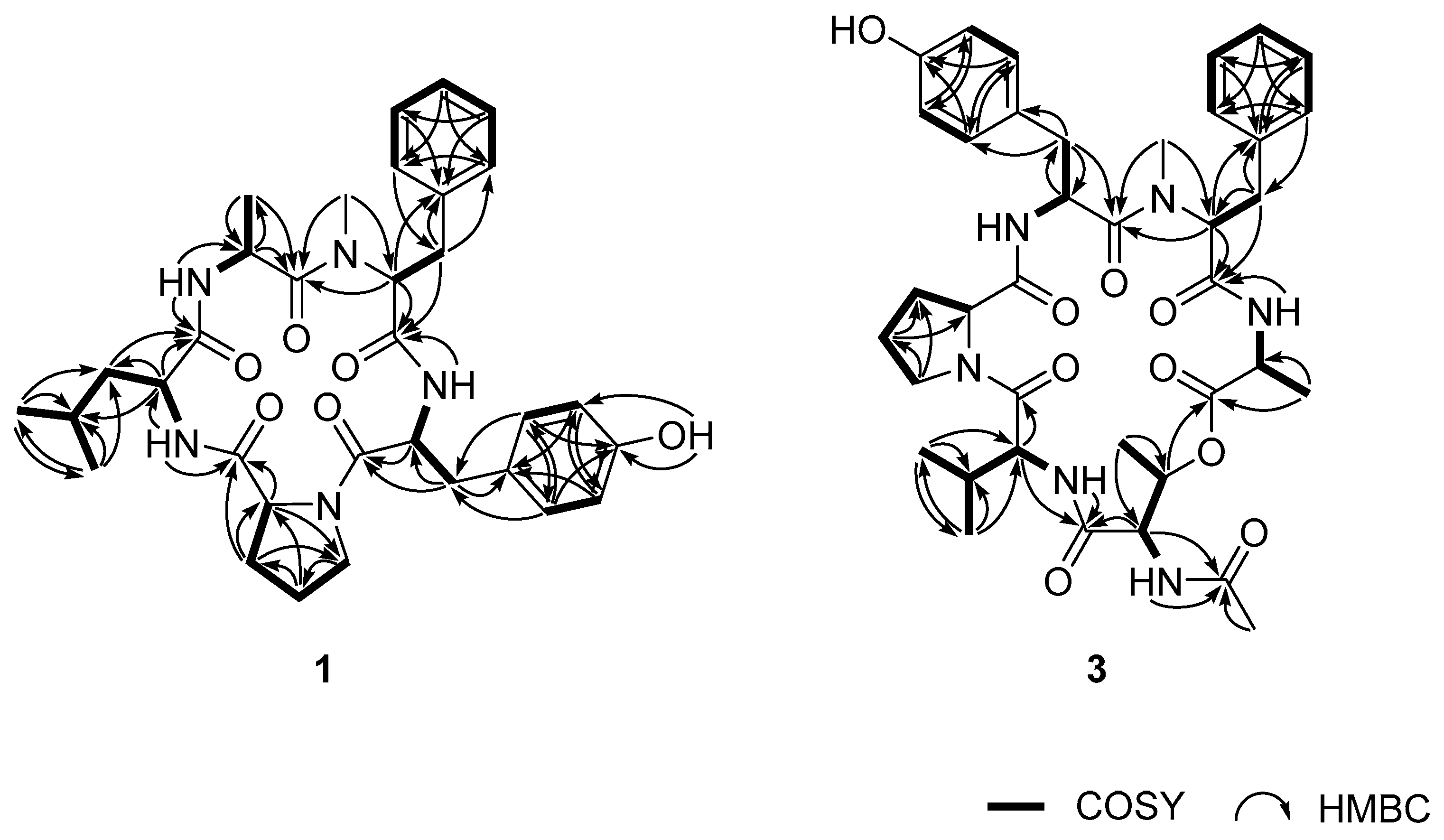
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.2.1. Aspergillus allahabadii (Strain Number JG002)
3.2.2. Aspergillus ochraceopetaliformis (Strain Number FJ120)
3.3. Fermentation
3.3.1. JG002
3.3.2. FJ120
3.4. Extraction and Isolation
3.4.1. JG002
3.4.2. FJ120
3.5. Stereochemical Analysis of the Amino Acid Residues
3.6. Biological Assays
3.6.1. Antibacterial Activity Assay
3.6.2. Antifungal Activity Assay
3.6.3. ICL Inhibition Assay
3.6.4. SrtA Inhibition Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Phyo, Y.Z.; Ribeiro, J.; Fernandes, C.; Kijjoa, A.; Pinto, M.M.M. Marine natural peptides: Determination of absolute configuration using liquid chromatography methods and evaluation of bioactivities. Molecules 2018, 23, 306. [Google Scholar] [CrossRef] [PubMed]
- Balkovec, J.M.; Hughes, D.L.; Masurekar, P.S.; Sable, C.A.; Schwartz, R.E.; Singh, S.B. Discovery and development of first in class antifungal caspofungin (CANCIDAS®)—A case study. Nat. Prod. Rep. 2014, 31, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, M.; Xu, D.; Lai, D.; Zhou, L. Structural diversity and biological activities of fungal cyclic peptides, excluding cyclodipeptides. Molecules 2017, 22, 2069. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Ansari, M.I.; Ahmad, A.; Mishra, M. Major bioactive metabolites from marine fungi: A review. Bioinformation 2015, 11, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wu, W.; Liu, X.; Zaleta-Pinet, D.A.; Clark, B.R. Bioactive compounds isolated from marine-derived microbes in China: 2009–2018. Mar. Drugs 2019, 17, 339. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine fungi: A source of potential anticancer compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef]
- Srivastava, A.; Mishra, V. Marine peptides act as novel chemotherapeutic agent. J. Microbiol. Exp. 2018, 6, 267–270. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef]
- Liao, L.; Bae, S.Y.; Won, T.H.; You, M.; Kim, S.H.; Oh, D.C.; Lee, S.K.; Oh, K.B.; Shin, J. Asperphenin A and B, lipopeptidyl benzophenones from a marine-derived Aspergillus sp. fungus. Org. Lett. 2017, 19, 2066–2069. [Google Scholar] [CrossRef]
