Zinc-Chelating Mechanism of Sea Cucumber (Stichopus japonicus)-Derived Synthetic Peptides
Abstract
1. Introduction
2. Results and Discussion
2.1. Determination of Zinc Chelating Capacity of Synthetic ZCPs
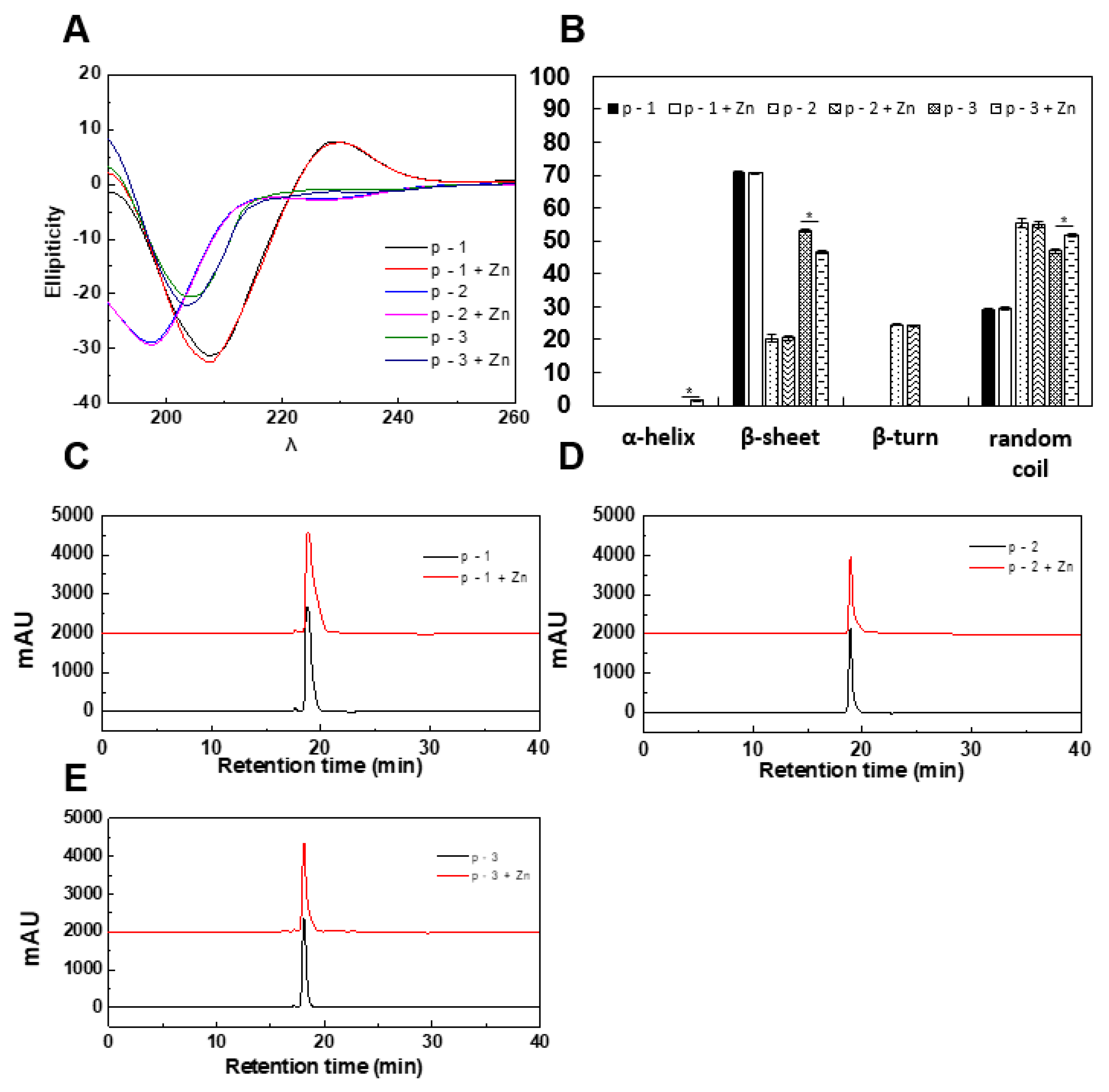
2.2. Evaluation of Structural Conformation of ZCPs after Chelation with Zinc
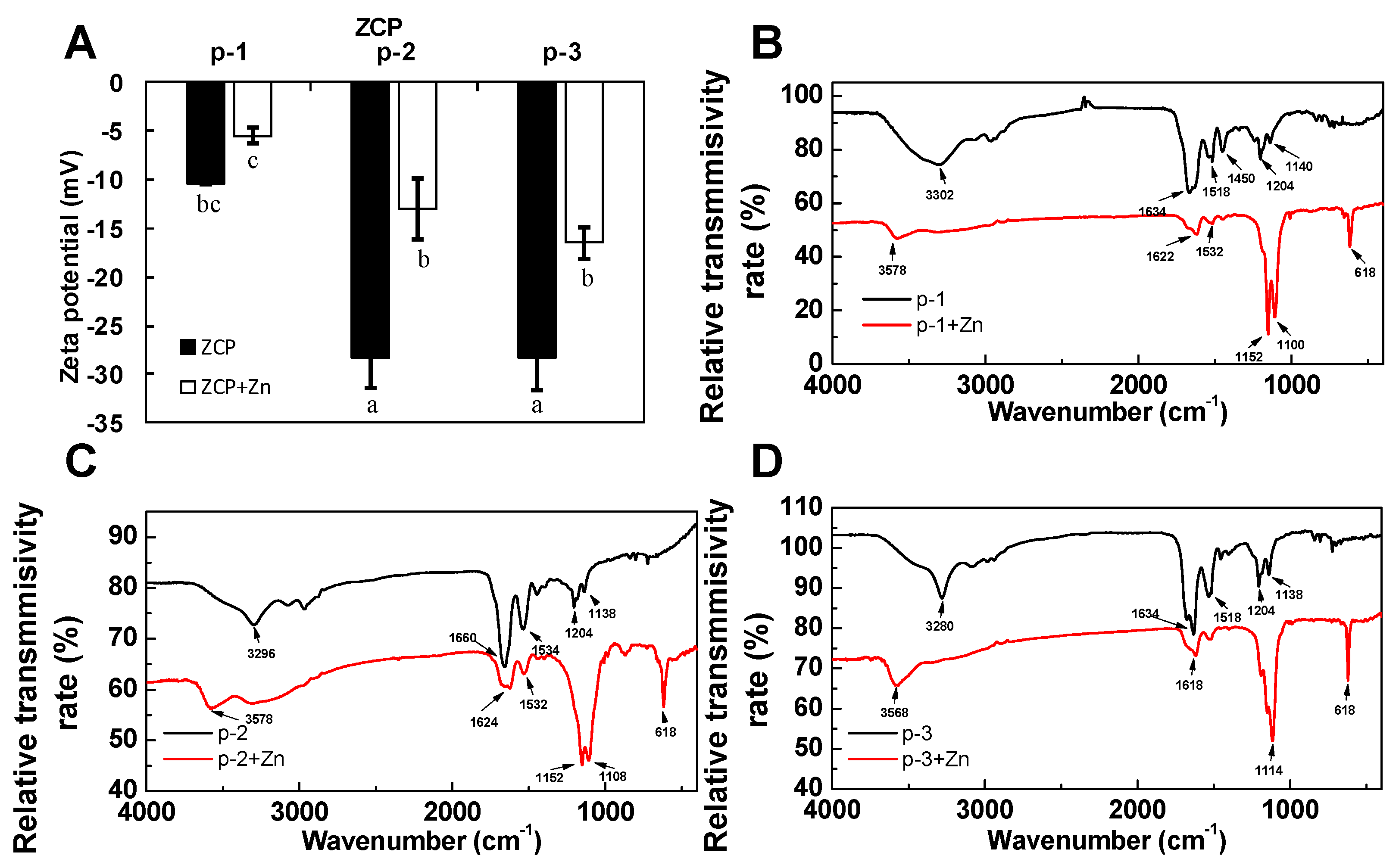
2.3. Evaluation of Chelation by Zeta-Potential Analysis and FTIR
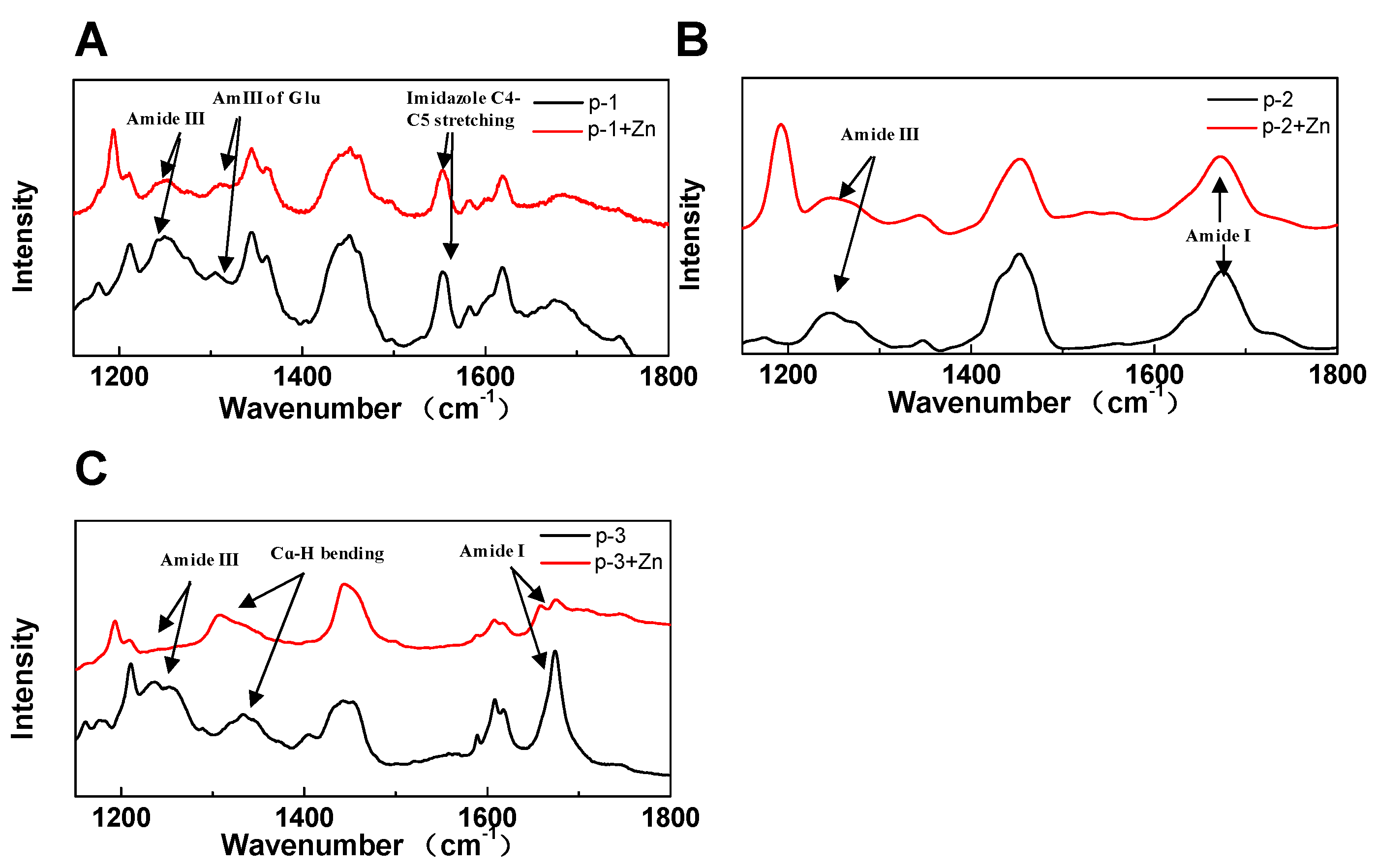
2.4. Raman Spectroscopy
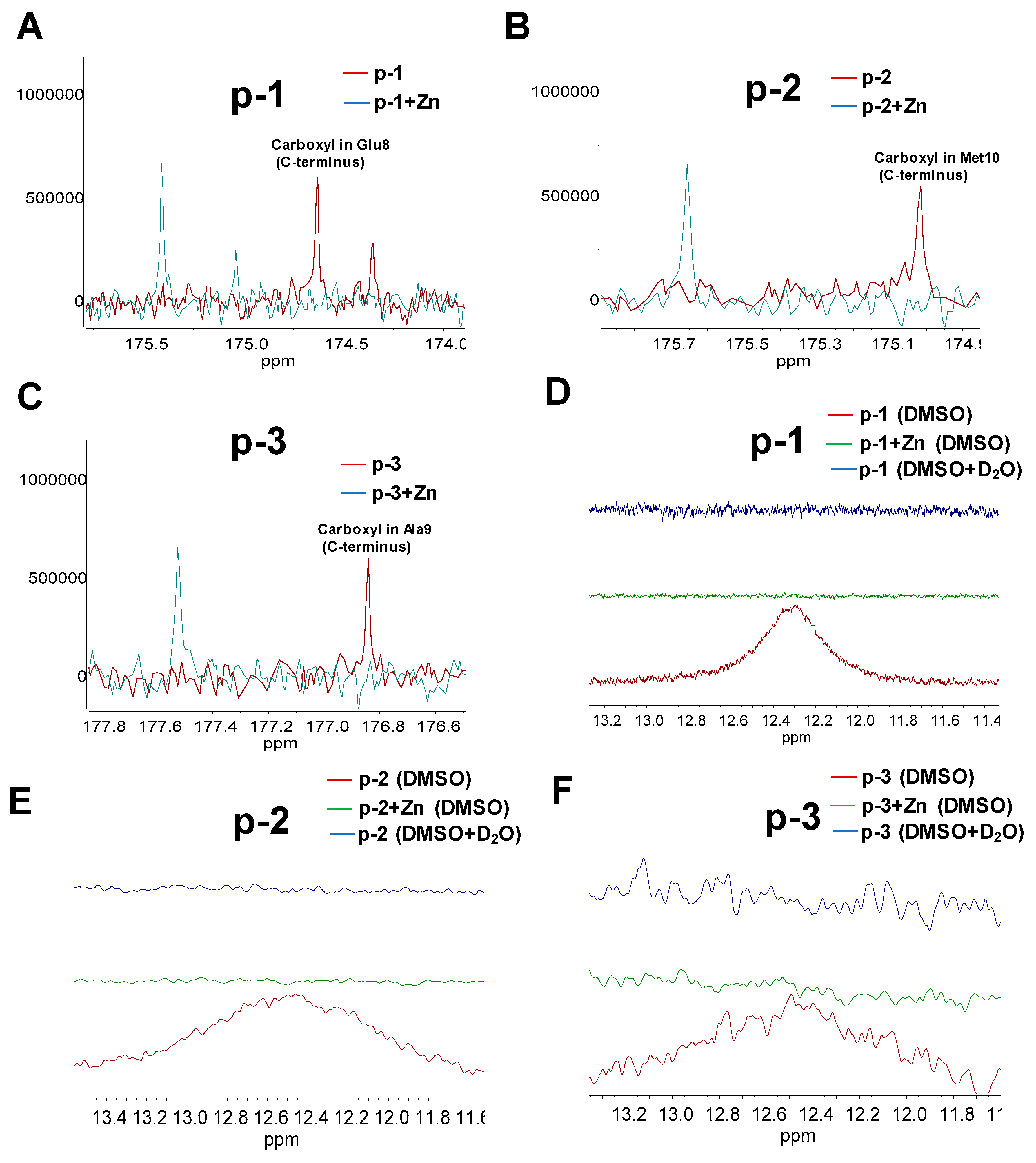
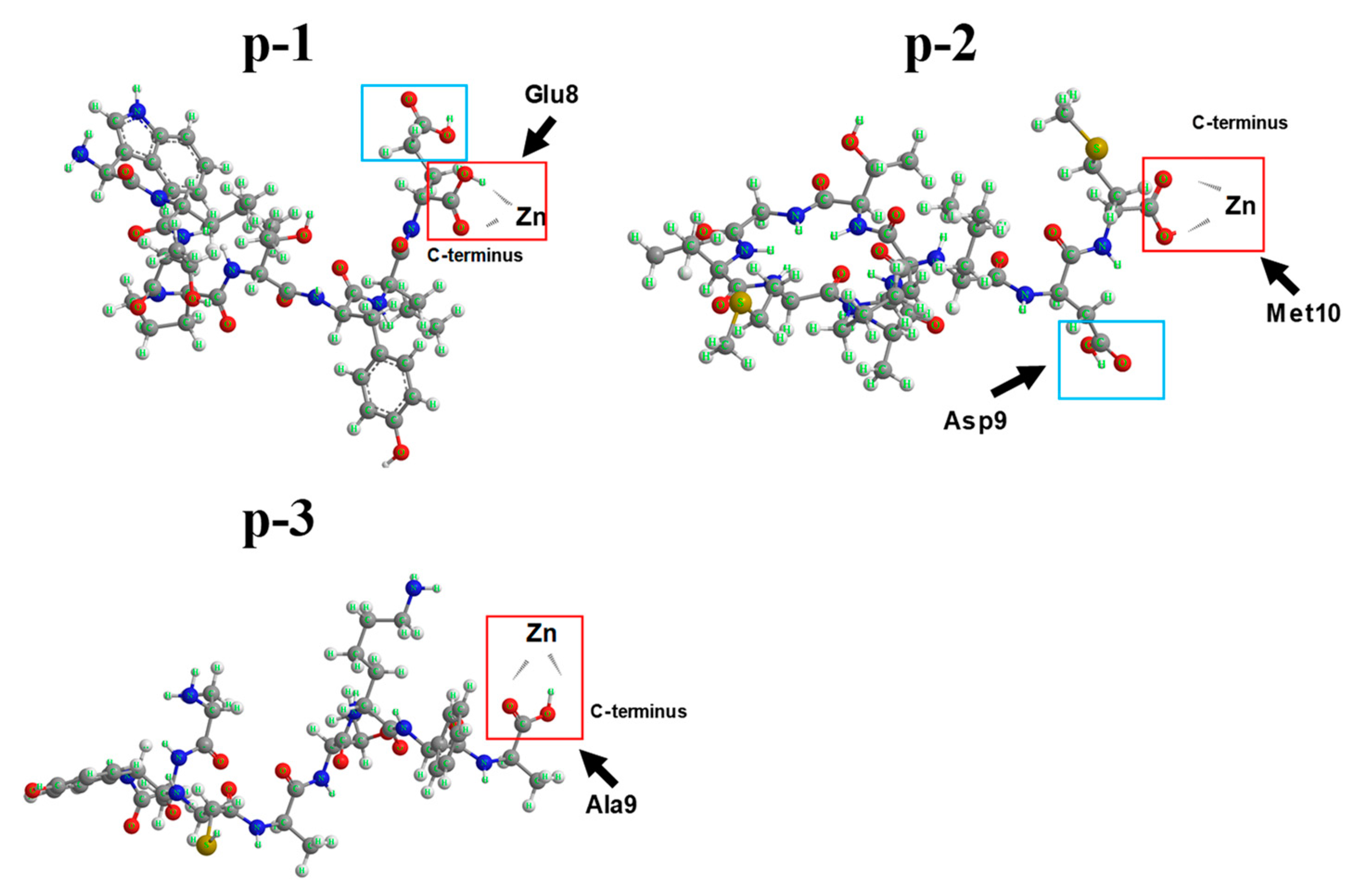
2.5. NMR
3. Materials and Methods
3.1. Materials
3.2. Determination of Zinc-Chelating Capacity
3.3. Flame Atomic Absorption Spectroscopy
3.4. Circular Dichroism (CD) Spectroscopy
3.5. Size Exclusion Chromatography (SEC)
3.6. Zeta-Potential Measurement
3.7. FTIR Spectroscopy
3.8. Raman Spectroscopy
3.9. NMR
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ZCP | zinc-chelating peptide |
| DI water | Distilled water |
| CSD | Chemical shift deviation |
| PAR | 4-(2-pyridylazo) |
| FAAS | Flame Atomic Absorption Spectroscopy |
| CD | Circular Dichroism |
| SEC | Size Exclusion Chromatography |
| FTIR | Fourier Transform infrared spectroscopy |
| NMR | Nuclear Magnetic Resonance |
| MW | molecular weight |
References
- Bozalioǧlu, S.; Özkan, Y.; Turan, M.; Şimşek, B. Prevalence of zinc deficiency and immune response in short-term hemodialysis. J. Trace Elem. Med. Biol. 2005, 18, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Sandström, B.; Lönnerdal, B. The effect of casein phosphopeptides on zinc and calcium absorption from high phytate infant diets assessed in rat pups and Caco-2 cells. Pediatr. Res. 1996, 40, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sinha, A.K.; Makkar, H.P.S.; Becker, K. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem. 2010, 120, 945–959. [Google Scholar] [CrossRef]
