Marine Bacterium Vibrio sp. CB1-14 Produces Guanidine Alkaloid 6-epi-Monanchorin, Previously Isolated from Marine Polychaete and Sponges
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Microorganisms
2.2. Preliminary Identification of Potential Microorganism-Producers by Monitoring of the Compounds Giving Ion Peaks at m/z 212.17 by MALDI MS, Characteristic of 1 and 2 MS
2.3. Identification of Compounds 1 or 2 by HRESIMS
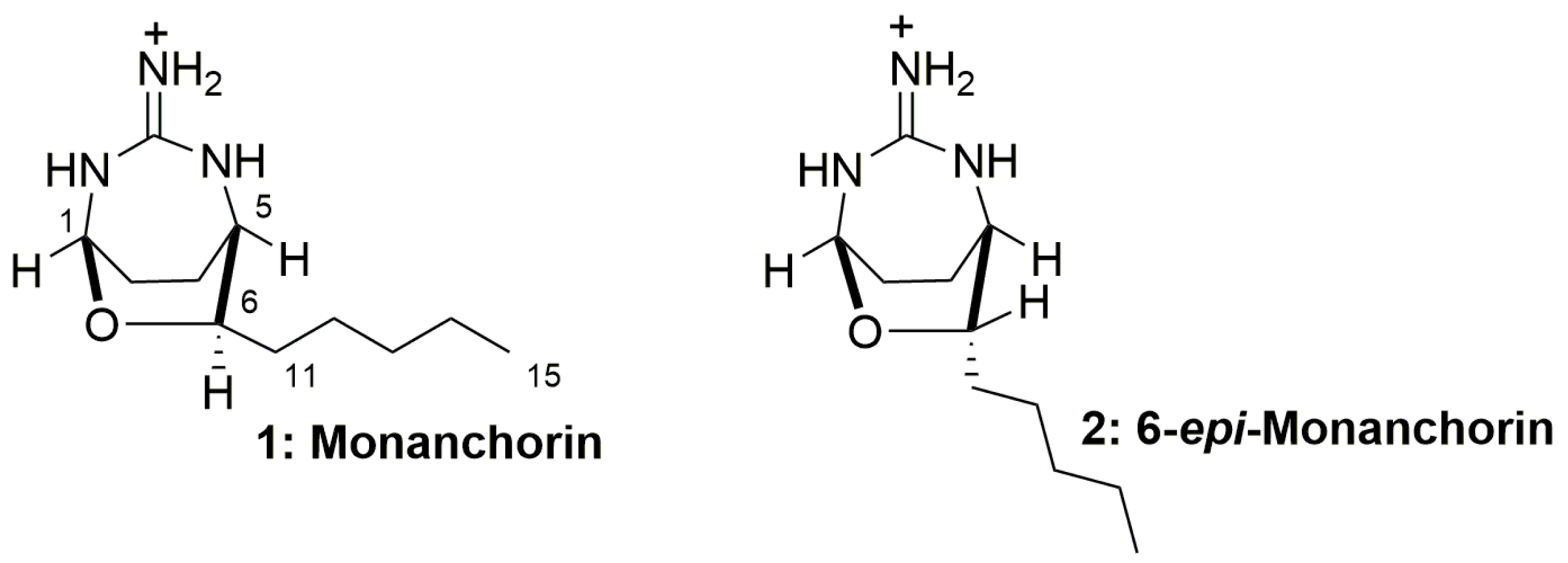
2.4. Isolation and Identification of 6-epi-Monanchorin by 1H NMR Spectroscopy
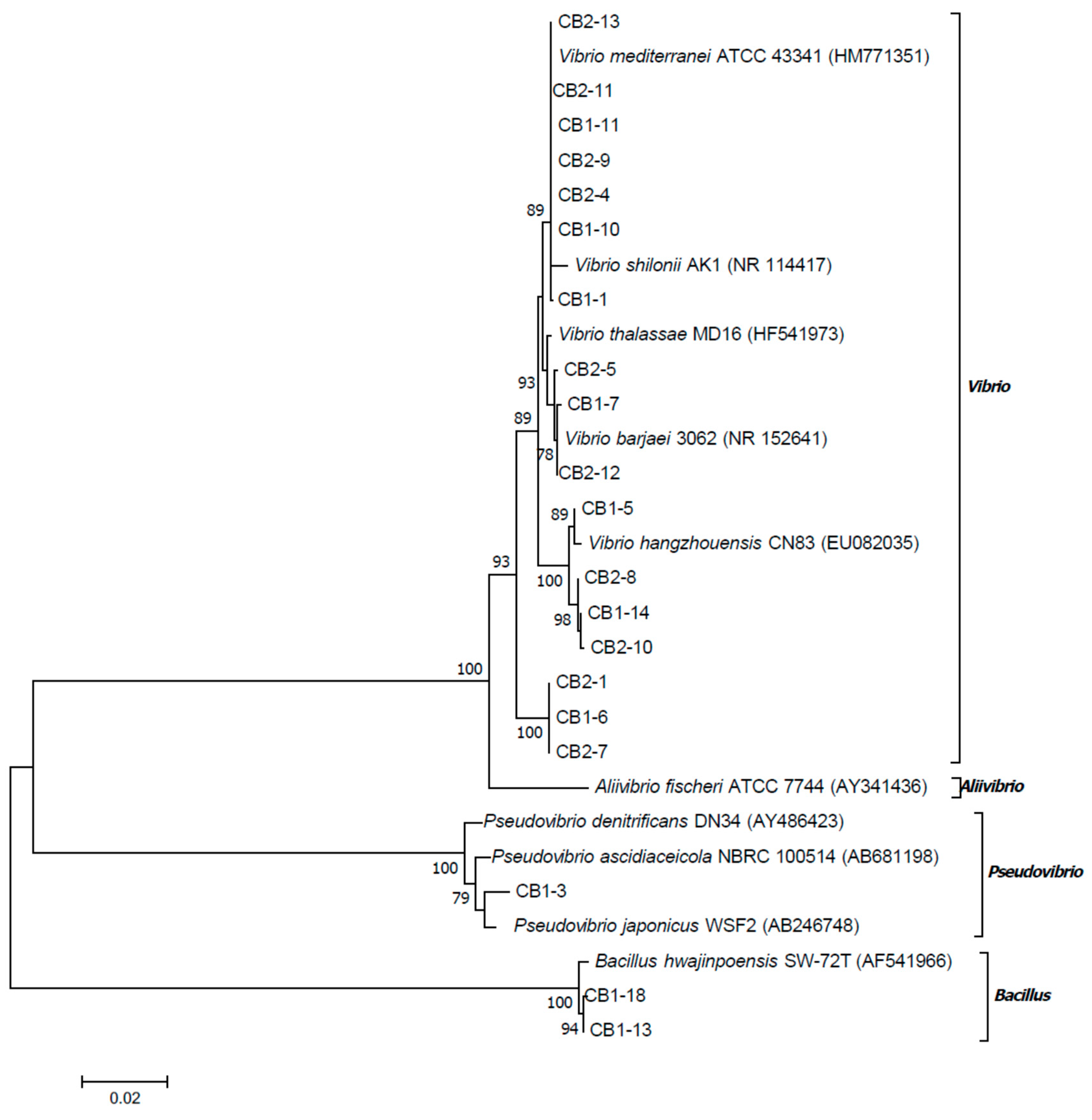
2.5. 16S rRNA Gene Sequence Analysis of Bacterial Isolates
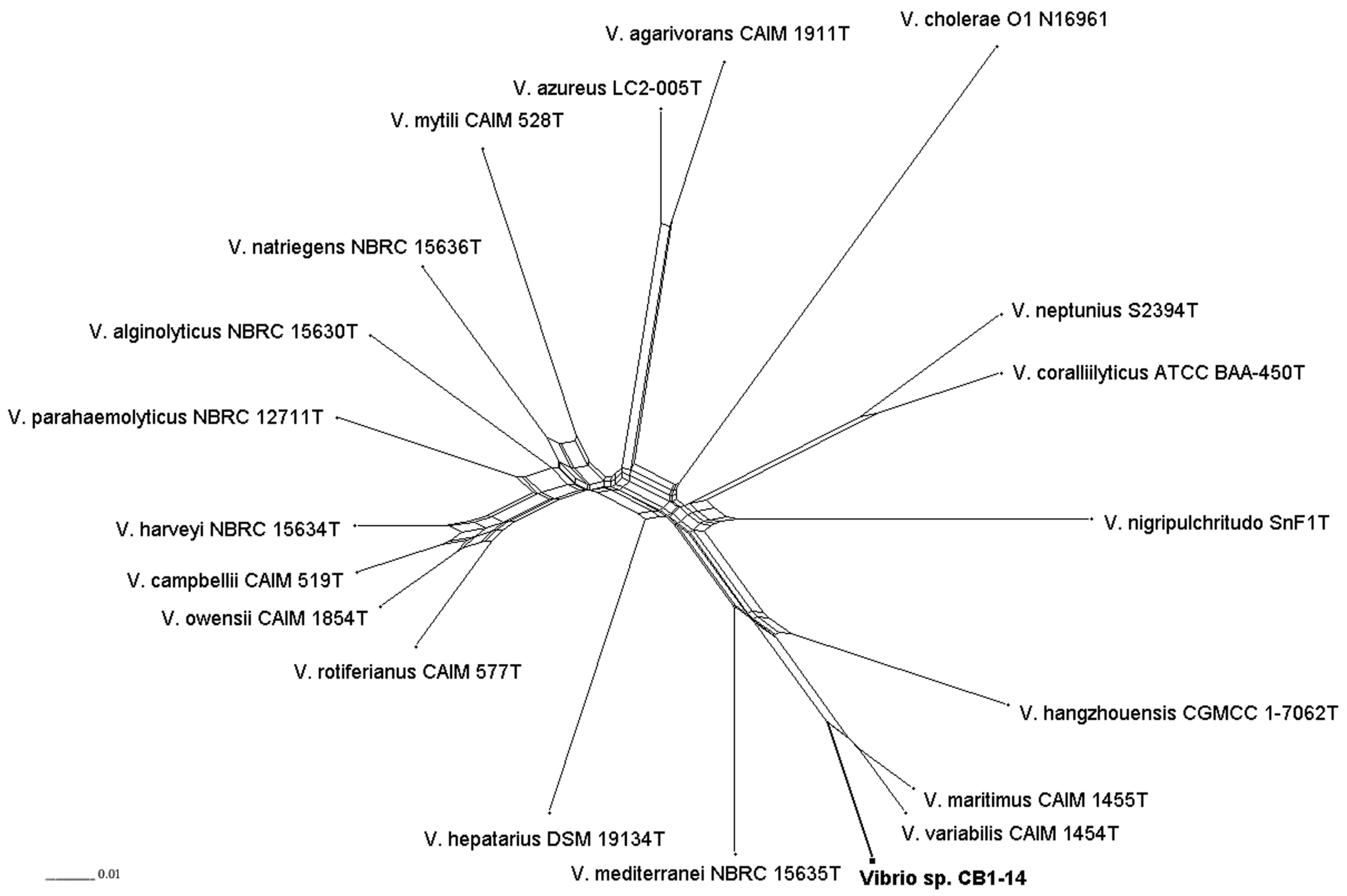
2.6. Multilocus Sequence Analysis of CB1-14
3. Materials and Methods
3.1. General
3.2. Animal Material
3.3. Isolation of Microorganisms
3.4. Identification of Microorganisms by MALDI MS
3.5. Incubation of Microorganisms for HRESI MS Analysis of Compounds
3.6. Isolation and Structure Identification of 6-epi-Monanchorin
3.7. DNA Isolation and Amplification and Phylogenetic Analysis of 16S rDNA Gene
3.8. Genome Sequencing and Multilocus Sequence Analysis of CB1-14
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berlinck, R.G.S.; Bertonha, A.F.; Takaki, M.; Rodriguez, J.P.G. The chemistry and biology of guanidine natural products. Nat. Prod. Rep. 2017, 34, 1264–1301. [Google Scholar] [CrossRef]
- Meragelman, K.M.; McKee, T.C.; McMahon, J.B. Monanchorin, a bicyclic alkaloid from the sponge Monanchora ungiculata. J. Nat. Prod. 2004, 67, 1165–1167. [Google Scholar] [CrossRef]