- Hu, G.P.; Yuan, J.; Sun, L.; She, Z.G.; Wu, J.H.; Lan, X.J.; Zhu, X.; Lin, Y.C.; Chen, S.P. Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar. Drugs 2011, 9, 514–525. [Google Scholar] [CrossRef]
- Fremlin, L.J.; Piggott, A.M.; Lacey, E.; Capon, R.J. Cottoquinazoline A and Cotteslosins A and B, Metabolites from an Australian Marine-Derived Strain of Aspergillus versicolor. J. Nat. Prod. 2009, 72, 666–670. [Google Scholar] [CrossRef]
- Capon, R.J.; Skene, C.; Stewart, M.; Ford, J.; O’Hair, R.A.J.; Williams, L.; Lacey, E.; Gill, J.H.; Heiland, K.; Friedel, T. Aspergillicins A–E: Five novel depsipeptides from the marine-derived fungus Aspergillus carneus. Org. Biomol. Chem. 2003, 1, 1856–1862. [Google Scholar] [CrossRef]
- Chen, M.; Shao, C.L.; Fu, X.M.; Kong, C.J.; She, Z.G.; Wang, C.Y. Lumazine peptides penilumamides B–D and the cyclic pentapeptide asperpeptide A from a gorgonian-derived Aspergillus sp. fungus. J. Nat. Prod. 2014, 77, 1601–1606. [Google Scholar] [CrossRef]
- Nakamura, I.; Yoshimura, S.; Masaki, T.; Takase, S.; Ohsumi, K.; Hashimoto, M.; Furukawa, S.; Fujie, A. ASP2397: A novel antifungal agent produced by Acremonium persicinum MF-347833. J. Antibiot. 2017, 70, 45–51. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, G.; Jensen, E.R.; Lenoy, E.; Schneewind, O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 2000, 97, 5510–5515. [Google Scholar] [CrossRef]
- Dunn, M.F.; Ramirez-Trujillo, J.A.; Hernández-Lucas, I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 2009, 155, 3166–3175. [Google Scholar] [CrossRef]
- McKinney, J.D.; Höner Zu Bentrup, K.; Muñoz-Elías, E.J.; Miczak, A.; Chen, B.; Chan, W.T.; Swenson, D.; Sacchettini, J.C.; Jacobs, W.R., Jr.; Russell, D.G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 2000, 406, 735–738. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, J.Y.; Shin, J.; Oh, K.B. Inhibitory effects of diketopiperazines from marine-derived Streptomyces puniceus on the isocitrate lyase of Candida albicans. Molecules 2019, 24, 2111. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Samson, R.A.; Peterson, S.W.; Frisvad, J.C.; Varga, J. New species in Aspergillus section Terrei. Stud. Mycol. 2011, 69, 39–55. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; M07; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Methods for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 3rd ed.; M38; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Oh, K.B.; Kim, S.H.; Lee, J.; Cho, W.J.; Lee, T.; Kim, S. Discovery of diarylacrylonitriles as a novel series of small molecule sortase A inhibitors. J. Med. Chem. 2004, 47, 2418–2421. [Google Scholar] [CrossRef]
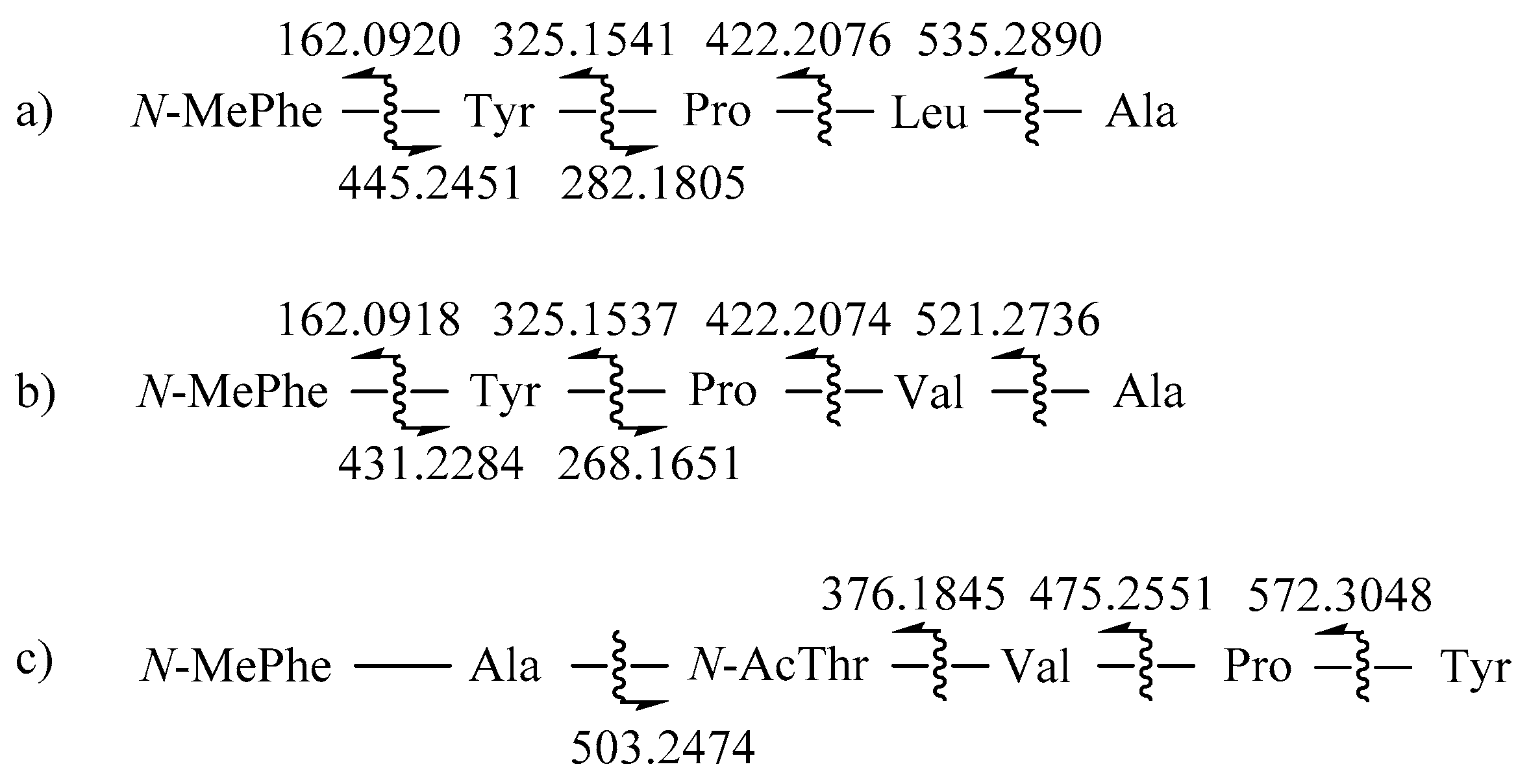
| 1 | 2 | ||||
|---|---|---|---|---|---|
| Unit | Position | δC, Type | δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) |
| Ala | CO | 171,2, C | 171.2, C | ||
| α | 44.2, CH | 3.54, dd (7.9, 6.8) | 43.9, CH | 3.60, dd (8.0, 6.8) | |
| β | 17.1, CH3 | 0.70, d (6.4) | 17.1 CH3 | 0.71, d (6.4) | |
| NH | 8.29, d (8.4) | 8.32, d (8.3) | |||
| N-Me-Phe | CO | 168.1, C | 168.2, C | ||
| α | 61.7, CH | 4.17, dd (11.6, 3.6) | 61.6, CH | 4.16, dd (11.4, 3.4) | |
| β | 33.9, CH2 | 3.24, dd (14.3, 3.3) | 33.9, CH2 | 3.25, dd (14.3, 3.3) | |
| 2.70, dd (14.4, 11.8) | 2.72, dd (14.4, 11.6) | ||||
| γ | 137.5, C | 137.5, C | |||
| ortho | 129.0, CH | 7.09, d (7.2) | 129.0, CH | 7.10, d (7.2) | |
| meta | 128.5, CH | 7.28, d (7.6) | 128.5, CH | 7.28, d (7.6) | |
| para | 126.7, CH | 7.22, t (7.4) | 126.7, CH | 7.22, t (7.4) | |
| N-CH3 | 30.5, CH3 | 2.61, s | 30.5, CH3 | 2.63, s | |
| Tyr | CO | 168.9, C | 168.9, C | ||
| α | 52.6, CH | 4.78, td (8.5, 5.2) | 52.7, CH | 4.76, td (8.5, 5.2) | |
| β | 37.3, CH2 | 3.04, dd (13.4, 8.8) | 37.3, CH2 | 3.02, dd (13.4, 9.0) | |
| 2.73, dd (13.5, 5.7) | 2.74, dd (13.5, 4.7) | ||||
| γ | 127.4, C | 127.4, C | |||
| ortho | 130.2, CH | 7.04, d (8.4) | 130.2, CH | 7.04, d (8.4) | |
| meta | 114.9, CH | 6.66, d (8.4) | 114.9, CH | 6.66, d (8.4) | |
| para | 155.7, C | 155.8, C | |||
| OH | 9.19, s | 9.21, s | |||
| NH | 7.27, m | 7.26, m | |||
