Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation
Abstract
1. Introduction
2. Results and Discussion
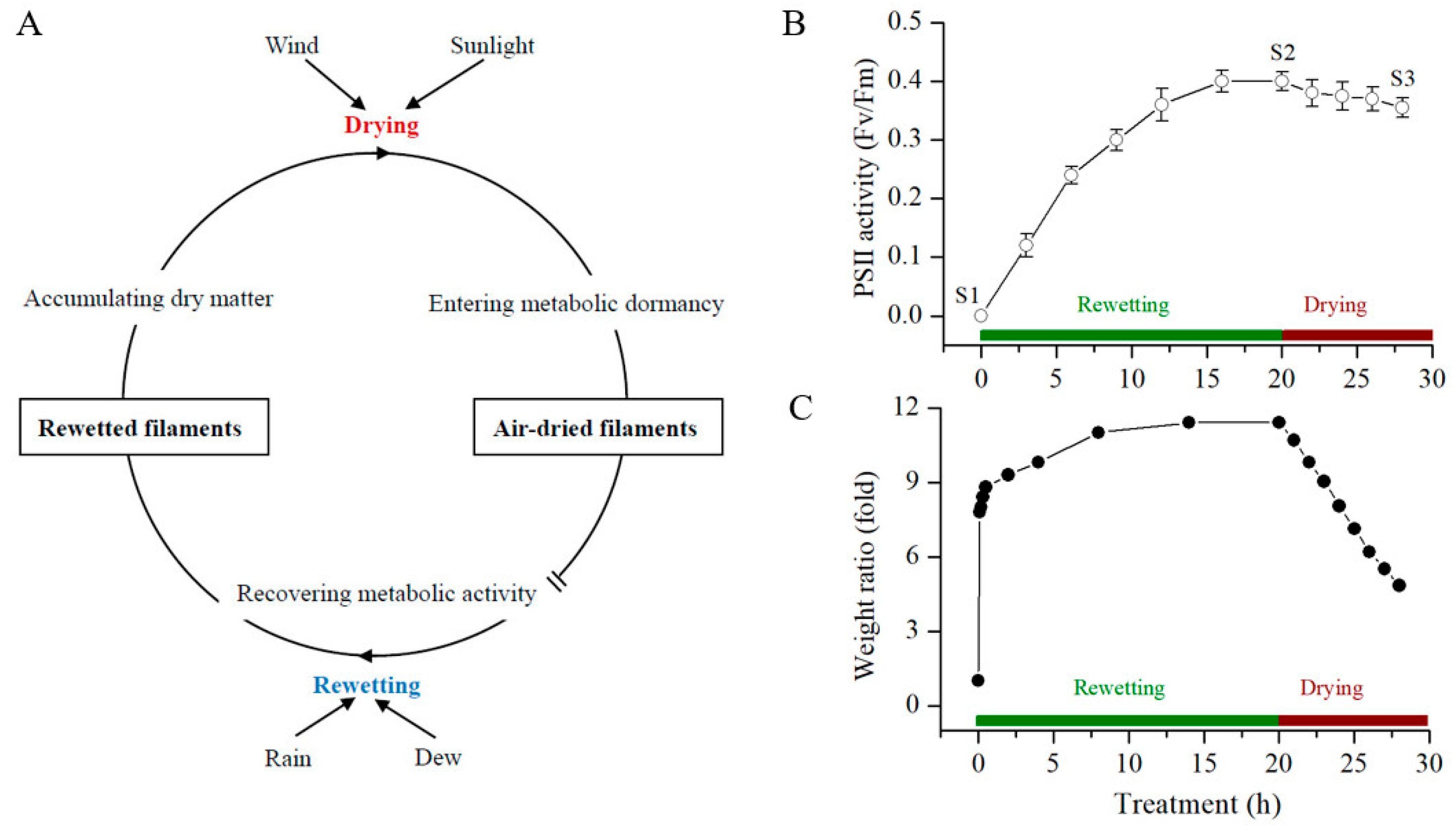
2.1. Overview of the Rehydration and Dehydration Processes
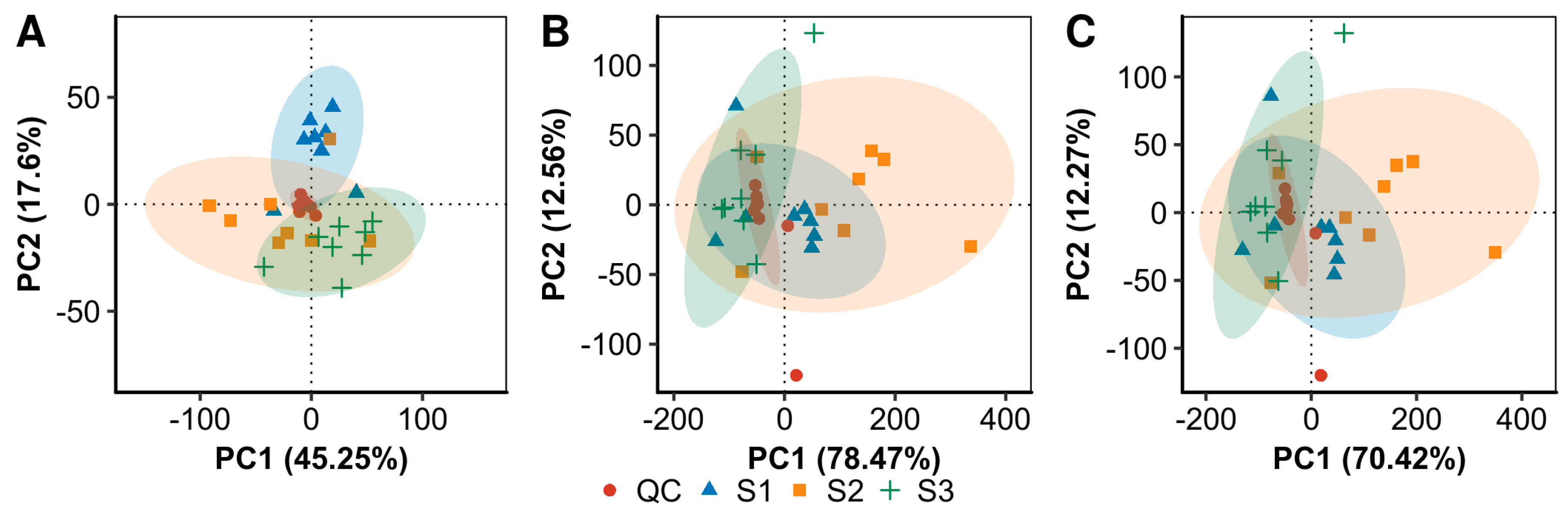
2.2. LC-MS-Based Metabolomic Analysis
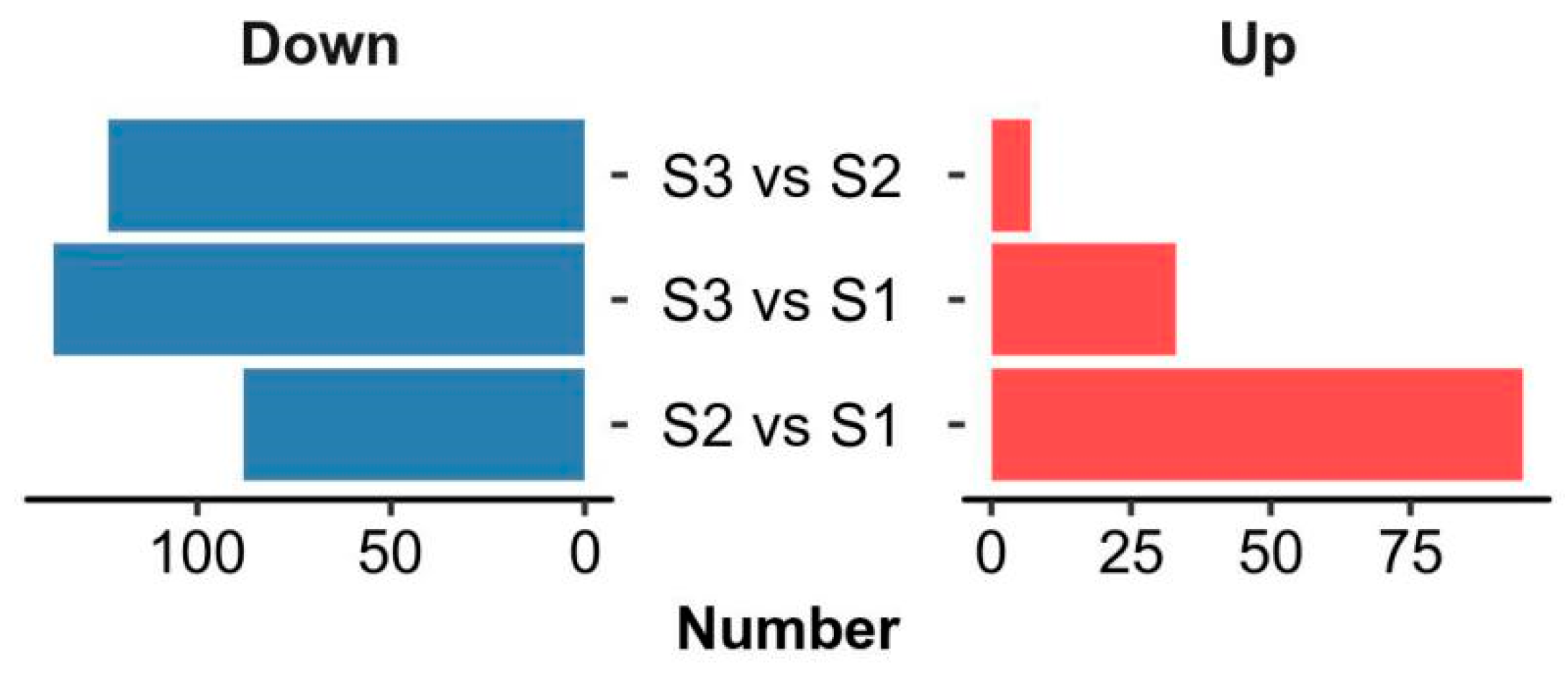
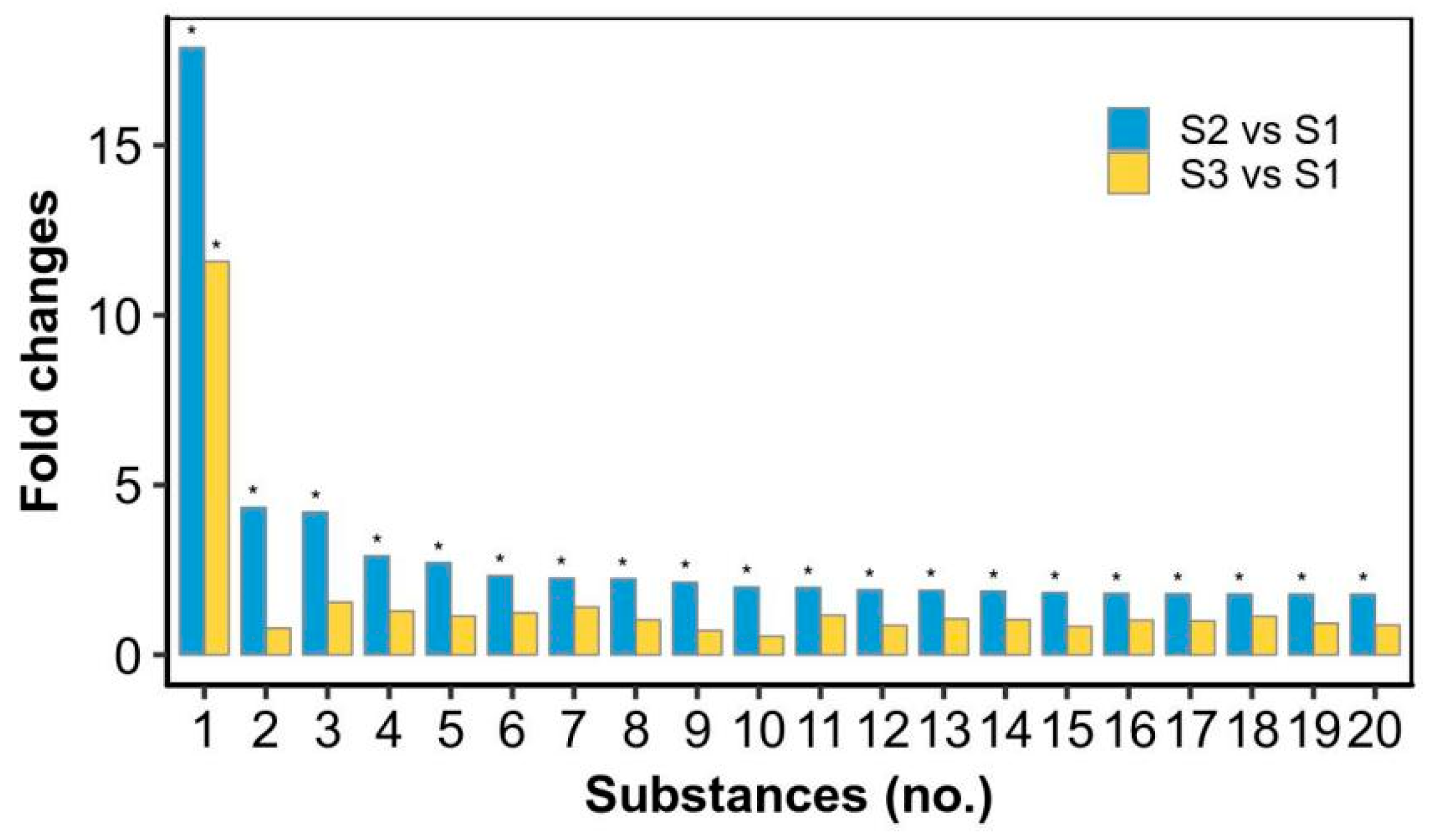
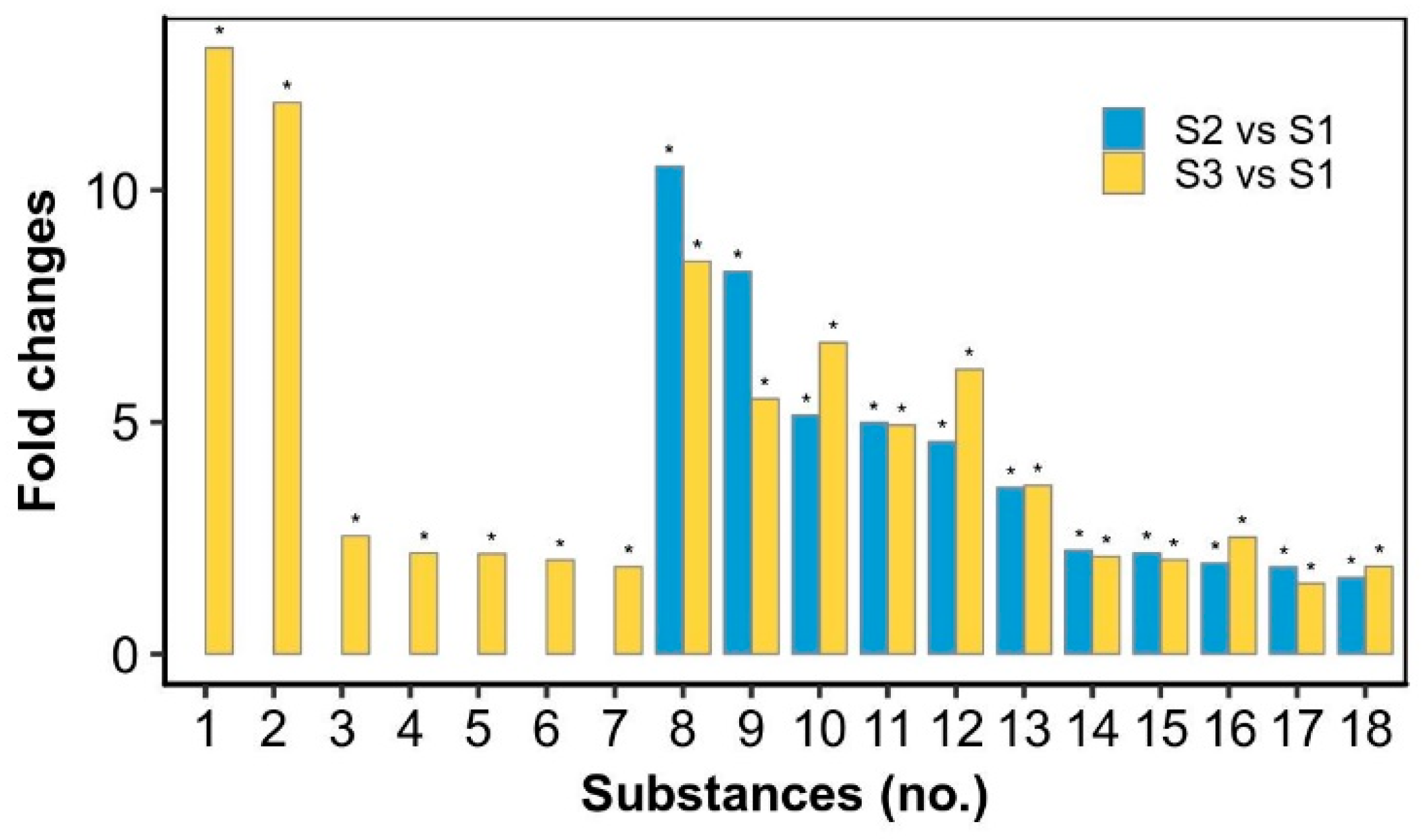
2.3. Metabolic Signatures in the Rehydration and Dehydration Processes
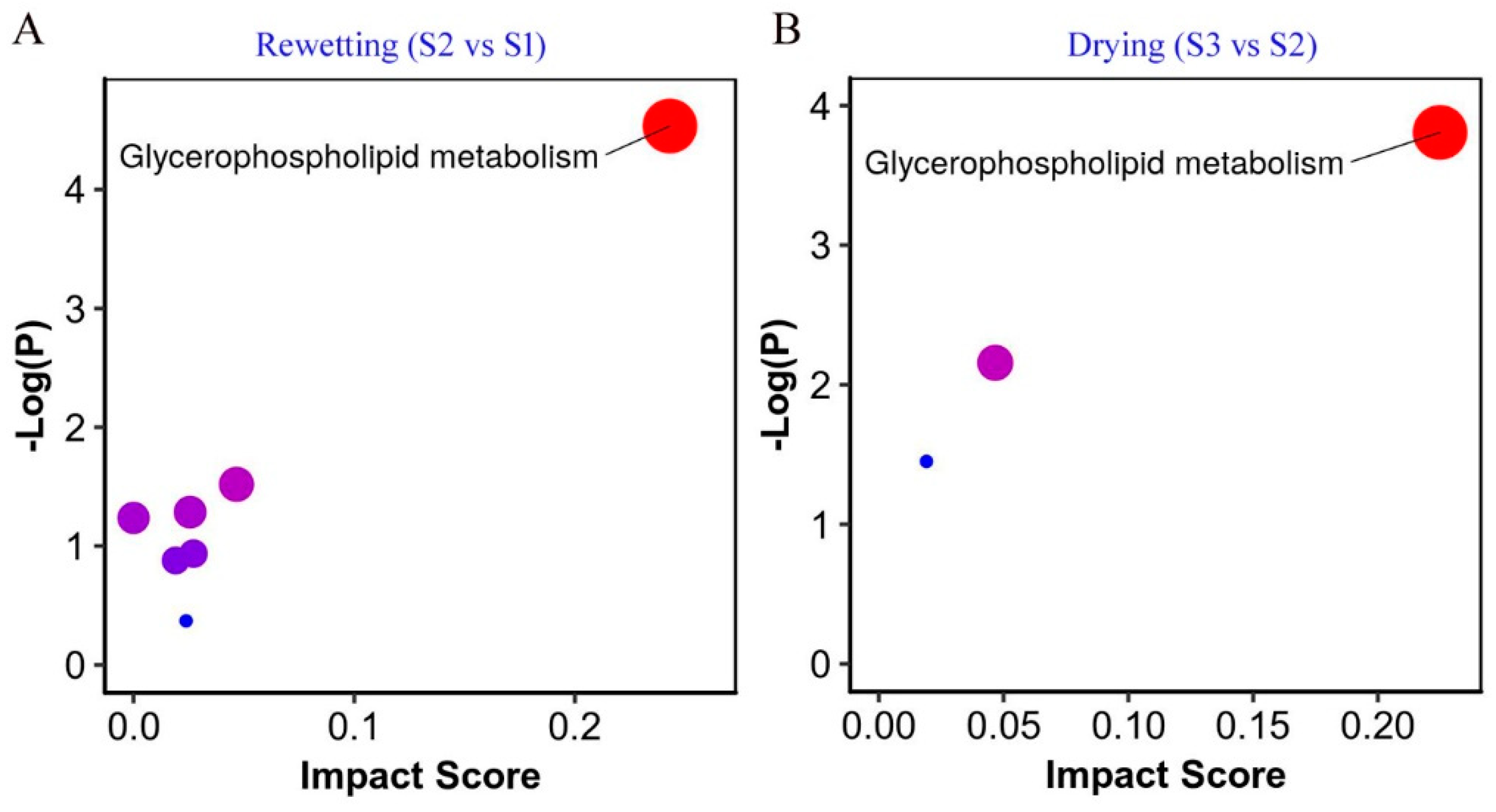
2.4. Metabolic Pathway Analysis
3. Material and Methods
3.1. Organisms, Culture Conditions, Sampling and Treatments
3.2. Metabolite Extraction
3.3. LC-MS Analysis
3.4. Metabolomic Analysis
3.5. Identification of Differential Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Schwarz, D.; Orf, I.; Kopka, J.; Hagemann, M. Recent applications of metabolomics toward cyanobacteria. Metabolites 2013, 3, 72–100. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, C.; Mitra, M.; Bhayani, K.; Bharadwaj, S.V.V.; Ghosh, T.; Dubey, S.; Mishra, S. Abiotic stresses as tools for metabolites in microalgae. Bioresour. Technol. 2017, 244, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Goulitquer, S.; Potin, P.; Tonon, T. Mass spectrometry-based metabolomics to elucidate functions in marine organisms and ecosystems. Mar. Drugs 2012, 10, 849–880. [Google Scholar] [CrossRef]
- Baran, R.; Ivanova, N.N.; Jose, N.; Garcia-Pichel, F.; Kyrpides, N.C.; Gugger, M.; Northen, T.R. Functional genomics of novel secondary metabolites from diverse cyanobacteria using untargeted metabolomics. Mar. Drugs 2013, 11, 3617–3631. [Google Scholar] [CrossRef] [PubMed]