- Etcheverry, P.; Grusak, M.A.; Fleige, L.E. Application of in vitro bioaccessibility and bioavailability methods for calcium, carotenoids, folate, iron, magnesium, polyphenols, zinc, and vitamins B6, B12, D, and E. Front. Physiol. 2012, 3, 317. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Harnedy, P.A.; Li, B.; Hou, H.; Zhang, Z.; Zhao, X.; FitzGerald, R.J. Food protein-derived chelating peptides: Biofunctional ingredients for dietary mineral bioavailability enhancement. Trends Food Sci. Technol. 2014, 37, 92–105. [Google Scholar] [CrossRef]
- Sóvágó, I.; Kállay, C.; Várnagy, K. Peptides as complexing agents: Factors influencing the structure and thermodynamic stability of peptide complexes. Coord. Chem. Rev. 2012, 256, 2225–2233. [Google Scholar] [CrossRef]
- Raics, M.; Sanna, D.; Sóvágó, I.; Kállay, C. Copper(II), nickel(II) and zinc(II) complexes of hexapeptides containing separate aspartyl and histidyl residues. Inorga. Chim. Acta 2015, 426, 99–106. [Google Scholar] [CrossRef]
- Pinkaew, S.; Winichagoon, P.; Hurrell, R.F.; Wegmuller, R. Extruded rice grains fortified with zinc, iron, and vitamin A increase zinc status of Thai school children when incorporated into a school lunch program. J. Nutr. 2013, 143, 362–368. [Google Scholar] [CrossRef]
- Chen, D.; Liu, Z.; Huang, W.; Zhao, Y.; Dong, S.; Zeng, M. Purification and characterisation of a zinc-binding peptide from oyster protein hydrolysate. J. Funct. Foods 2013, 5, 689–697. [Google Scholar] [CrossRef]
- Glahn, R.P.; Van, C.D.R. Iron uptake is enhanced in Caco-2 cell monolayers by cysteine and reduced cysteinyl glycine. J. Nutr. 1997, 127, 642–647. [Google Scholar] [CrossRef]