- Abdjul, D.B.; Yamazaki, H.; Kanno, S.; Takahashi, O.; Kirikoshi, R.; Ukai, K.; Namikoshi, M. Haliclonadiamine derivatives and 6-epi-monanchorin from the marine sponge Halichondria panicea collected at Iriomote Island. J. Nat. Prod. 2016, 79, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Shubina, L.K.; Makarieva, T.N.; Denisenko, V.A.; Dmitrenok, P.S.; Dyshlovoy, S.A.; von Amsberg, G.; Glazunov, V.P.; Silchenko, A.S.; Stonik, I.V.; Lee, H.S.; et al. Absolute configuration and body part distribution of alkaloid 6-epi-monanchorin from the marine polychaete Chaetopterus variopedatus. Nat. Prod. Commun. 2016, 11, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Enders, H.E. A study of the life history and habits of Chaetopterus variopedatus. J. Morphol. 1909, 20, 479–531. [Google Scholar] [CrossRef]
- MacGinitie, G.E. The method of feeding in Chaetopterus. Biol. Bull. 1939, 77, 115–118. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Stanier, R.Y. Isolation and Growth of Cyanobacteria from Marine and Hypersaline Environments. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin, Germany, 1981; pp. 221–223. [Google Scholar]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Kita-Tsukamoto, K.; Oyaizu, H.; Nanba, K.; Simidu, U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int. J. Syst. Bacteriol. 1993, 43, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of Vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007, 189, 7932–7936. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Ogura, Y.; Matsumura, Y.; Gao, F.; Amin, A.K.M.; Mino, S.; Nakagawa, S.; Sawabe, T.; Kumar, R.; Fukui, Y.; Satomi, M. Updating the Vibrio clades defined by multilocus sequence phylogeny: Proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 2013, 4, 414. [Google Scholar] [CrossRef]
- Rizzo, C.; Lo Giudice, A. Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules. Diversity (Basel) 2018, 10, 52. [Google Scholar] [CrossRef]
- Schmitz, F.J.; Vanderah, D.J.; Hollenbeak, K.H.; Enwall, C.E.L.; Gopichand, Y.; Sengupta, P.K.; Hossain, M.B.; van der Helm, D. Metabolites from the marine sponge Tedania ignes―A new atisanediol and several known diketopiperazines. J. Org. Chem. 1983, 48, 3941–3945. [Google Scholar] [CrossRef]
- Nicacio, K.J.; Ioca, L.P.; Froes, A.M.; Leomil, L.; Appolinario, L.R.; Thompson, C.C.; Thompson, F.L.; Ferreira, A.G.; Williams, D.E.; Andersen, R.J.; et al. Cultures of the Marine Bacterium Pseudovibrio denitrificans Ab134 Produce Bromotyrosine-Derived Alkaloids Previously Only Isolated from Marine Sponges. J. Nat. Prod. 2018, 80, 235–240. [Google Scholar] [CrossRef]
- Stierle, A.C.; Cardellina, J.H., II; Singleton, F.L. A marine micrococus produces metabolites ascribed to the sponge Tedania ignis. Experientia 1988, 44, 1021. [Google Scholar] [CrossRef]
- Elyakov, G.B.; Kuznetsova, T.A.; Mikhailov, V.V.; Maltsev, I.I.; Voinov, V.G.; Fedoreyev, S.A. Brominated diphenyl ethers from a marine bacterium associated with the sponge Dysidea sp. Experientia 1991, 47, 632–633. [Google Scholar] [CrossRef]