- Dodds, W.K.; Gudder, D.A.; Mollenhauer, D. The ecology of Nostoc. J. Phycol. 1995, 31, 2–18. [Google Scholar] [CrossRef]
- Mazard, S.; Penesyan, A.; Ostrowski, M.; Paulsen, I.T.; Egan, S. Tiny microbes with a big impact: The role of cyanobacteria and their metabolites in shaping our future. Mar. Drugs 2016, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotechnol. 2010, 21, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Sakamoto, T.; Matsugo, S. Multiple roles of photosynthetic and sunscreen pigments in cyanobacteria focusing on the oxidative stress. Metabolites 2013, 3, 463–483. [Google Scholar] [CrossRef] [PubMed]
- França, M.B.; Panek, A.D.; Eleutherio, E.C.A. Oxidative stress and its effects during dehydration. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Gao, K. Chinese studies on the edible blue-green alga, Nostoc flagelliforme: A review. J. Appl. Phycol. 1998, 10, 37–49. [Google Scholar] [CrossRef]
- Gao, X.; Yang, Y.; Ai, Y.; Luo, H.; Qiu, B. Quality evaluation of the edible blue-green alga Nostoc flagelliforme using a chlorophyll fluorescence parameter and several biochemical markers. Food Chem. 2014, 143, 307–312. [Google Scholar] [CrossRef]
- Liang, W.; Zhou, Y.; Wang, L.; You, X.; Zhang, Y.; Cheng, C.-L.; Chen, W. Ultrastructural, physiological and proteomic analysis of Nostoc flagelliforme in response to dehydration and rehydration. J. Proteom. 2012, 75, 5604–5627. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, L.; Zhang, Y.; Chen, S.; Gao, X.; Wan, C. Investigation of the dynamical expression of Nostoc flagelliforme proteome in response to rehydration. J. Proteom. 2019, 192, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ai, Y.; Qiu, B. Drought adaptation of a terrestrial macroscopic cyanobacterium, Nostoc flagelliforme, in arid areas: A review. Afr. J. Microbiol. Res. 2012, 6, 5728–5735. [Google Scholar]
- Ye, C.P.; Zhang, M.C.; Yang, Y.F.; Thirumaran, G. Photosynthetic performance in aquatic and terrestrial colonies of Nostoc flagelliforme (Cyanophyceae) under aquatic and aerial conditions. J. Arid Environ. 2012, 85, 56–61. [Google Scholar] [CrossRef]
- Zhao, X.M.; Bi, Y.H.; Chen, L.; Hu, S.; Hu, Z.Y. Responses of photosynthetic activity in the drought-tolerant cyanobacterium, Nostoc flagelliforme to rehydration at different temperature. J. Arid Environ. 2008, 72, 370–377. [Google Scholar] [CrossRef]
- Bartsch, H.; Nair, J. New DNA-based biomarkers for oxidative stress and cancer chemoprevention studies. Eur. J. Cancer 2000, 36, 1229–1234. [Google Scholar] [CrossRef]
- Hanaoka, T.; Nair, J.; Takahashi, Y.; Sasaki, S.; Bartsch, H.; Tsugane, S. Urinary level of 1,N6-ethenodeoxyadenosine, a marker of oxidative stress, is associated with salt excretion and ω6-polyunsaturated fatty acid intake in postmenopausal Japanese women. Int. J. Cancer 2002, 100, 71–75. [Google Scholar] [CrossRef]