- Torres Fuentes, C.; Alaiz, M.; Vioque, J. Affinity purification and characterisation of chelating peptides from chickpea protein hydrolysates. Food Chem. 2011, 129, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H. Isolation of a calcium-binding peptide from enzymatic hydrolysates of porcine blood plasma protein. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 290–294. [Google Scholar] [CrossRef]
- Wang, C.; Li, B.; Ao, J. Separation and identification of zinc-chelating peptides from sesame protein hydrolysate using IMAC-Zn2+and LC-MS/MS. Food Chem. 2012, 134, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Torres Fuentes, C.; Alaiz, M.; Vioque, J. Iron-chelating activity of chickpea protein hydrolysate peptides. Food Chem. 2012, 134, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Ren, L.; Jiang, J. Purification of a histidine-containing peptide with calcium binding activity from shrimp processing byproducts hydrolysate. Eur. Food Res. Technol. 2011, 232, 281–287. [Google Scholar] [CrossRef]
- Lv, Y.; Bao, X.L.; Yang, B.C.; Ren, C.G.; Guo, S.T. Effect of soluble soybean protein hydrolysate-calcium complexes on calcium uptake by Caco-2 cells. J. Food Sci. 2008, 73, 1750–3841. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Mahoney, R.R. Binding of iron by chicken muscle protein digests: The size of the iron-binding peptides. J. Sci. Food Agric. 2000, 80, 1595–1600. [Google Scholar] [CrossRef]
- Khanal, R.C.; Nemere, I. Regulation of intestinal calcium transport. Annu. Rev. Nutr. 2008, 28, 179–196. [Google Scholar] [CrossRef]
- Yang, D.S.; Mclaurin, J.; Qin, K.; Westaway, D.; Fraser, P.E. Examining the zinc binding site of the amyloid-β peptide. Eur. J. Biochem. 2000, 267, 6692–6698. [Google Scholar] [CrossRef]
- Worthington, M.T.; Amann, B.T.; Nathans, D.; Berg, J.M. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc. Natl. Acad. Sci. USA 1996, 93, 13754–13759. [Google Scholar] [CrossRef]
