Sea Anemone Toxins: A Structural Overview
Abstract
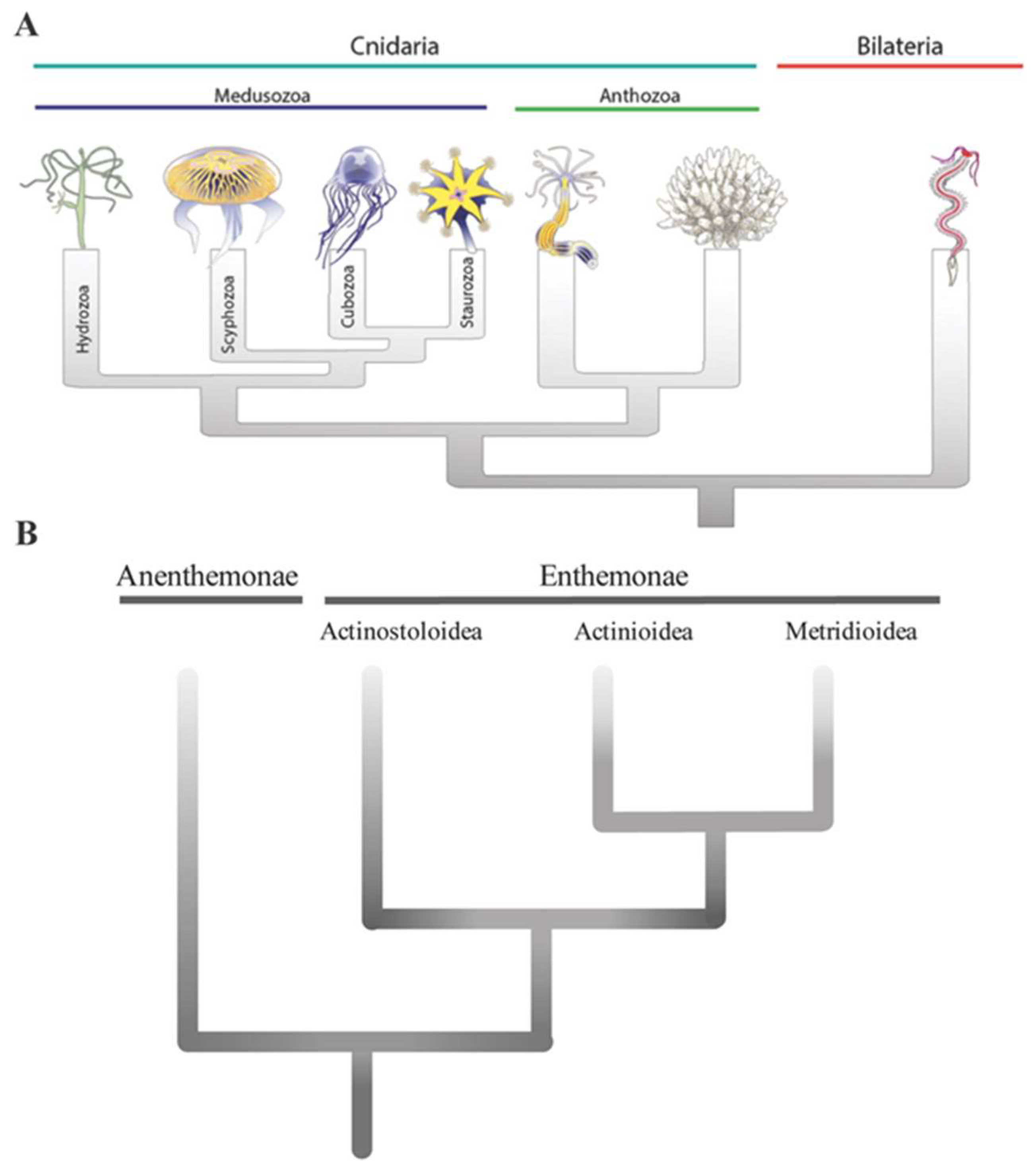
1. Introduction
2. Venom Tissue
3. Venom Composition
3.1. Non-Proteinaceous Venom Components
3.2. Enzymes
3.3. Non-Enzymatic Proteins – Cytotoxins
3.4. Non-Enzymatic Proteins – Neurotoxins
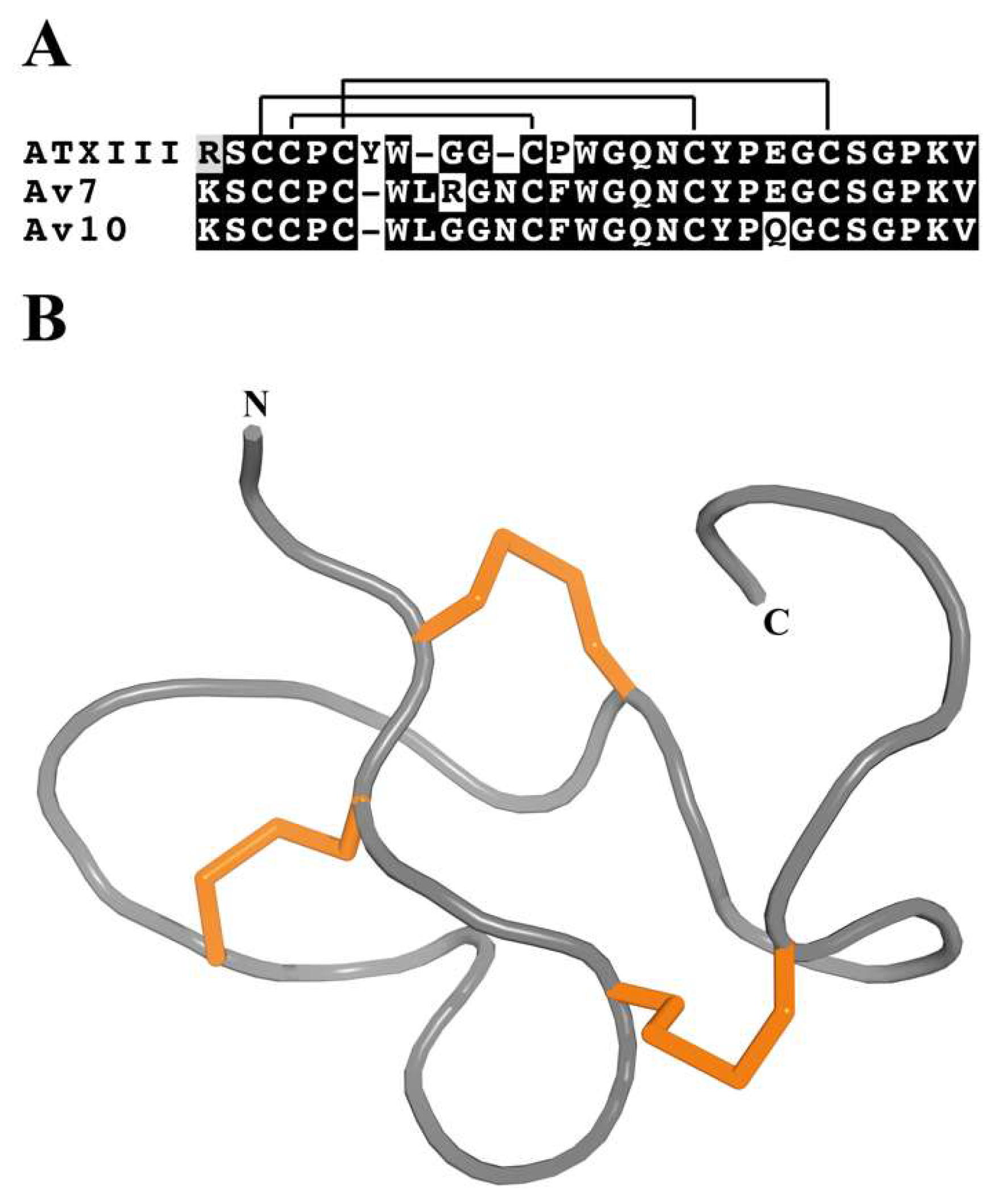
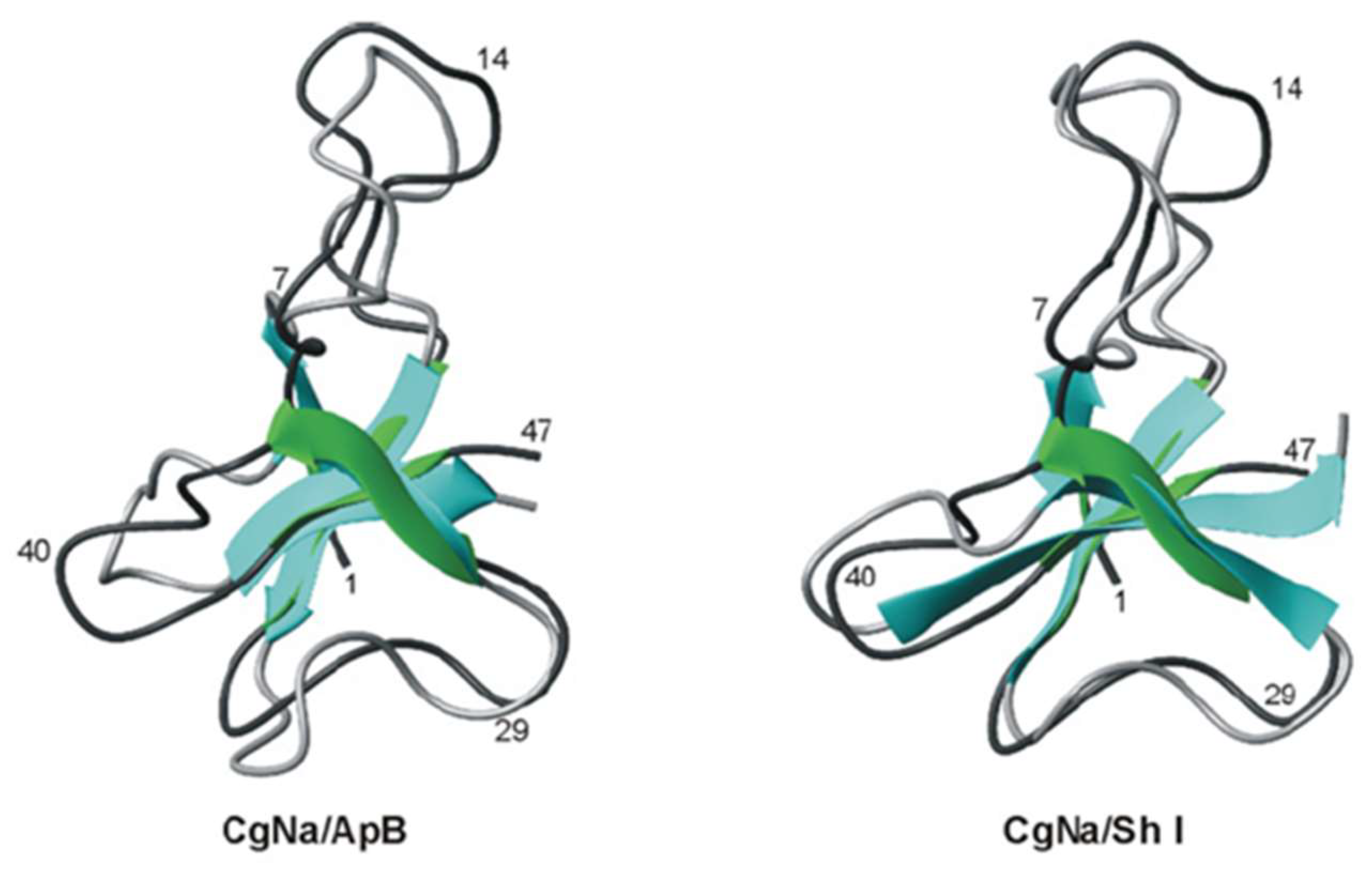
3.4.1. ATX III
3.4.2. β-Defensins
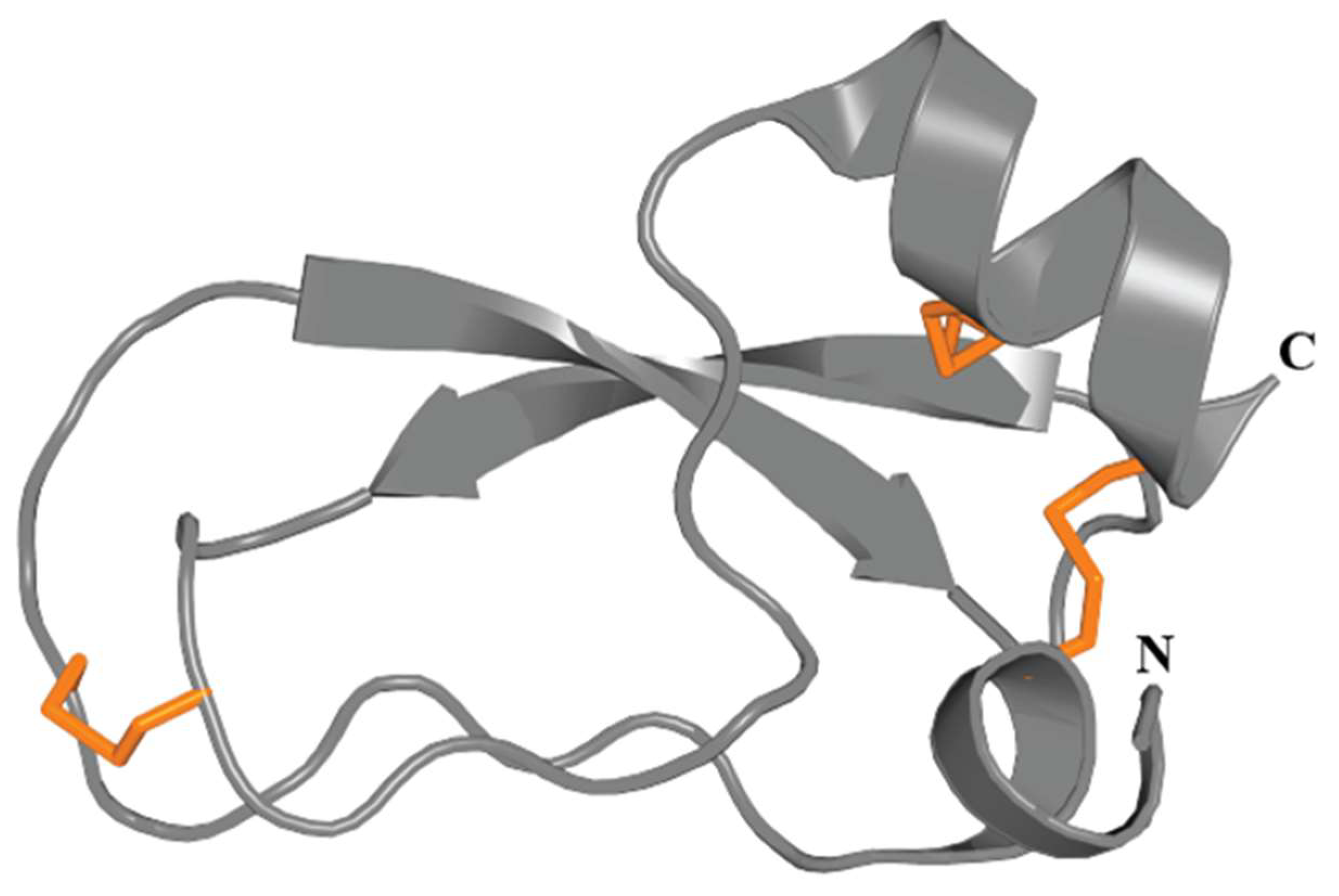
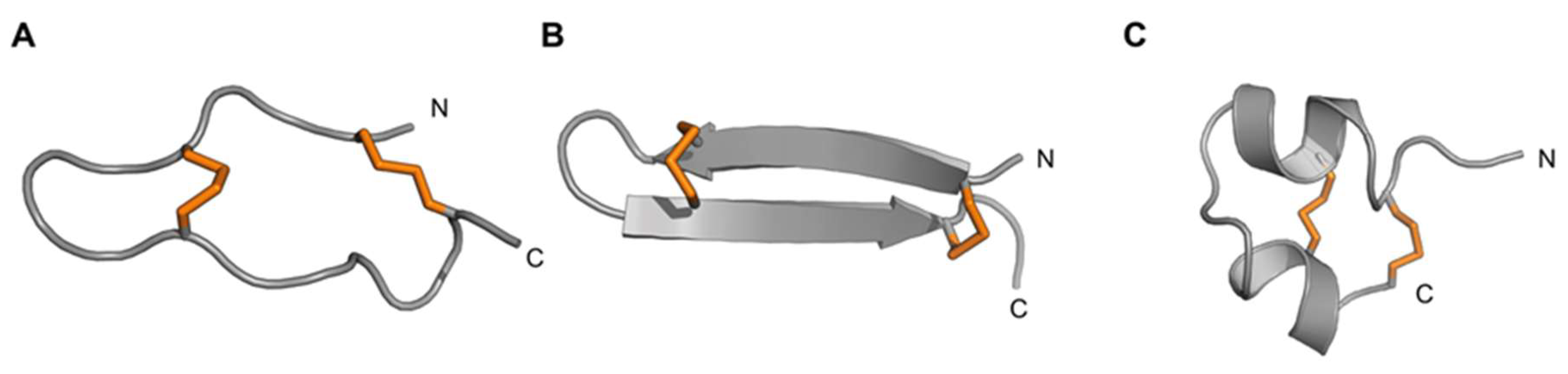
3.4.3. Boundless β-Hairpin
3.4.4. EGF-Like Peptides
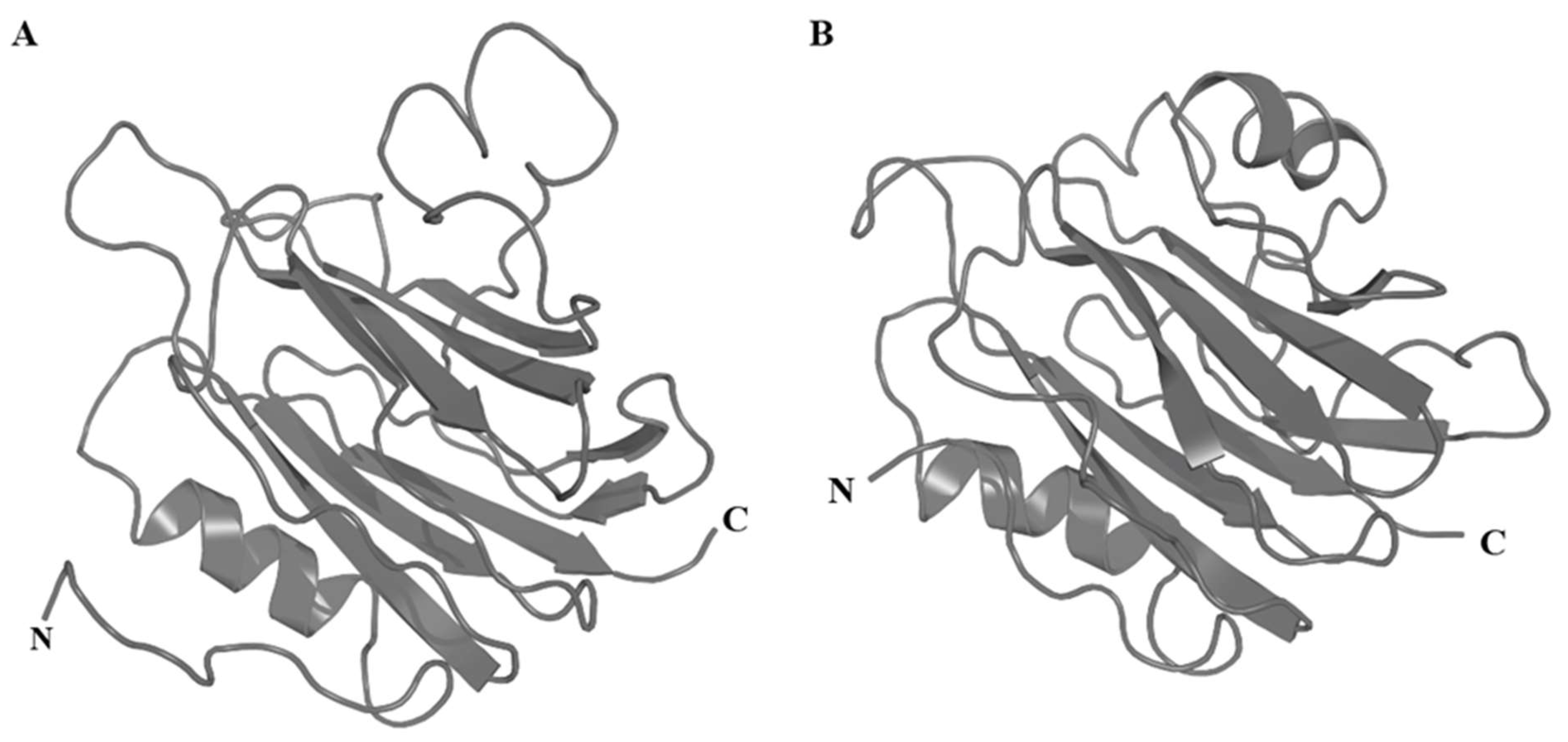
3.4.5. Inhibitor Cystine Knot Fold
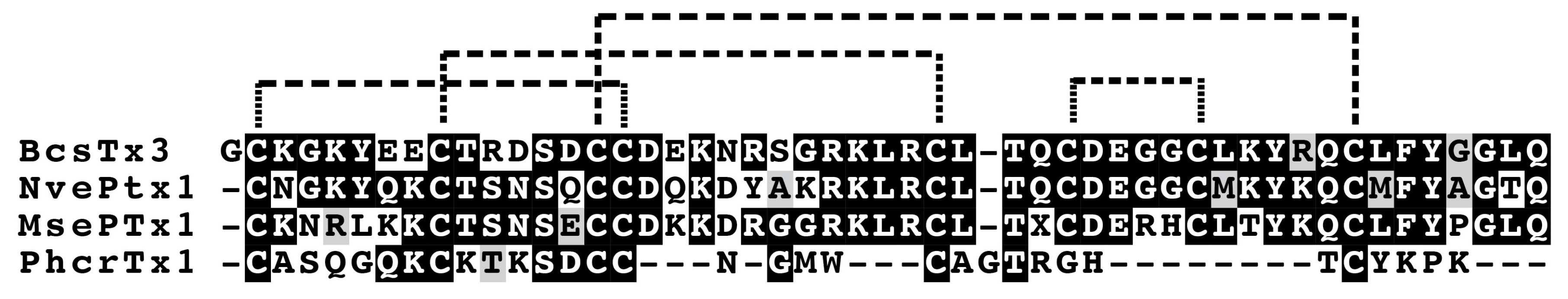
3.4.6. Kunitz-Domain
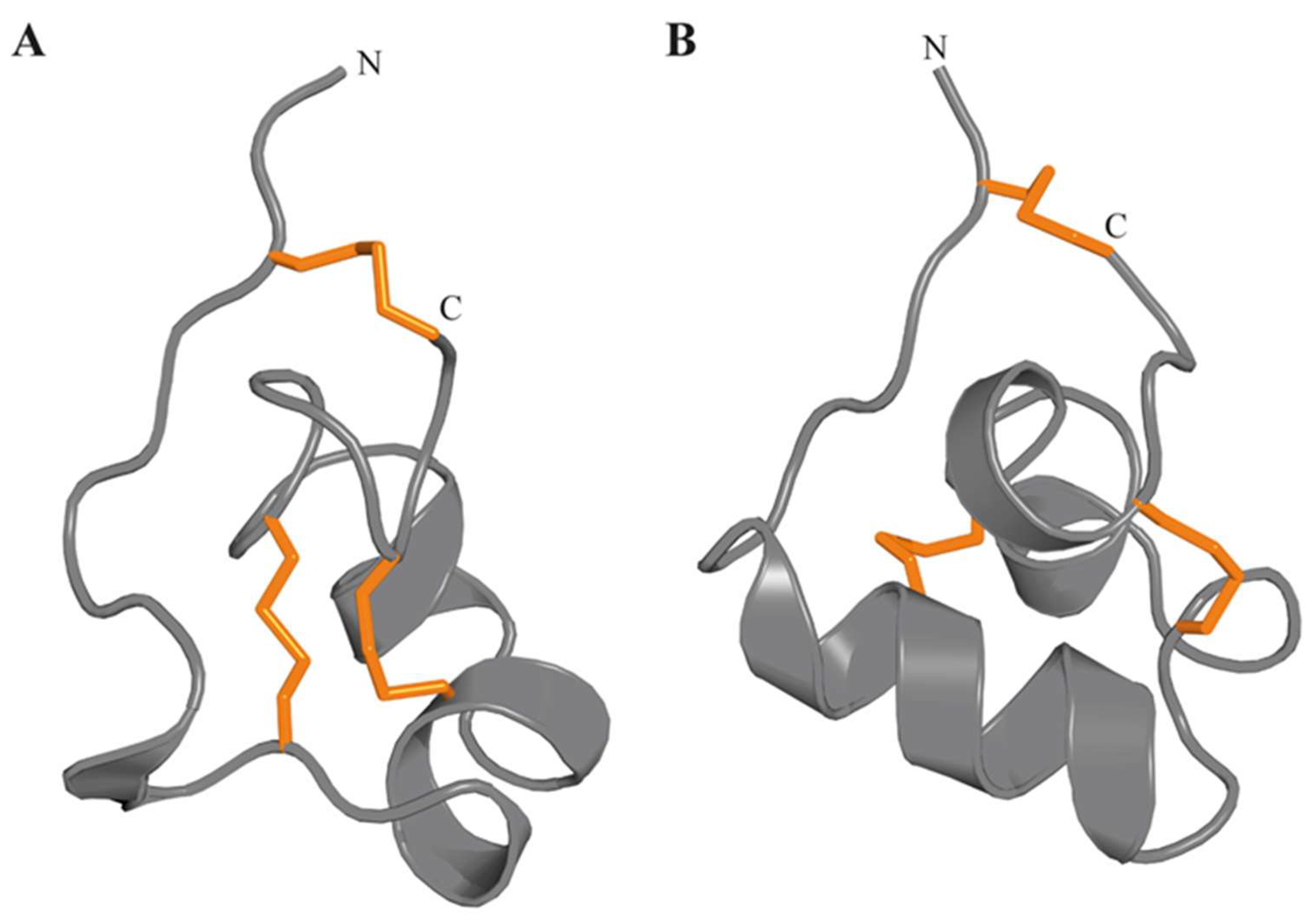
3.4.7. Proline-Hinged Asymmetric β-Hairpin (PHAB) Fold
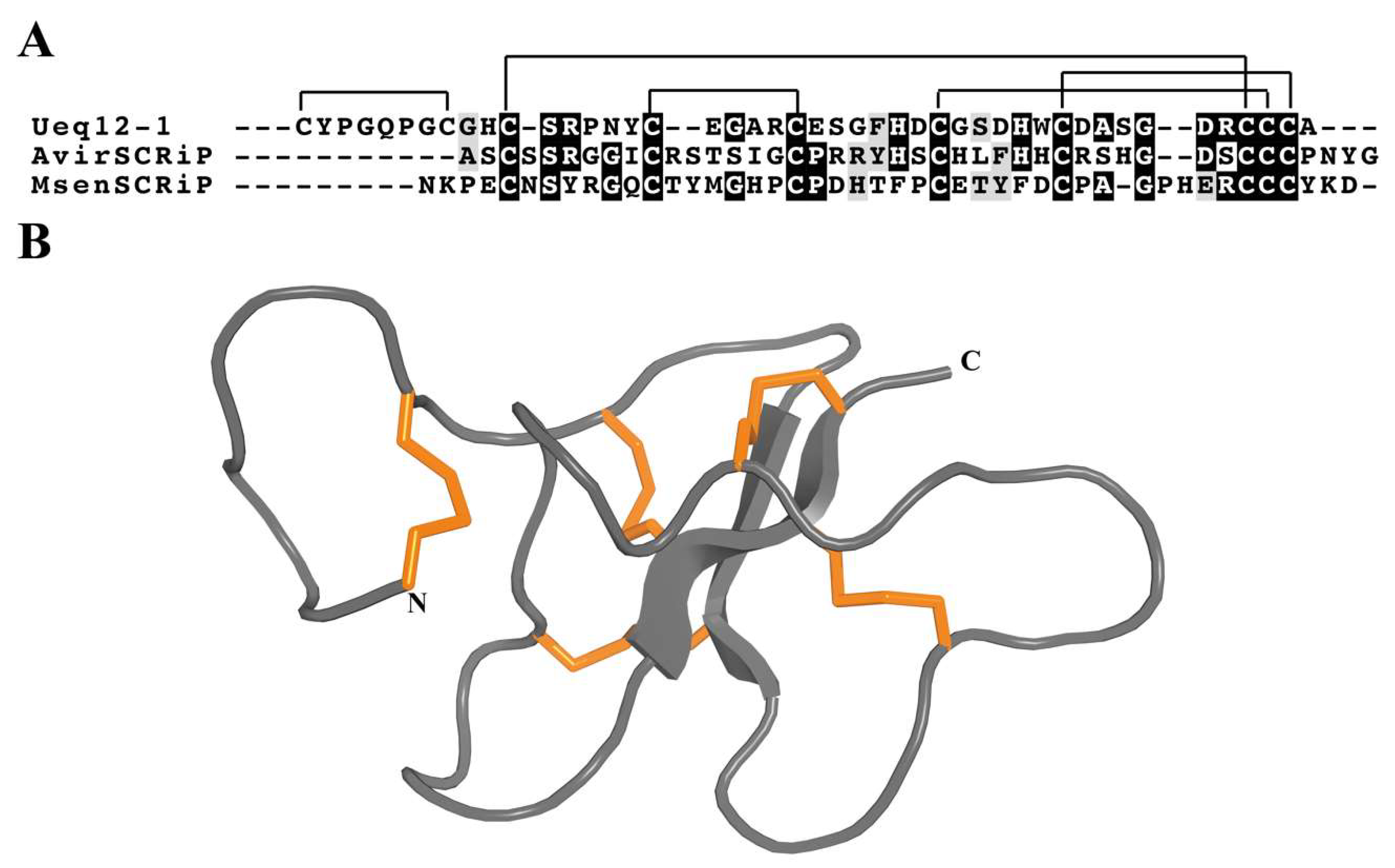
3.4.8. SCRiPs
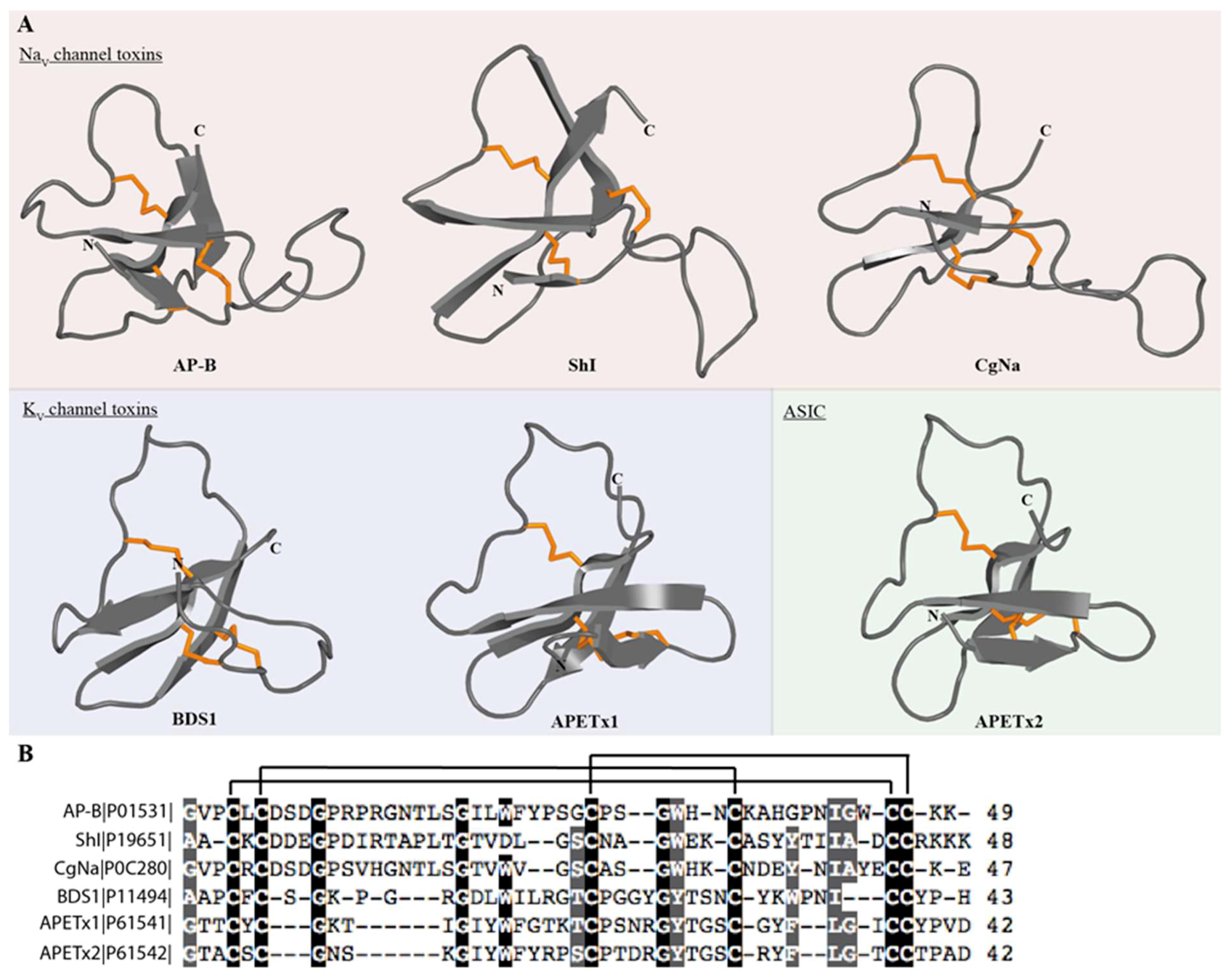
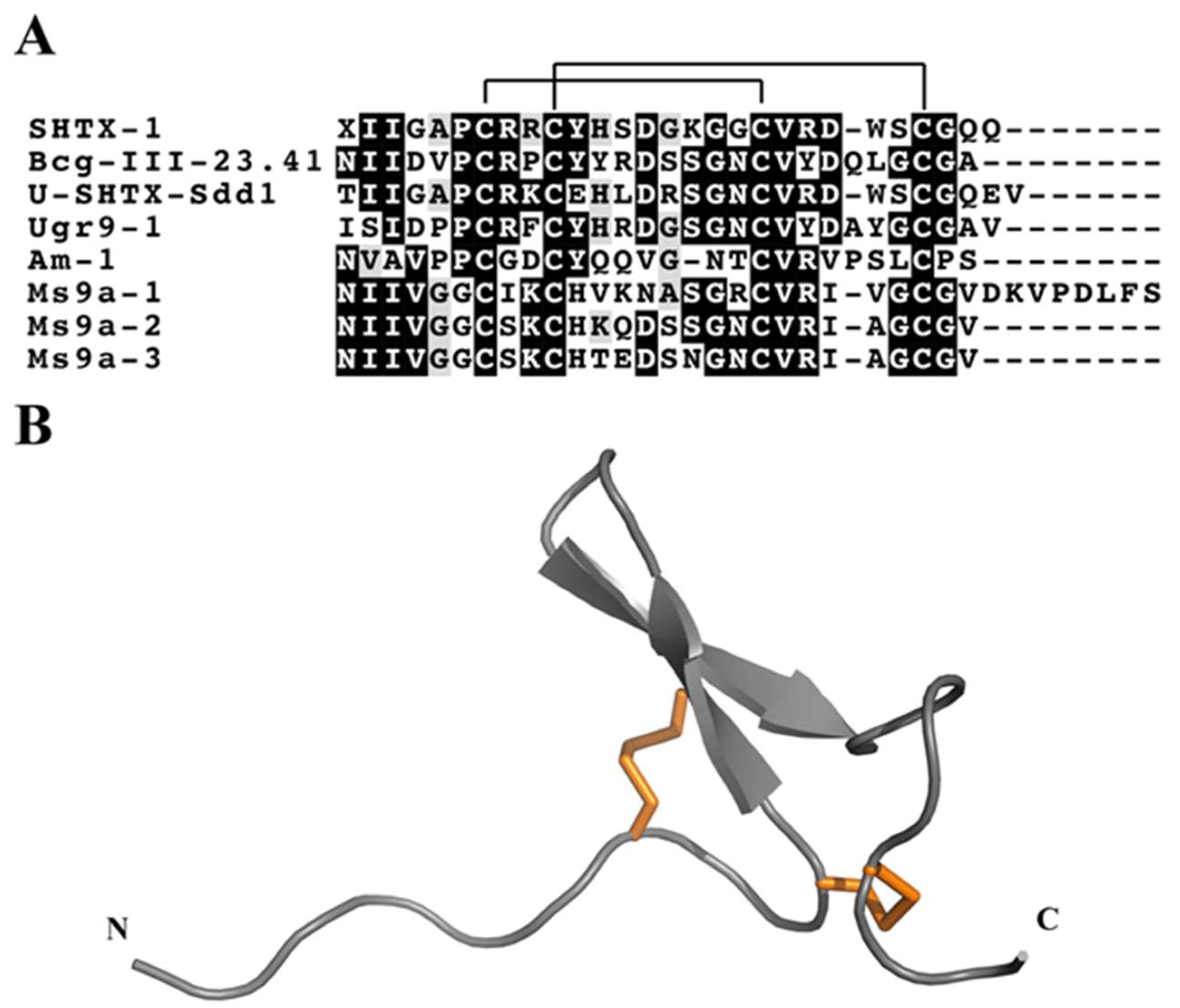
3.4.9. ShK Motif
3.5. Others
4. Conclusions
Funding
Conflicts of Interest
References
- Brusca, R.C.; Brusca, G.J.; Haver, N. Invertebrates, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Stewart, Z.K.; Pavasovic, A.; Hock, D.H.; Prentis, P.J. Transcriptomic investigation of wound healing and regeneration in the cnidarian Calliactis polypus. Sci. Rep. 2017, 7, 41458. [Google Scholar] [CrossRef] [PubMed]
- Galliot, B.; Schmid, V. Cnidarians as a model system for understanding evolution and regeneration. Int J. Dev. Biol. 2002, 46, 39–48. [Google Scholar] [PubMed]
- Rodriguez, E.; Barbeitos, M.S.; Brugler, M.R.; Crowley, L.M.; Grajales, A.; Gusmao, L.; Haussermann, V.; Reft, A.; Daly, M. Hidden among sea anemones: the first comprehensive phylogenetic reconstruction of the order Actiniaria (Cnidaria, Anthozoa, Hexacorallia) reveals a novel group of hexacorals. PLoS One 2014, 9, e96998. [Google Scholar] [CrossRef] [PubMed]
- Shick, J.M. A Functional Biology of Sea Anemones; Chapman & Hall: London, UK, 1991. [Google Scholar]
- Chintiroglou, C.; Koukouras, A. The feeding-habits of three mediterranean sea anemone species, Anemonia viridis (Forskål), Actinia equina (Linnaeus) and Cereus pedunculatus (Pennant). Helgoland Mar. Res. 1992, 46, 53–68. [Google Scholar] [CrossRef]
- Ruppert, E.E.; Fox, R.S.; Barnes, R.D. Invertebrate Zoology: A Functional Evolutionary Approach, 7th ed.; Thomson-Brooks/Cole: Belmont, CA, USA, 2004. [Google Scholar]
- Dubinsky, Z.; Stambler, N. Coral Reefs: An Ecosystem in Transition; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Technau, U.; Schwaiger, M. Recent advances in genomics and transcriptomics of cnidarians. Mar. Genomics 2015, 24, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Genikhovich, G.; Gordon, D.; Wienkoop, S.; Zenkert, C.; Ozbek, S.; Technau, U.; Gurevitz, M. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc. Biol. Sci. 2012, 279, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Reft, A.J.; Daly, M. Morphology, distribution, and evolution of apical structure of nematocysts in hexacorallia. J. Morphol. 2012, 273, 121–136. [Google Scholar] [CrossRef]
- David, C.N.; Ozbek, S.; Adamczyk, P.; Meier, S.; Pauly, B.; Chapman, J.; Hwang, J.S.; Gojobori, T.; Holstein, T.W. Evolution of complex structures: minicollagens shape the cnidarian nematocyst. Trends Genet. 2008, 24, 431–438. [Google Scholar] [CrossRef]
- Nuchter, T.; Benoit, M.; Engel, U.; Ozbek, S.; Holstein, T.W. Nanosecond-scale kinetics of nematocyst discharge. Cur.r Biol. 2006, 16, R316–R318. [Google Scholar] [CrossRef]
- Kass-Simon, G.; Scappaticci, A.A. The behavioral and developmental physiology of nematocysts. Can. J. Zool. 2002, 80, 1772–1794. [Google Scholar] [CrossRef]
- Beckmann, A.; Ozbek, S. The nematocyst: a molecular map of the cnidarian stinging organelle. Int. J. Dev. Biol. 2012, 56, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Daly, M. Functional and genetic diversity of toxins in sea anemones. In Evolution of Venomus Animals and Their Toxins; Gopalakrishnakone, P., Malhotra, A., Eds.; Spinger: Dordrecht, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, M. Envenomation by cnidarians and renal injuries. In The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters; Goffredo, S., Dubinsky, Z., Eds.; Springer: Berlin, Germany, 2016; pp. 623–636. [Google Scholar]
- Ardelean, A.; Fautin, D.G. A new species of the sea anemone Megalactis (Cnidaria: Anthozoa: Actiniaria: Actinodendridae) from Taiwan and designation of a neotype for the type species of the genus. Proc. Biol. Soc. Wash. 2004, 117, 488–504. [Google Scholar]
- Macrander, J.; Broe, M.; Daly, M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome. Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef] [PubMed]
- Kitatani, R.; Yamada, M.; Kamio, M.; Nagai, H. Length is associated with pain: jellyfish with painful sting have longer nematocyst tubules than harmless jellyfish. PLoS ONE 2015, 10, e0135015. [Google Scholar] [CrossRef] [PubMed]