- Meerang, M.; Nair, J.; Sirankapracha, P.; Thephinlap, C.; Srichairatanakool, S.; Fucharoen, S.; Bartsch, H. Increased urinary 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine excretion in thalassemia patients: Markers for lipid peroxidation-induced DNA damage. Free Radic. Biol. Med. 2008, 44, 1863–1868. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P.; Birincioglu, M.; Rodriguez, H. Free radical-induced damage to DNA: Mechanisms and measurement. Free Radic. Biol. Med. 2002, 32, 1102–1115. [Google Scholar] [CrossRef]
- Shirkey, B.; McMaster, N.J.; Smith, S.C.; Wright, D.J.; Rodriguez, H.; Jaruga, P.; Birincioglu, M.; Helm, R.F.; Potts, M. Genomic DNA of Nostoc commune (Cyanobacteria) becomes covalently modified during long-term (decades) desiccation but is protected from oxidative damage and degradation. Nucleic Acids Res. 2003, 31, 2995–3005. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abed, R.M.M.; Kohls, K.; Schoon, R.; Scherf, A.-K.; Rullkötter, J.; Schacht, M.; Palinska, K.A.; Al-Hassani, H.; Hamza, W.; Golubic, S. Lipid biomarkers, pigments and cyanobacterial diversity of microbial mats across intertidal flats of the arid coast of the Arabian Gulf (Abu Dhabi, UAE). Fems Microbiol. Ecol. 2008, 65, 449–462. [Google Scholar] [CrossRef]
- Rajendran, U.M.; Kathirvel, E.; Anand, N. Desiccation-induced changes in antioxidant enzymes, fatty Acids, and amino acids in the cyanobacterium Tolypothrix scytonemoides. World J. Microbiol. Biotechnol. 2007, 23, 251–257. [Google Scholar] [CrossRef]