- Gerbino, E.; Mobili, P.; Tymczyszyn, E.; Fausto, R.; Gómez Zavaglia, A. FTIR spectroscopy structural analysis of the interaction between Lactobacillus kefir S-layers and metal ions. J. Mol. Struct. 2011, 987, 186–192. [Google Scholar] [CrossRef]
- Kirin, S.I.; Ott, I.; Gust, R.; Mier, W.; Weyhermüller, T.; Metzler-Nolte, N. Cellular uptake quantification of metalated peptide and peptide nucleic acid bioconjugates by atomic absorption spectroscopy. Angew. Chem. Int. Ed. 2008, 47, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Udechukwu, M.C.; Downey, B.; Udenigwe, C.C. Influence of structural and surface properties of whey-derived peptides on zinc-chelating capacity, and in vitro gastric stability and bioaccessibility of the zinc-peptide complexes. Food Chem. 2018, 240, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.X.; Wang, X.P.; Guo, X.N. Isolation and characterization of zinc-chelating peptides from wheat germ protein hydrolysates. J. Funct. Foods 2015, 12, 23–32. [Google Scholar] [CrossRef]
- Curtain, C.C.; Ali, F.; Volitakis, I.; Cherny, R.A.; Norton, R.S.; Beyreuther, K.; Barrow, C.J.; Masters, C.L.; Bush, A.I.; Barnham, K.J. Alzheimer’s disease amyloid-β binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J. Biol. Chem. 2001, 276, 20466–20473. [Google Scholar] [CrossRef] [PubMed]
- Cnudde, S.E.; Prorok, M.; Jia, X.; Castellino, F.J.; Geiger, J.H. The crystal structure of the calcium-bound con-G[Q6A] peptide reveals a novel metal-dependent helical trimer. J. Biol. Inorg. Chem. 2011, 16, 257–266. [Google Scholar] [CrossRef]
- Grøndahl, L.; Sokolenko, N.; Abbenante, G.; Fairlie, D.P.; Hanson, G.R.; Gahan, L.R. Interaction of zinc(II) with the cyclic octapeptides, cyclo[Ile(Oxn)-D-Val(Thz)]2 and ascidiacyclamide, a cyclic peptide from Lissoclinum patella. J. Chem. Soc. Dalt. Trans. 1999, 578, 1227–1234. [Google Scholar] [CrossRef]