- Logashina, Y.A.; Solstad, R.G.; Mineev, K.S.; Korolkova, Y.V.; Mosharova, I.V.; Dyachenko, I.A.; Palikov, V.A.; Palikova, Y.A.; Murashev, A.N.; Arseniev, A.S.; et al. New disulfide-stabilized fold provides sea anemone peptide to exhibit both antimicrobial and TRPA1 potentiating properties. Toxins (Basel) 2017, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins (Basel) 2018, 10, 36. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in cnidaria. Comp. Biochem. Physiol. Part. B Biochem Mol. Biol 2004, 139, 731–735. [Google Scholar] [CrossRef]
- Anderluh, G.; Macek, P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria). Toxicon 2002, 40, 111–124. [Google Scholar] [CrossRef]
- Monastyrnaya, M.; Peigneur, S.; Zelepuga, E.; Sintsova, O.; Gladkikh, I.; Leychenko, E.; Isaeva, M.; Tytgat, J.; Kozlovskaya, E. Kunitz-Type Peptide HCRG21 from the sea anemone Heteractis crispa is a full antagonist of the TRPV1 receptor. Mar. Drugs 2016, 14, 229. [Google Scholar] [CrossRef]
- Andreev, Y.A.; Kozlov, S.A.; Koshelev, S.G.; Ivanova, E.A.; Monastyrnaya, M.M.; Kozlovskaya, E.P.; Grishin, E.V. Analgesic compound from sea anemone Heteractis crispa is the first polypeptide inhibitor of vanilloid receptor 1 (TRPV1). J. Biol. Chem. 2008, 283, 23914–23921. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, E.; Peigneur, S.; Debaveye, S.; Shiomi, K.; Tytgat, J. TRPV1 channel as new target for marine toxins: example of gigantoxin I, a sea anemone toxin acting via modulation of the PLA2 pathway. Acta Chim. Slov. 2011, 58, 735–741. [Google Scholar] [PubMed]
- Norton, R.S. Structure and structure function-relationships of sea anemone proteins that interact with the sodium channel. Toxicon 1991, 29, 1051–1084. [Google Scholar] [CrossRef]
- Honma, T.; Shiomi, K. Peptide toxins in sea anemones: structural and functional aspects. Mar. Biotech. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Grishin, E. Convenient nomenclature of cysteine-rich polypeptide toxins from sea anemones. Peptides 2012, 33, 240–244. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Fuentes-Silva, D.; King, G.F. Development of a rational nomenclature for naming peptide and protein toxins from sea anemones. Toxicon 2012, 60, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S. Sea anemone peptides. In Handbook of Biologically Active Peptides, 2nd ed.; Ribeiro, S.M., Porto, W.F., Silva, O.N., Santos, M.D.O., Dias, S.C., Franco, O.L., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 430–436. [Google Scholar] [CrossRef]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Jones, A.; Clauser, K.R.; Holland, J.W.; Pineda, S.S.; King, G.F.; Fry, B.G. Clawing through evolution: toxin diversification and convergence in the ancient lineage Chilopoda (centipedes). Mol. Biol. Evol. 2014, 31, 2124–2148. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Sunagar, K.; Jackson, T.N.; Undheim, E.A.; Ali, S.A.; Antunes, A.; Fry, B.G. Three-fingered RAVERs: rapid accumulation of variations in exposed residues of snake venom toxins. Toxins (Basel) 2013, 5, 2172–2208. [Google Scholar] [CrossRef]
- de Freitas, J.C.; Sawaya, M.I. Anomalies in sea-urchin egg development induced by a novel purine isolated from the sea anemone Bunodosoma caissarum. Toxicon 1986, 24, 751–755. [Google Scholar] [CrossRef]
- de Freitas, J.C.; Sawaya, M.I. Increase of mammalian intestinal motility by the iminopurine caissarone isolated from the sea anemone Bunodosoma caissarum. Toxicon 1990, 28, 1029–1037. [Google Scholar] [CrossRef]
- Garateix, A.; Flores, A.; Garcia-Andrade, J.M.; Palmero, A.; Aneiros, A.; Vega, R.; Soto, E. Antagonism of glutamate receptors by a chromatographic fraction from the exudate of the sea anemone Phyllactis flosculifera. Toxicon 1996, 34, 443–450. [Google Scholar] [CrossRef]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteomics 2017, 166, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Kordis, D. Evolution of phospholipase A2 toxins in venomous animals. Acta Chim Slov 2011, 58, 638–646. [Google Scholar] [PubMed]
- Murakami, M.; Taketomi, Y.; Miki, Y.; Sato, H.; Yamamoto, K.; Lambeau, G. Emerging roles of secreted phospholipase A2 enzymes: The 3rd edition. Biochimie 2014, 107, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Razpotnik, A.; Krizaj, I.; Sribar, J.; Kordis, D.; Macek, P.; Frangez, R.; Kem, W.R.; Turk, T. A new phospholipase A2 isolated from the sea anemone Urticina crassicornis - its primary structure and phylogenetic classification. FEBS J. 2010, 277, 2641–2653. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.W.; Feil, S.C. Pore-forming protein toxins: from structure to function. Progr. Biophys. Mol. Biol. 2005, 88, 91–142. [Google Scholar] [CrossRef]
- Elliott, R.C.; Konya, R.S.; Vickneshwara, K. The isolation of a toxin from the dahlia sea anemone, Tealia felina L. Toxicon 1986, 24, 117–122. [Google Scholar] [CrossRef]