- Liu, X.-J.; Chen, F.; Jiang, Y. Differentiation of Nostoc flagelliforme and its neighboring species using fatty-acid profiling as a chemotaxonomic tool. Curr. Microbiol. 2003, 47, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Eklund, M.; Bauer, E.; Wamatu, J.; Mosenthin, R. Potential nutritional and physiological functions of betaine in livestock. Nutr. Res. Rev. 2005, 18, 31–48. [Google Scholar] [CrossRef]
- Inoue, S.; Kitajima, K. KDN (Deaminated neuraminic acid): Dreamful past and exciting future of the newest member of the sialic acid family. Glycoconj. J. 2006, 23, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-H.; Chen, Y.-Y.; Pan, H.-H.; Wang, F.-P.; Cheng, C.-H.; Lee, W.-C. Production of N-acetyl-D-neuraminic acid using two sequential enzymes overexpressed as double-tagged fusion proteins. BMC Biotechnol. 2009, 9, 63. [Google Scholar] [CrossRef]
- Punginelli, C.; Wilson, A.; Routaboul, J.-M.; Kirilovsky, D. Influence of zeaxanthin and echinenone binding on the activity of the Orange Carotenoid Protein. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 280–288. [Google Scholar] [CrossRef]
- Ukibe, K.; Hashida, K.; Yoshida, N.; Takagi, H. Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Appl. Environ. Microbiol. 2009, 75, 7205–7211. [Google Scholar] [CrossRef]
- Mochimaru, M.; Masukawa, H.; Takaichi, S. The cyanobacterium Anabaena sp. PCC 7120 has two distinct β-carotene ketolases: CrtO for echinenone and CrtW for ketomyxol synthesis. Febs Lett. 2005, 579, 6111–6114. [Google Scholar] [CrossRef]
- Zolman, B.K.; Martinez, N.; Millius, A.; Adham, A.R.; Bartel, B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 2008, 180, 237–251. [Google Scholar] [CrossRef]