- Agarwal, V.; El Gamal, A.A.; Yamanaka, K.; Poth, D.; Kersten, R.D.; Schorn, M.; Allen, E.E.; Moore, B.S. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 2014, 10, 640–647. [Google Scholar] [CrossRef]
- Unson, M.D.; Faulkner, D.J. Cyanobacterial symbiont biosynthesis of chlorinated metabolites from Dysidea herbacea (Porifera). Experientia 1993, 49, 349–353. [Google Scholar] [CrossRef]
- Oclarit, J.M.; Okada, H.; Ohta, S.; Kaminura, K.; Yamaoka, Y.; Iizuka, T.; Miyashiro, S.; Ikegami, S. Anti-bacillus substance in the marine sponge Hyatella species, prodiced by an associated Vibrio species bacterium. Microbios 1994, 78, 7–16. [Google Scholar]
- Schmidt, E.W.; Donia, M.S.; McIntosh, J.A.; Fricke, W.F.; Ravel, J. Origin and Variation of Tunicate Secondary Metabolites. J. Nat. Prod. 2012, 75, 95–304. [Google Scholar] [CrossRef]
- Rizzo, C.; Michaud, L.; Hormann, B.; Gerce, B.; Syldatk, C.; Hausmann, R.; De Domenico, E.; Lo Giudice, A. Bacteria associated with sabellids (Polychaeta: Annelida) as a novel source of surface active compounds. Mar. Pollut. Bull. 2013, 70, 125–133. [Google Scholar] [CrossRef]
- Lemos, M.L.; Balado, M.; Osorio, C.R. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: Evolution and ecology of a fish pathogen. Environ. Microbiol. Rep. 2010, 2, 19–26. [Google Scholar] [CrossRef]
- Noguchi, T.; Hwang, D.F.; Arakawa, O.; Sugita, H.; Deguchi, Y.; Shida, Y.; Hashimoto, K. Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu-Vermicularis vermicularis. Mar. Biol. 1987, 94, 625–630. [Google Scholar] [CrossRef]