- Broncel, M.; Wagner, S.C.; Paul, K.; Hackenberger, C.P.R.; Koksch, B. Towards understanding secondary structure transitions: Phosphorylation and metal coordination in model peptides. Org. Biomol. Chem. 2010, 8, 2575–2579. [Google Scholar] [CrossRef]
- Liu, M.; Tang, Y.C.; Fan, K.Q.; Jiang, X.; Lai, L.H.; Ye, Y.H. Cyclization of several linear penta and heptapeptides with different metal ions studied by CD spectroscopy. J. Pept. Res. 2005, 65, 55–64. [Google Scholar] [CrossRef]
- Su, G.; Ren, J.; Zhao, M.; Sun, D.W. Comparison of superdex peptide HR 10/30 column and TSK gel G2000 SWxl column for molecular weight distribution analysis of protein hydrolysates. Food Bioprocess. Technol. 2013, 6, 3620–3626. [Google Scholar] [CrossRef]
- Koscielski, T.; Sybilska, D.; Jljrczak, J. Separation of hydrophobic peptide polymers by size-exclusion and reversed-phase high-performance liquid chromatography. J. Chromatogr. 1984, 280, 131–134. [Google Scholar]
- Mayo, K.H.; Ilyina, E. A folding pathway for betapep-4 peptide 33mer: from unfolded monomers and beta-sheet sandwich dimers to well-structured tetramers. Protein Sci. 1998, 7, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Cabibbo, A.; Pagani, M.; Fabbri, M.; Rocchi, M.; Farmery, M.R.; Bulleid, N.J.; Sitia, R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 2000, 275, 4827–4833. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Rajarathnam, K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR 2000, 18, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Zidane, F.; Matéos, A.; Cakir-Kiefer, C.; Miclo, L.; Rahuel-Clermont, S.; Girardet, J.M.; Corbier, C. Binding of divalent metal ions to 1-25 β-caseinophosphopeptide: An isothermal titration calorimetry study. Food Chem. 2012, 132, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Schwaminger, S.P.; Blank-Shim, S.A.; Scheifele, I.; Fraga-García, P.; Berensmeier, S. Peptide binding to metal oxide nanoparticles. Faraday Discuss. 2017, 204, 233–250. [Google Scholar] [CrossRef]