- Zykova, T.A.; Monastyrnaia, M.M.; Apalikova, O.V.; Shvets, T.V.; Kozlovskaia, E.P. Low-molecular cytolysins and trypsin inhibitors from sea anemone Radianthus macrodactylus. Isolation and partial characterization. Bioorg. Khim. 1998, 24, 509–516. [Google Scholar]
- Monastyrnaya, M.; Leychenko, E.; Isaeva, M.; Likhatskaya, G.; Zelepuga, E.; Kostina, E.; Trifonov, E.; Nurminski, E.; Kozlovskaya, E. Actinoporins from the sea anemones, tropical Radianthus macrodactylus and northern Oulactis orientalis: comparative analysis of structure-function relationships. Toxicon 2010, 56, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Suput, D. In vivo effects of cnidarian toxins and venoms. Toxicon 2009, 54, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.; Mancheno, J.M.; Martinez, D.; Tejuca, M.; Pazos, F.; Lanio, M.E. Sticholysins, two pore-forming toxins produced by the Caribbean Sea anemone Stichodactyla helianthus: their interaction with membranes. Toxicon 2009, 54, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Sepcic, K.; Turk, T.; Macek, P. Cytolytic proteins from cnidarians—An overview. Acta Chim. Slov. 2011, 58, 724–729. [Google Scholar] [PubMed]
- Malovrh, P.; Barlic, A.; Podlesek, Z.; MaCek, P.; Menestrina, G.; Anderluh, G. Structure-function studies of tryptophan mutants of equinatoxin II, a sea anemone pore-forming protein. Biochem. J. 2000, 346, 223–232. [Google Scholar] [PubMed]
- Hong, Q.; Gutierrez-Aguirre, I.; Barlic, A.; Malovrh, P.; Kristan, K.; Podlesek, Z.; Macek, P.; Turk, D.; Gonzalez-Manas, J.M.; Lakey, J.H.; et al. Two-step membrane binding by Equinatoxin II, a pore-forming toxin from the sea anemone, involves an exposed aromatic cluster and a flexible helix. J. Biol. Chem. 2002, 277, 41916–41924. [Google Scholar] [CrossRef] [PubMed]
- Morante, K.; Caaveiro, J.M.; Viguera, A.R.; Tsumoto, K.; Gonzalez-Manas, J.M. Functional characterization of Val60, a key residue involved in the membrane-oligomerization of fragaceatoxin C, an actinoporin from Actinia fragacea. FEBS Lett. 2015, 589, 1840–1846. [Google Scholar] [CrossRef]
- Bakrac, B.; Gutierrez-Aguirre, I.; Podlesek, Z.; Sonnen, A.F.; Gilbert, R.J.; Macek, P.; Lakey, J.H.; Anderluh, G. Molecular determinants of sphingomyelin specificity of a eukaryotic pore-forming toxin. J. Biol. Chem. 2008, 283, 18665–18677. [Google Scholar] [CrossRef]
- Garcia-Linares, S.; Richmond, R.; Garcia-Mayoral, M.F.; Bustamante, N.; Bruix, M.; Gavilanes, J.G.; Martinez-Del-Pozo, A. The sea anemone actinoporin (Arg-Gly-Asp) conserved motif is involved in maintaining the competent oligomerization state of these pore-forming toxins. FEBS J. 2014, 281, 1465–1478. [Google Scholar] [CrossRef]
- Wang, Y.; Yap, L.L.; Chua, K.L.; Khoo, H.E. A multigene family of Heteractis magnificalysins (HMgs). Toxicon 2008, 51, 1374–1382. [Google Scholar] [CrossRef]
- Leychenko, E.; Isaeva, M.; Tkacheva, E.; Zelepuga, E.; Kvetkina, A.; Guzev, K.; Monastyrnaya, M.; Kozlovskaya, E. Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa. Mar. Drugs 2018, 16, 183. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ortega, L.; Alegre-Cebollada, J.; Garcia-Linares, S.; Bruix, M.; Martinez-Del-Pozo, A.; Gavilanes, J.G. The behavior of sea anemone actinoporins at the water-membrane interface. Biochim. Biophys. Acta 2011, 1808, 2275–2288. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Linares, S.; Castrillo, I.; Bruix, M.; Menendez, M.; Alegre-Cebollada, J.; Martinez-del-Pozo, A.; Gavilanes, J.G. Three-dimensional structure of the actinoporin sticholysin I. Influence of long-distance effects on protein function. Arch. Biochem. Biophys. 2013, 532, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Basulto, A.; Perez, V.M.; Noa, Y.; Varela, C.; Otero, A.J.; Pico, M.C. Immunohistochemical targeting of sea anemone cytolysins on tentacles, mesenteric filaments and isolated nematocysts of Stichodactyla helianthus. J. Exp. Zool. Part. A Comp. Exp. Biol. 2006, 305, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Valle, A.; Alvarado-Mesen, J.; Lanio, M.E.; Alvarez, C.; Barbosa, J.A.; Pazos, I.F. The multigene families of actinoporins (part I): Isoforms and genetic structure. Toxicon 2015, 103, 176–187. [Google Scholar] [CrossRef]
- Mancheno, J.M.; Martin-Benito, J.; Martinez-Ripoll, M.; Gavilanes, J.G.; Hermoso, J.A. Crystal and electron microscopy structures of sticholysin II actinoporin reveal insights into the mechanism of membrane pore formation. Structure 2003, 11, 1319–1328. [Google Scholar] [CrossRef]
- Malovrh, P.; Viero, G.; Serra, M.D.; Podlesek, Z.; Lakey, J.H.; Macek, P.; Menestrina, G.; Anderluh, G. A novel mechanism of pore formation: membrane penetration by the N-terminal amphipathic region of equinatoxin. J. Biol. Chem. 2003, 278, 22678–22685. [Google Scholar] [CrossRef]