- Jin, H.; Du, F.; Xie, J.; Zhao, J.; Wu, P. Culture of Nostoc flagelliforme Born. et Flah. J. Inn. Mong. Inst. Agric. Anim. Husb. 1999, 20, 43–48. (in Chinese). [Google Scholar]
- El-Gaied, L.F.; Abu El-Heba, G.A.; El-Sherif, N.A. Effect of growth hormones on some antioxidant parameters and gene expression in tomato. GM Crop. Food 2013, 4, 67–73. [Google Scholar] [CrossRef]
- Hong, S.-M.; Min, Z.W.; Mok, C.; Kwon, H.; Kim, T.; Kim, D. Aqueous degradation of imidacloprid and fenothiocarb using contact glow discharge electrolysis: Degradation behavior and kinetics. Food Sci. Biotechnol. 2013, 22, 1773–1778. [Google Scholar] [CrossRef]
- Luck, K.; Jia, Q.; Huber, M.; Handrick, V.; Wong, G.K.-S.; Nelson, D.R.; Chen, F.; Gershenzon, J.; Köllner, T.G. CYP79 P450 monooxygenases in gymnosperms: CYP79A118 is associated with the formation of taxiphyllin in Taxus Baccata. Plant Mol. Biol. 2017, 95, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.E.; Chutuape, M.A.; Strain, E.C.; Walsh, S.L.; Stitzer, M.L.; Bigelow, G.E. A Comparison of levomethadyl acetate, buprenorphine, and methadone for opioid dependence. N. Engl. J. Med. 2000, 343, 1290–1297. [Google Scholar] [CrossRef]
- Zhao, G.; Winkler, M.E. A novel alpha-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J. Bacteriol. 1996, 178, 232–239. [Google Scholar] [CrossRef]