- Kaasalainen, M.; Mäkilä, E.; Riikonen, J.; Kovalainen, M.; Järvinen, K.; Herzig, K.H.; Lehto, V.P.; Salonen, J. Effect of isotonic solutions and peptide adsorption on zeta potential of porous silicon nanoparticle drug delivery formulations. Int. J. Pharm. 2012, 431, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kosmulski, M. Surface charging and points of zero charge. In Surface Charging and Points of Zero Charge; CRC Press: Boca Raton, FL, USA, 2009; pp. 1–1065. [Google Scholar]
- Lupaescu, A.V.; Jureschi, M.; Ciobanu, C.I.; Ion, L.; Zbancioc, G.; Petre, B.A.; Drochioiu, G. FTIR and MS evidence for heavy metal binding to anti-amyloidal NAP-like peptides. Int. J. Pept. Res. Ther. 2018, 25, 303–309. [Google Scholar] [CrossRef]
- Murariu, M.; Habasescu, L.; Ciobanu, C.I.; Gradinaru, R.V.; Pui, A.; Drochioiu, G.; Mangalagiu, I. Interaction of amyloid Aβ(9–16) peptide fragment with metal ions: CD, FT-IR, and fluorescence spectroscopic studies. Int. J. Pept. Res. Ther. 2018, 1–13. [Google Scholar] [CrossRef]
- Murariu, M.; Dragan, E.S.; Drochioiu, G. Model peptide-based system used for the investigation of metal ions binding to histidine-containing polypeptides. Biopolymers 2010, 93, 497–508. [Google Scholar] [CrossRef]
- Mikhonin, A.V.; Ahmed, Z.; Ianoul, A.; Asher, S.A. Assignments and conformational dependencies of the amide III peptide backbone UV resonance Raman bands. J. Phys. Chem. B 2004, 108, 19020–19028. [Google Scholar] [CrossRef]