- Gutierrez-Aguirre, I.; Barlic, A.; Podlesek, Z.; Macek, P.; Anderluh, G.; Gonzalez-Manas, J.M. Membrane insertion of the N-terminal alpha-helix of equinatoxin II, a sea anemone cytolytic toxin. Biochem. J. 2004, 384, 421–428. [Google Scholar] [CrossRef]
- Belmonte, G.; Pederzolli, C.; Macek, P.; Menestrina, G. Pore formation by the sea anemone cytolysin equinatoxin-II in red blood cells and model lipid membranes. J. Membr. Biol. 1993, 131, 11–22. [Google Scholar] [CrossRef]
- Tejuca, M.; Serra, M.D.; Ferreras, M.; Lanio, M.E.; Menestrina, G. Mechanism of membrane permeabilization by sticholysin I, a cytolysin isolated from the venom of the sea anemone Stichodactyla helianthus. Biochemistry 1996, 35, 14947–14957. [Google Scholar] [CrossRef]
- Alegre-Cebollada, J.; Martinez del Pozo, A.; Gavilanes, J.G.; Goormaghtigh, E. Infrared spectroscopy study on the conformational changes leading to pore formation of the toxin sticholysin II. Biophys. J. 2007, 93, 3191–3201. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Macek, P. Dissecting the actinoporin pore-forming mechanism. Structure 2003, 11, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Valcarcel, C.A.; Dalla Serra, M.; Potrich, C.; Bernhart, I.; Tejuca, M.; Martinez, D.; Pazos, F.; Lanio, M.E.; Menestrina, G. Effects of lipid composition on membrane permeabilization by sticholysin I and II, two cytolysins of the sea anemone Stichodactyla helianthus. Biophys. J. 2001, 80, 2761–2774. [Google Scholar] [CrossRef]
- Mechaly, A.E.; Bellomio, A.; Gil-Carton, D.; Morante, K.; Valle, M.; Gonzalez-Manas, J.M.; Guerin, D.M. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure 2011, 19, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Subburaj, Y.; Ros, U.; Hermann, E.; Tong, R.; Garcia-Saez, A.J. Toxicity of an α-pore-forming toxin depends on the assembly mechanism on the target membrane as revealed by single molecule imaging. J. Biol Chem 2015, 290, 4856–4865. [Google Scholar] [CrossRef]
- Tanaka, K.; Caaveiro, J.M.; Morante, K.; Gonzalez-Manas, J.M.; Tsumoto, K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 2015, 6, 6337. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.; Ros, U.; Valle, A.; Pedrera, L.; Soto, C.; Hervis, Y.P.; Cabezas, S.; Valiente, P.A.; Pazos, F.; Lanio, M.E. Biophysical and biochemical strategies to understand membrane binding and pore formation by sticholysins, pore-forming proteins from a sea anemone. Biophys. Rev. 2017, 9, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Podobnik, M.; Anderluh, G. Pore-forming toxins in Cnidaria. Semin. Cell Dev. Biol 2017, 72, 133–141. [Google Scholar] [CrossRef]
- Lenhoff, D.A.H.H.M. Assay and properties of the hemolysis activity of pure venom from the nematocysts of the acontia of the sea anemone Aiptasia pallida. Arch. Biochem. Biophys. 1973, 159, 629–638. [Google Scholar]
- Razpotnik, A.; Krizaj, I.; Kem, W.R.; Macek, P.; Turk, T. A new cytolytic protein from the sea anemone Urticina crassicornis that binds to cholesterol- and sphingomyelin-rich membranes. Toxicon 2009, 53, 762–769. [Google Scholar] [CrossRef]
- Cline, E.I.; Wiebe, L.I.; Young, J.D.; Samuel, J. Toxic effects of the novel protein UpI from the sea anemone Urticina piscivora. Pharmacol. Res. 1995, 32, 309–314. [Google Scholar] [CrossRef]
- Hessinger, D.A.; Lenhoff, H.M. Binding of active and inactive hemolytic factor of sea anemone nematocyst venom to red blood cells. Biochem. Biophy.s Res. Commun 1973, 53, 475–481. [Google Scholar] [CrossRef]
- Avigad, A.W.B.L.S. A cholesterol-inhibitable cytolytic from the sea anemone Metridium senile. Biochim. Biophys. Act. A—Gen. Subjects 1978, 541, 96–106. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Avigad, L.S.; Kim, K. Comparison fo metridiolysin from the sea anemone with thiol-activated cytolysins from bacteria. Toxicon 1979, 17, 69–75. [Google Scholar] [CrossRef]
- Muller-Eberhard, H.J. The membrane attack complex of complement. Annu. Rev. Immunol. 1986, 4, 503–528. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Walsh, C.M.; Young, J.D. Perforin: structure and function. Immunology today 1995, 16, 194–201. [Google Scholar] [CrossRef]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Rees, B.; Bilwes, A. Three-dimensional structures of neurotoxins and cardiotoxins. Chem. Res. Toxicol 1993, 6, 385–406. [Google Scholar] [CrossRef]
- Cristofori-Armstrong, B.; Rash, L.D. Acid-sensing ion channel (ASIC) structure and function: Insights from spider, snake and sea anemone venoms. Neuropharmacology 2017. [Google Scholar] [CrossRef]
- Rodriguez, A.A.; Salceda, E.; Garateix, A.G.; Zaharenko, A.J.; Peigneur, S.; Lopez, O.; Pons, T.; Richardson, M.; Diaz, M.; Hernandez, Y.; et al. A novel sea anemone peptide that inhibits acid-sensing ion channels. Peptides 2014, 53, 3–12. [Google Scholar] [CrossRef]