| Pro | CO | 170.6, C | 170.6, C | ||
| α | 60.9, CH | 4.10, dd (7.9, 1.7) | 60.9, CH | 4.10, dd (7.3, 2.6) | |
| β | 31.5, CH2 | 1.95, m | 31.5, CH2 | 1.93, m | |
| 1.88, m | |||||
| γ | 21.4, CH2 | 1.77, m | 21.5, CH2 | 1.78, m | |
| 1.58, m | 1.61, dq (12.3, 9.1) | ||||
| δ | 46.4, CH2 | 3.47, m | 46.2, CH2 | 3.49, m | |
| 3.37, m | 3.35, m | ||||
| Leu | CO | 170.4, C | |||
| α | 53.3, CH | 4.19, m | |||
| β | 41.3, CH2 | 1.37, m | |||
| 1.34, m | |||||
| γ | 24.5, CH | 1.37, m | |||
| δ | 22.4, CH3 | 0.88, d (6.1) | |||
| 21.7, CH3 | 0.80, d (6.2) | ||||
| NH | 6.96, d (9.0) | ||||
| Val | CO | 169.2, C | |||
| α | 61.0, CH | 3.82, t (9.6) | |||
| β | 30.7, CH | 1.69, m | |||
| γ | 19.3, CH3 | 0.80, d (6.6) | |||
| 18.9, CH3 | 0.78, d (6.7) | ||||
| NH | 7.00, d (8.7) | ||||
| 3 a | 4 b | ||||
|---|---|---|---|---|---|
| Unit | Position | δC, Type | δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) |
| Val | CO | 172.0, C | 172.8, C | ||
| α | 57.1, CH | 4.43, t (9.8) | 59.4, CH | 4.10, d (9.0) | |
| β | 30.3, CH | 1.89, m | 30.8, CH | 1.97, m | |
| γ | 19.2, CH3 | 0.86, d (6.5) | 19.4, CH3 | 1.02, d (6.6) | |
| 18.8, CH3 | 0.92, d (6.5) | 19.2, CH3 | 0.91, d (6.8) | ||
| NH | 6.43, d (9.1) | ||||
| Pro | CO | 173.3, C | 173.7, C | ||
| α | 59.5, CH | 4.31, d (5.8) | 61.7, CH | 4.35, dd (8.3, 2.4) | |
| β | 29.7, CH2 | 2.13, m | 30.5, CH2 | 1.88, m | |
| 1.74, m | |||||
| γ | 24.7, CH2 | 1.88, m | 24.2, CH2 | 1.63, m | |
| 1.04, m | |||||
| δ | 48.1, CH2 | 3.93, m | 48.6, CH2 | 3.72, m | |
| 3.52, m | 3.46, m | ||||
| Tyr | CO | 172.4, C | 173.9, C | ||
| α | 50.6, CH | 4.54, dd (13.3, 6.6) | 51.2, CH | 4.62, dd (11.2, 4.0) | |
| β | 35.9, CH2 | 2.42, dd (13.6, 9.2) | 36.0, CH2 | 2.63, dd (13.1, 11.5) | |
| 1.87, dd (13.2, 6.2) | 1.43, dd (13.2, 3.5) | ||||
| γ | 126.6, C | 129.2, C | |||
| ortho | 130.2, CH | 6.77, d (7.8) | 131.5, CH | 6.92, d (8.3) | |
| meta | 115.9, CH | 6.66, d (8.0) | 115.9, CH | 6.64, d (8.4) | |
| para | 155.9, C | 157.3, C | |||
| NH | 6.44, d (9.1) | ||||
| N-Me-Phe | CO | 168.1, C | 170.5, C | ||
| α | 63.2, CH | 4.82, t (6.8) | 63.7, CH | 5.28, dd (10.9, 3.4) | |