- Miura, T.; Mitani, S.; Takanashi, C.; Mochizuki, N. Copper selectively triggers β-sheet assembly of an N-terminally truncated amyloid β-peptide beginning with Glu3. J. Inorg. Biochem. 2004, 98, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Yasutake, A.; Aisawa, S.; Takahashi, S.; Hirahara, H.; Narita, E. Synthesis of biopolymer intercalated inorganic-layered materials: Intercalation of collagen peptide and soybean peptide into Zn-Al layered double hydroxide and layered zinc hydroxide. J. Phys. Chem. Solids 2008, 69, 1542–1546. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, Y.; Guo, S.; Lin, N.; Deng, L.; Yue, T.; Huang, F. Identifying Cu(II)-amyloid peptide binding intermediates in the early stages of aggregation by resonance Raman spectroscopy: A simulation study. Phys. Chem. Chem. Phys. 2017, 19, 31103–31112. [Google Scholar] [CrossRef] [PubMed]
- Razmiafshari, M.; Kao, J.; D’Avignon, A.; Zawia, N.H. NMR identification of heavy metal-binding sites in a synthetic zinc finger peptide: Toxicological implications for the interactions of xenobiotic metals with zinc finger proteins. Toxicol. Appl. Pharmacol. 2001, 172, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, J.; Pierattelli, R.; Banci, L.; Gräslund, A. High-resolution NMR studies of the zinc-binding site of the Alzheimer’s amyloid β-peptide. FEBS J. 2007, 274, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.P.; Hansch, C. Use of 13C NMR chemical shift as QSAR/QSPR descriptor. Chem. Rev. 2011, 111, 2865–2899. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef]
- Zheng, A.; Yang, M.; Yue, Y.; Ye, C.; Deng, F. 13C NMR shielding tensors of carboxyl carbon in amino acids calculated by ONIOM method. Chem. Phys. Lett. 2004, 399, 172–176. [Google Scholar] [CrossRef]
- Azócar, M.I.; Aldabaldetrecu, M.; Levin, P.; Tamayo, L.; Guerrero, J.; Páez, M.A. Correlating light and thermal stability of silver carboxylate complexes by infrared and 13C NMR spectroscopy. J. Coord. Chem. 2016, 69, 3472–3479. [Google Scholar] [CrossRef]