- Moran, Y.; Gordon, D.; Gurevitz, M. Sea anemone toxins affecting voltage-gated sodium channels — molecular and evolutionary features. Toxicon 2009, 54, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Jouiaei, M.; Sunagar, K.; Federman Gross, A.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, F.; Tytgat, J. Sea anemone venom as a source of insecticidal peptides acting on voltage-gated Na+ channels. Toxicon 2007, 49, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Deuis, J.R.; Mueller, A.; Israel, M.R.; Vetter, I. The pharmacology of voltage-gated sodium channel activators. Neuropharmacology 2017, 127, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Lazdunski, M. Sea anemone toxins affecting potassium channels. Prog. Mol. Subcell Biol. 2009, 46, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.B.; Moran, Y.; Cologna, C.T.; Peigneur, S.; Madio, B.; Praher, D.; Quinton, L.; De Pauw, E.; Bicudo, J.E.P.W.; Tytgat, J.; et al. BcsTx3 is a founder of a novel sea anemone toxin family of potassium channel blocker. FEBS J. 2013, 280, 4839–4852. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, O.; Harvey, A.L. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 2009, 54, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic. Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Manoleras, N.; Norton, R.S. Three-dimensional structure in solution of neurotoxin III from the sea anemone Anemonia sulcata. Biochemistry 1994, 33, 11051–11061. [Google Scholar] [CrossRef] [PubMed]
- Beress, L.; Beress, R.; Wunderer, G. Isolation and characterisation of three polypeptides with neurotoxic activity from Anemonia sulcata. FEBS Lett. 1975, 50, 311–314. [Google Scholar] [CrossRef]
- Schweitz, H.; Vincent, J.P.; Barhanin, J.; Frelin, C.; Linden, G.; Hugues, M.; Lazdunski, M. Purification and pharmacological properties of eight sea anemone toxins from Anemonia sulcata, Anthopleura xanthogrammica, Stoichactis giganteus, and Actinodendron plumosum. Biochemistry 1981, 20, 5245–5252. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Kahn, R.; Cohen, L.; Gur, M.; Karbat, I.; Gordon, D.; Gurevitz, M. Molecular analysis of the sea anemone toxin Av3 reveals selectivity to insects and demonstrates the heterogeneity of receptor site-3 on voltage-gated Na+ channels. Biochem. J. 2007, 406, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Carmona, M.; Hubert, P.; Delvenne, P.; Herfs, M. Defensins: “Simple” antimicrobial peptides or broad-spectrum molecules? Cytokine Growth Factor Rev. 2015, 26, 361–370. [Google Scholar] [CrossRef]
- Shafee, T.M.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol. Biol. Evol. 2016, 33, 2345–2356. [Google Scholar] [CrossRef]
- Chagot, B.; Diochot, S.; Pimentel, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx1 from the sea anemone Anthopleura elegantissima: a new fold for an HERG toxin. Proteins 2005, 59, 380–386. [Google Scholar] [CrossRef]
- Chagot, B.; Escoubas, P.; Diochot, S.; Bernard, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx2, a specific peptide inhibitor of ASIC3 proton-gated channels. Protein Sci. 2005, 14, 2003–2010. [Google Scholar] [CrossRef]
- Smith, J.J.; Blumenthal, K.M. Site-3 sea anemone toxins: molecular probes of gating mechanisms in voltage-dependent sodium channels. Toxicon 2007, 49, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Baron, A.; Rash, L.D.; Deval, E.; Escoubas, P.; Scarzello, S.; Salinas, M.; Lazdunski, M. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 2004, 23, 1516–1525. [Google Scholar] [CrossRef]
- Diochot, S.; Loret, E.; Bruhn, T.; Beress, L.; Lazdunski, M. APETx1, a new toxin from the sea anemone Anthopleura elegantissima, blocks voltage-gated human ether-a-go-go-related gene potassium channels. Mol. Pharmacol. 2003, 64, 59–69. [Google Scholar] [CrossRef]
- Diochot, S.; Schweitz, H.; Beress, L.; Lazdunski, M. Sea anemone peptides with a specific blocking activity against the fast inactivating potassium channel KV3.4. J. Biol. Chem. 1998, 273, 6744–6749. [Google Scholar] [CrossRef]
- Beress, L.; Doppelfeld, I.-S.; Etschenberg, E.; Graf, E.; Henschen, A.; Zwick, J. Polypeptides, Process for Their Preparation, and Their Use As Hypotensive Active Compounds. US Patent DE 3324689 A1, 17 January 1985. [Google Scholar]
- Yeung, S.Y.; Thompson, D.; Wang, Z.; Fedida, D.; Robertson, B. Modulation of KV3 subfamily potassium currents by the sea anemone toxin BDS: significance for CNS and biophysical studies. J. Neurosci. 2005, 25, 8735–8745. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jo, S.; Bean, B.P. Modulation of neuronal sodium channels by the sea anemone peptide BDS-I. J. Neurophysiol. 2012, 107, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; Beress, L.; Moller, C.; Mari, F.; Forssmann, W.G.; Tytgat, J. A natural point mutation changes both target selectivity and mechanism of action of sea anemone toxins. FASEB J. 2012, 26, 5141–5151. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Yokoyama, A.; Shimakura, K.; Nagashima, Y.; Shiomi, K. Halcurin, a polypeptide toxin from the sea anemone Halcurias sp., with a structural resemblance to type 1 and 2 toxins. Toxicon 1997, 35, 537–544. [Google Scholar] [CrossRef]