- Rodgers, G.A. Inhibition of soil urease activity by aminocresols. Plant Soil 1984, 79, 155–157. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Hu, S.; Yu, Y.; Wu, X.; Xia, X.; Xiao, X.; Wu, H. Comparative proteomic analysis of Cronobacter sakazakii by iTRAQ provides insights into response to desiccation. Food Res. Int. 2017, 100, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Moyankova, D.; Lin, C.-T.; Mladenov, P.; Sun, R.-Z.; Djilianov, D.; Deng, X. Transcriptome reprogramming during severe dehydration contributes to physiological and metabolic changes in the resurrection plant Haberlea Rhodopensis. BMC Plant Biol. 2018, 18, 351. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yang, G.; Zhang, L.; Yang, X.; Zhao, S.; Ji, Z.; Zhou, Q.; Hu, M.; Wang, Y.; Chen, M.; et al. The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proc. Natl. Acad. Sci. USA 2015, 112, 5833–5837. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.C.; Sinha, R.P.; Häider, D.P. Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 2002, 41, 297–308. [Google Scholar]
- Cui, L.; Liu, Y.; Yang, Y.; Ye, S.; Luo, H.; Qiu, B.; Gao, X. The drnf1 gene from the drought-adapted cyanobacterium Nostoc flagelliforme improved salt tolerance in transgenic Synechocystis and Arabidopsis plant. Genes 2018, 9, 441. [Google Scholar] [CrossRef]
- Loftus, N.; Barnes, A.; Ashton, S.; Michopoulos, F.; Theodoridis, G.; Wilson, I.; Ji, C.; Kaplowitz, N. Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J. Proteome Res. 2011, 10, 705–713. [Google Scholar] [CrossRef] [PubMed]
| Substances | Formula | S1 | S2 | S3 |
|---|---|---|---|---|
| 4-Amino-o-cresol | C7H9NO | - | + | ++ |