- Kieffer, B.; Mer, G.; Mann, A.; Lefèvre, J.F. Structural studies of two antiaggregant RGDW peptides by lH and 13C NMR. Int. J. Pept. Protein Res. 1994, 44, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, K.L.; Tubergen, P.J.; Swartz, M.A.; DeVries, J.T.; Tatko, C.D. Zinc binding with L-dopa peptides. Inorga. Chim. Acta 2017, 461, 120–126. [Google Scholar] [CrossRef]
- Yang, J.T.; Wu, C.S.C.; Martinez, H.M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986, 130, 208–269. [Google Scholar] [PubMed]

| ZCP:Zinc (c:c) | p-1 (ug mg−1) | p-2 (ug mg−1) | p-3 (ug mg−1) |
|---|---|---|---|
| 1:0.5 | 1.10 ± 0.01a | 0.88 ± 0.01a | 1.54 ± 0.19a |
| 1:1 | 2.34 ± 0.03a | 2.03 ± 0.07a | 2.01 ± 0.27ab |
| 1:2 | 2.27 ± 0.05b | 1.91 ± 0.11b | 2.03 ± 0.03b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wang, Z.; Yin, F.; Liu, Y.; Qin, N.; Nakamura, Y.; Shahidi, F.; Yu, C.; Zhou, D.; Zhu, B. Zinc-Chelating Mechanism of Sea Cucumber (Stichopus japonicus)-Derived Synthetic Peptides. Mar. Drugs 2019, 17, 438. https://doi.org/10.3390/md17080438
Liu X, Wang Z, Yin F, Liu Y, Qin N, Nakamura Y, Shahidi F, Yu C, Zhou D, Zhu B. Zinc-Chelating Mechanism of Sea Cucumber (Stichopus japonicus)-Derived Synthetic Peptides. Marine Drugs. 2019; 17(8):438. https://doi.org/10.3390/md17080438
Chicago/Turabian StyleLiu, Xiaoyang, Zixu Wang, Fawen Yin, Yuxin Liu, Ningbo Qin, Yoshimasa Nakamura, Fereidoon Shahidi, Chenxu Yu, Dayong Zhou, and Beiwei Zhu. 2019. "Zinc-Chelating Mechanism of Sea Cucumber (Stichopus japonicus)-Derived Synthetic Peptides" Marine Drugs 17, no. 8: 438. https://doi.org/10.3390/md17080438
APA StyleLiu, X., Wang, Z., Yin, F., Liu, Y., Qin, N., Nakamura, Y., Shahidi, F., Yu, C., Zhou, D., & Zhu, B. (2019). Zinc-Chelating Mechanism of Sea Cucumber (Stichopus japonicus)-Derived Synthetic Peptides. Marine Drugs, 17(8), 438. https://doi.org/10.3390/md17080438