- Moran, Y.; Gurevitz, M. When positive selection of neurotoxin genes is missing—The riddle of the sea anemone Nematostella vectensis. FEBS J. 2006, 273, 3886–3892. [Google Scholar] [CrossRef] [PubMed]
- Widmer, H.; Billeter, M.; Wüthrich, K. Three-dimensional structure of the neurotoxin ATX Ia from Anemonia sulcata in aqueous solution determined by nuclear magnetic resonance spectroscopy. Proteins 1989, 6, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Fogh, R.H.; Kem, W.R.; Norton, R.S. Solution structure of neurotoxin I from the sea anemone Stichodactyla helianthus. A nuclear magnetic resonance, distance geometry, and restrained molecular dynamics study. J. Biol. Chem. 1990, 265, 13016–13028. [Google Scholar] [PubMed]
- Pallaghy, P.K.; Scanlon, M.J.; Monks, S.A.; Norton, R.S. Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A. Biochemistry 1995, 34, 3782–3794. [Google Scholar] [CrossRef]
- Monks, S.A.; Pallaghy, P.K.; Scanlon, M.J.; Norton, R.S. Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A. Structure 1995, 3, 791–803. [Google Scholar] [CrossRef]
- Salceda, E.; Perez-Castells, J.; Lopez-Mendez, B.; Garateix, A.; Salazar, H.; Lopez, O.; Aneiros, A.; Standker, L.; Beress, L.; Forssmann, W.G.; et al. CgNa, a type I toxin from the giant Caribbean sea anemone Condylactis gigantea shows structural similarities to both type I and II toxins, as well as distinctive structural and functional properties. Biochem. J. 2007, 406, 67–76. [Google Scholar] [CrossRef]
- Catterall, W.A.; Beress, L. Sea anemone toxin and scorpion toxin share a common receptor site associated with the action potential sodium ionophore. J. Biol. Chem. 1978, 253, 7393–7396. [Google Scholar] [PubMed]
- Kruger, L.M.; Griffiths, C.L. Sources of nutrition in intertidal sea anemones from the south-western Cape, South Africa. South. Afr. J. Zool. 1996, 31, 110–119. [Google Scholar] [CrossRef]
- Norton, T.R.; Shibata, S.; Kashiwagi, M.; Bentley, J. Isolation and characterization of the cardiotonic polypeptide anthopleurin-A from the sea anemone Anthopleura xanthogrammica. J. Pharm. Sci. 1976, 65, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Cariello, L.; de Santis, A.; Fiore, F.; Piccoli, R.; Spagnuolo, A.; Zanetti, L.; Parente, A. Calitoxin, a neurotoxic peptide from the sea anemone Calliactis parasitica: amino acid sequence and electrophysiological properties. Biochemistry 1989, 28, 2484–2489. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, A.; Zanetti, L.; Cariello, L.; Piccoli, R. Isolation and characterization of two genes encoding calitoxins, neurotoxic peptides from Calliactis parasitica (Cnidaria). Gene 1994, 138, 187–191. [Google Scholar] [CrossRef]
- Torres, A.M.; Kuchel, P.W. The β-defensin-fold family of polypeptides. Toxicon 2004, 44, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Kalina, R.; Gladkikh, I.; Dmitrenok, P.; Chernikov, O.; Koshelev, S.; Kvetkina, A.; Kozlov, S.; Kozlovskaya, E.; Monastyrnaya, M. New APETx-like peptides from sea anemone Heteractis crispa modulate ASIC1a channels. Peptides 2018, 104, 41–49. [Google Scholar] [CrossRef]
- Osmakov, D.I.; Kozlov, S.A.; Andreev, Y.A.; Koshelev, S.G.; Sanamyan, N.P.; Sanamyan, K.E.; Dyachenko, I.A.; Bondarenko, D.A.; Murashev, A.N.; Mineev, K.S.; et al. Sea anemone peptide with uncommon β-hairpin structure inhibits acid-sensing ion channel 3 (ASIC3) and reveals analgesic activity. J. Biol. Chem. 2013, 288, 23116–23127. [Google Scholar] [CrossRef]
- Zaharenko, A.J.; Ferreira, W.A., Jr.; Oliveira, J.S.; Richardson, M.; Pimenta, D.C.; Konno, K.; Portaro, F.C.; de Freitas, J.C. Proteomics of the neurotoxic fraction from the sea anemone Bunodosoma cangicum venom: novel peptides belonging to new classes of toxins. Comp. Biochem Physiol Part. D Genomics Proteomics 2008, 3, 219–225. [Google Scholar] [CrossRef]
- Honma, T.; Kawahata, S.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 2008, 29, 536–544. [Google Scholar] [CrossRef]
- Cassoli, J.S.; Verano-Braga, T.; Oliveira, J.S.; Montandon, G.G.; Cologna, C.T.; Peigneur, S.; Pimenta, A.M.; Kjeldsen, F.; Roepstorff, P.; Tytgat, J.; et al. The proteomic profile of Stichodactyla duerdeni secretion reveals the presence of a novel O-linked glycopeptide. J. Proteomics 2013, 87, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Logashina, Y.A.; Mosharova, I.V.; Korolkova, Y.V.; Shelukhina, I.V.; Dyachenko, I.A.; Palikov, V.A.; Palikova, Y.A.; Murashev, A.N.; Kozlov, S.A.; Stensvag, K.; et al. Peptide from sea anemone Metridium senile affects transient receptor potential ankyrin-repeat 1 (TRPA1) function and produces analgesic effect. J. Biol. Chem. 2017, 292, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Honma, T.; Ide, M.; Nagashima, Y.; Ishida, M.; Chino, M. An epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Toxicon 2003, 41, 229–236. [Google Scholar] [CrossRef]