| β | 34.5, CH2 | 3.34, dd (14.0, 5.7) | 35.4, CH2 | 3.16, dd (14.3, 3.5) | |
| 2.52, dd (13.9, 7.8) | 2.97, dd (14.1, 9.3) | ||||
| γ | 137.9, C | 139.4, C | |||
| ortho | 129.4, CH | 7.13, d (7.4) | 130.7, CH | 7.28, d (7.2) | |
| meta | 129.1, CH | 7.24, d (7.5) | 130.1, CH | 7.30, d (7.3) | |
| para | 127.3, CH | 7.23, d (7.3) | 128.1, CH | 7.17, d (7.5) | |
| N-CH3 | 29.5, CH3 | 2.87, s | 30.2, CH3 | 2.90, s | |
| Ala | CO | 170.9, C | 180.8, C | ||
| α | 48.5, CH | 4.67, d (7.9) | 52.1, CH | 4.19, m | |
| β | 18.6, CH3 | 1.32, d (7.3) | 18.5, CH3 | 1.34, d (6.5) | |
| NH | 8.16, d (8.3) | ||||
| N-Ac-Thr | CO | 169.7, C | 173.0, C | ||
| α | 55.4, CH | 4.69, m | 59.7, CH | 4.47, d (5.2) | |
| β | 70.1, CH | 5.53, q (5.9) | 68.6, CH | 4.17, q (5.9) | |
| γ | 17.0, CH3 | 1.18, d (6.5) | 19.7, CH3 | 1.21, d (6.4) | |
| NH | 6.29, d (9.0) | ||||
| CO | 171.8, C | 174.1, C | |||
| α | 23.1, CH3 | 1.83, s | 22.8, CH3 | 2.09, s | |
| IC50 (μM) | ||
|---|---|---|
| Compound | SrtA Inhibition | ICL Inhibition |
| 1 | 70.0 | >128 |
| 2 | 53.1 | 116.8 |
| 3 | 131.9 | >128 |
| 4 | 77.0 | >128 |
| Berberine chloride a | 104.3 | |
| Curcumin a | 47.8 | |
| 3-Nitropropionic acid a | 18.5 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.-Y.; Lee, J.-H.; Park, S.C.; Lee, J.; Oh, D.-C.; Oh, K.-B.; Shin, J. New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Mar. Drugs 2019, 17, 488. https://doi.org/10.3390/md17090488
Hwang J-Y, Lee J-H, Park SC, Lee J, Oh D-C, Oh K-B, Shin J. New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Marine Drugs. 2019; 17(9):488. https://doi.org/10.3390/md17090488
Chicago/Turabian StyleHwang, Ji-Yeon, Jung-Ho Lee, Sung Chul Park, Jayho Lee, Dong-Chan Oh, Ki-Bong Oh, and Jongheon Shin. 2019. "New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis" Marine Drugs 17, no. 9: 488. https://doi.org/10.3390/md17090488
APA StyleHwang, J.-Y., Lee, J.-H., Park, S. C., Lee, J., Oh, D.-C., Oh, K.-B., & Shin, J. (2019). New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Marine Drugs, 17(9), 488. https://doi.org/10.3390/md17090488