| (2’S)-Deoxymyxol 2’-(2,4-di-O-methyl-α-l-fucoside) | C48H70O6 | - | + | + |
| PE(15:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | C42H70NO8P | - | + | + |
| MGDG(18:5(3Z,6Z,9Z,12Z,15Z)/18:5(3Z,6Z,9Z,12Z,15Z)) | C45H66O10 | - | + | + |
| Asparaginyl-Threonine | C7H9NO | - | - | + |
| Tropine | C8H15NO | - | - | + |
| Coronene | C24H12 | - | - | + |
| PC(18:0/19:3(9Z,12Z,15Z))[U] | C45H84NO8P | ++ | + | - |
| PS(19:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | C47H78NO10P | + | - | - |
| DG(13:0/18:3(9Z,12Z,15Z)/0:0)[iso2] | C34H60O5 | + | - | - |
| PC(18:4(6Z,9Z,12Z,15Z)/20:5(5Z,8Z,11Z,14Z,17Z)) | C46H74NO8P | + | - | - |
| 9-hydroperoxy-12,13-epoxy-10-octadecenoic acid | C18H32O5 | + | - | - |
| CDP-DG(18:0/18:0) | C48H89N3O15P2 | + | - | - |
| 4-Ketonostoxanthin 3-sulfate | C40H53NaO8S | + | - | - |
| TG(12:0/12:0/18:1(9Z))[iso3] | C45H84O6 | + | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Liu, B.; Ji, B. Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation. Mar. Drugs 2019, 17, 298. https://doi.org/10.3390/md17050298
Gao X, Liu B, Ji B. Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation. Marine Drugs. 2019; 17(5):298. https://doi.org/10.3390/md17050298
Chicago/Turabian StyleGao, Xiang, Bin Liu, and Boyang Ji. 2019. "Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation" Marine Drugs 17, no. 5: 298. https://doi.org/10.3390/md17050298
APA StyleGao, X., Liu, B., & Ji, B. (2019). Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation. Marine Drugs, 17(5), 298. https://doi.org/10.3390/md17050298