- Undheim, E.A.; Mobli, M.; King, G.F. Toxin structures as evolutionary tools: Using conserved 3D folds to study the evolution of rapidly evolving peptides. Bioessays 2016, 38, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Antuch, W.; Berndt, K.D.; Chavez, M.A.; Delfin, J.; Wuthrich, K. The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus. Eur. J. Biochem. 1993, 212, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Wunderer, G.; Beress, L.; Machleidt, W.; Fritz, H. Broad-specificity inhibitors from sea anemones. Methods Enzymol. 1976, 45, 881–888. [Google Scholar]
- Fritz, H.; Brey, B.; Beress, L. Polyvalent isoinhibitors for trypsin, chymotrypsin, plasmin and kallikreins of sea anemones (Anemonia sulcata), isolation, inhibitory behavior and amino acid composition. Hoppe Seylers Z Physiol. Chem. 1972, 353, 19–30. [Google Scholar] [CrossRef]
- Minagawa, S.; Sugiyama, M.; Ishida, M.; Nagashima, Y.; Shiomi, K. Kunitz-type protease inhibitors from acrorhagi of three species of sea anemones. Comp. Biochem. Phys. B 2008, 150, 240–245. [Google Scholar] [CrossRef]
- Frazao, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef]
- Peigneur, S.; Billen, B.; Derua, R.; Waelkens, E.; Debaveye, S.; Beress, L.; Tytgat, J. A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochem. Pharmacol. 2011, 82, 81–90. [Google Scholar] [CrossRef]
- Mourao, C.B.F.; Schwartz, E.F. Protease inhibitors from marine venomous animals and their counterparts in terrestrial venomous animals. Mar. Drugs 2013, 11, 2069–2112. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.M.; Beress, L.; Lazdunski, M. Kalicludines and Kaliseptine—two different classes of sea anemone toxins for voltage-sensitive K+ channels. J. Bio.l Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef] [PubMed]
- Zykova, T.A.; Vinokurov, L.M.; Markova, L.F.; Kozlovskaya, E.P.; Elyakov, G.B. Amino acid sequence of trypsin inhibitor-IV from Radianthus macrodactylus. Bioorg. Khim. 1985, 11, 293–301. [Google Scholar]
- Sokotun, I.N.; Leichenko, E.V.; Vakorina, T.I.; Es’kov, A.A.; Il’ina, A.P.; Monastyrnaya, M.M.; Kozlovskaya, E.P. A serine protease inhibitor from the anemone Radianthus macrodactylus: isolation and physicochemical characteristics. Russ. J. Bioorg. Chem. 2007, 33, 415–422. [Google Scholar] [CrossRef]
- Sokotun, I.N.; Gnedenko, O.V.; Leichenko, E.V.; Monastyrnaia, M.M.; Kozlovskaia, E.P.; Mol’nar, A.A.; Ivanov, A.C. Interaction investigation of trypsin inhibitor from sea anemone Radianthus macrodactylus with proteases. Biomed. Khim. 2006, 52, 595–600. [Google Scholar] [PubMed]
- Kozlov, S.A.; Andreev Ia, A.; Murashev, A.N.; Skobtsov, D.I.; D’Iachenko I, A.; Grishin, E.V. New polypeptide components from the Heteractis crispa sea anemone with analgesic activity. Bioorg. Khim. 2009, 35, 789–798. [Google Scholar] [CrossRef]
- Andreev, Y.A.; Kozlov, S.A.; Kozlovskaya, E.P.; Grishin, E.V. Analgesic effect of a polypeptide inhibitor of the TRPV1 receptor in noxious heat pain models. Doki. Biochem. Biophy.s 2009, 424, 46–48. [Google Scholar] [CrossRef]
- Cuypers, E.; Yanagihara, A.; Karlsson, E.; Tytgat, J. Jellyfish and other cnidarian envenomations cause pain by affecting TRPV1 channels. FEBS Lett. 2006, 580, 5728–5732. [Google Scholar] [CrossRef]
- Andreev, Y.A.; Kozlov, S.A.; Korolkova, Y.V.; Dyachenko, I.A.; Bondarenko, D.A.; Skobtsov, D.I.; Murashev, A.N.; Kotova, P.D.; Rogachevskaja, O.A.; Kabanova, N.V.; et al. Polypeptide modulators of TRPV1 produce analgesia without hyperthermia. Mar. Drugs 2013, 11, 5100–5115. [Google Scholar] [CrossRef]
- Madio, B.; Peigneur, S.; Chin, Y.K.Y.; Hamilton, B.R.; Henriques, S.T.; Smith, J.J.; Cristofori-Armstrong, B.; Dekan, Z.; Boughton, B.A.; Alewood, P.F.; et al. PHAB toxins: a unique family of predatory sea anemone toxins evolving via intra-gene concerted evolution defines a new peptide fold. Cell Mol. Life Sci. 2018, 75, 4511–4524. [Google Scholar] [CrossRef]
- Orts, D.J.B.; Peigneur, S.; Silva-Goncalves, L.C.; Arcisio-Miranda, M.; JE, P.W.B.; Tytgat, J. AbeTx1 is a novel sea anemone toxin with a dual mechanism of action on Shaker-type K+ channels activation. Mar. Drugs 2018, 16, 360. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Balandin, S.V.; Ivanov, V.T.; Ovchinnikova, T.V. A Therapeutic potential of animal β-hairpin antimicrobial peptides. Curr Med. Chem 2017, 24, 1724–1746. [Google Scholar] [CrossRef] [PubMed]
- Chagot, B.; Pimentel, C.; Dai, L.; Pil, J.; Tytgat, J.; Nakajima, T.; Corzo, G.; Darbon, H.; Ferrat, G. An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis. Biochem. J. 2005, 388, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; DeSalvo, M.K.; Voolstra, C.R.; Reyes-Bermudez, A.; Medina, M. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS ONE 2009, 4, e4865. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, I.; Ooki, S.; Fujita, T.; Murayama, E.; Nagasawa, H.; Isa, Y.; Watanabe, T. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem. Biophys. Res. Commun. 2003, 304, 11–17. [Google Scholar] [CrossRef]
- Tudor, J.E.; Pallaghy, P.K.; Pennington, M.W.; Norton, R.S. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat. Struct Biol. 1996, 3, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, O.; Sotolongo, V.; Amor, A.M.; Stocklin, R.; Anderson, A.J.; Harvey, A.L.; Engstrom, A.; Wernstedt, C.; Karlsson, E. Characterization of a potassium channel toxin from the Caribbean Sea anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef]
- Chi, V.; Pennington, M.W.; Norton, R.S.; Tarcha, E.J.; Londono, L.M.; Sims-Fahey, B.; Upadhyay, S.K.; Lakey, J.T.; Iadonato, S.; Wulff, H.; et al. Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 2012, 59, 529–546. [Google Scholar] [CrossRef] [PubMed]
- Dauplais, M.; Lecoq, A.; Song, J.; Cotton, J.; Jamin, N.; Gilquin, B.; Roumestand, C.; Vita, C.; de Medeiros, C.L.; Rowan, E.G.; et al. On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 1997, 272, 4302–4309. [Google Scholar] [CrossRef]
- Gasparini, S.; Danse, J.M.; Lecoq, A.; Pinkasfeld, S.; Zinn-Justin, S.; Young, L.C.; de Medeiros, C.C.; Rowan, E.G.; Harvey, A.L.; Menez, A. Delineation of the functional site of α-dendrotoxin. The functional topographies of dendrotoxins are different but share a conserved core with those of other KV1 potassium channel-blocking toxins. J. Biol. Chem. 1998, 273, 25393–25403. [Google Scholar] [CrossRef]
- Kalman, K.; Pennington, M.W.; Lanigan, M.D.; Nguyen, A.; Rauer, H.; Mahnir, V.; Paschetto, K.; Kem, W.R.; Grissmer, S.; Gutman, G.A.; et al. ShK-Dap22, a potent KV1.3-specific immunosuppressive polypeptide. J. Biol. Chem. 1998, 273, 32697–32707. [Google Scholar] [CrossRef] [PubMed]
- Lanigan, M.D.; Kalman, K.; Lefievre, Y.; Pennington, M.W.; Chandy, K.G.; Norton, R.S. Mutating a critical lysine in ShK toxin alters its binding configuration in the pore-vestibule region of the voltage-gated potassium channel, KV1.3. Biochemistry 2002, 41, 11963–11971. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kuyucak, S. Affinity and selectivity of ShK toxin for the KV1 potassium channels from free energy simulations. J. Phys. Chem. B 2012, 116, 4812–4822. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Eckel-Mahan, K.L.; Mirbolooki, M.R.; Tjong, I.; Griffey, S.M.; Schmunk, G.; Koehne, A.; Halbout, B.; Iadonato, S.; Pedersen, B.; et al. Selective KV1.3 channel blocker as therapeutic for obesity and insulin resistance. Proc. Natl. Acad Sci. USA 2013, 110, E2239–E2248. [Google Scholar] [CrossRef] [PubMed]
- King, G.F. Venoms to Drugs: Venom As A Source for the Development of Human Therapeutics; Royal Society of Chemistry: Cambridge, UK, 2015. [Google Scholar]
- Beraud, E.; Viola, A.; Regaya, I.; Confort-Gouny, S.; Siaud, P.; Ibarrola, D.; Le Fur, Y.; Barbaria, J.; Pellissier, J.F.; Sabatier, J.M.; et al. Block of neural KV1.1 potassium channels for neuroinflammatory disease therapy. Ann. Neurol. 2006, 60, 586–596. [Google Scholar] [CrossRef] [PubMed]

| Protein Type | Structural Family | Pharmacological Group 1 |
|---|---|---|
| Enzymes | Endonuclease D | Unknown |
| Phospholipase type A2 (PLA2) | PLA2 Type III cytolysins | |
| Serine protease S1 | Unknown | |
| Non-enzymatic proteins | Actinoporins | Type II cytolysins |
| CAP | Unknown | |
| WSC domain proteins | Unknown | |
| Peptide neurotoxins | ATX-III | NaV type 3 |
| B-defensin-like | ASIC KV type 3 NaV type 1 NaV type 2 NaV type 4 | |
| Boundless β-hairpin (BBH) | ASIC KV type 4 | |
| Epidermal growth factor-like (EGF-like) | EGF activity TRPV1 | |
| Inhibitor cystine-knot (ICK) | ASIC KV type 5 | |
| Kunitz-domain | KV type 2 TRPV1 Protease inhibitor | |
| Proline-hinged asymmetric β-hairpin (PHAB) | KV type 6 | |
| Small cysteine-rich peptides (SCRiPs) | TRPA1 | |
| ShK | KV type 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madio, B.; King, G.F.; Undheim, E.A.B. Sea Anemone Toxins: A Structural Overview. Mar. Drugs 2019, 17, 325. https://doi.org/10.3390/md17060325
Madio B, King GF, Undheim EAB. Sea Anemone Toxins: A Structural Overview. Marine Drugs. 2019; 17(6):325. https://doi.org/10.3390/md17060325
Chicago/Turabian StyleMadio, Bruno, Glenn F. King, and Eivind A. B. Undheim. 2019. "Sea Anemone Toxins: A Structural Overview" Marine Drugs 17, no. 6: 325. https://doi.org/10.3390/md17060325
APA StyleMadio, B., King, G. F., & Undheim, E. A. B. (2019). Sea Anemone Toxins: A Structural Overview. Marine Drugs, 17(6), 325. https://doi.org/10.3390/md17060325