- Lee, M.J.; Jeong, D.Y.; Kim, W.S.; Kim, H.D.; Kim, C.H.; Park, W.W.; Park, Y.H.; Kim, K.S.; Kim, H.M.; Kim, D.S. A tetrodotoxin-producing Vibrio strain, LM-1, from the puffer fish Fugu Vermicularis radiatus. Appl. Environ. Microbiol. 2000, 66, 1698–1701. [Google Scholar] [CrossRef]
- Noguchi, T.; Ali, A.E.; Arakawa, O.; Miyazawa, K.; Kanoh, S.; Shida, Y.; Nishio, S.; Hashimoto, K. Tetrodonic acid-like substance―A possible precursor of tetrodotoxin. Toxicon 1991, 29, 845–855. [Google Scholar] [CrossRef]
- Noguchi, T.; Jeon, J.K.; Arakawa, O.; Sugita, H.; Deguchi, Y.; Shida, Y.; Hashimoto, K. Occurrence of tetrodotoxin and anhydrotetrodotoxin in Vibrio sp. isolated from the intestines of a xanthid crab, Atergatis floridus. J. Biochem. 1986, 99, 311–314. [Google Scholar] [CrossRef]
- Stenkova, A.M.; Isaeva, M.P.; Shubin, F.N.; Rasskazov, V.A.; Rakin, A.V. Trends of the Major Porin Gene (ompF) Evolution: Insight from the Genus Yersinia. PLoS ONE 2011, 6, e20546. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Zagnitko, O. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Dress, A.W.M. Split decomposition: A new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1992, 1, 242–252. [Google Scholar] [CrossRef]
| No | Strain | Taxon | m/z 212.17 * | No | Strain | Taxon | m/z 212.17 |
|---|---|---|---|---|---|---|---|
| 1 | CB1-3 | nd | nd | 13 | CB2-3 | nd | nd |
| 2 | CB1-5 | nd | nd | 14 | CB2-4 | Vibrio sp. | nd |
| 3 | CB1-6 | nd | nd | 15 | CB2-5 | Vibrio sp. | nd |
| 4 | CB-1-7 | Vibrio sp. | nd | 16 | CB2-6 | Vibrio sp. | + |
| 5 | CB1-8 | nd | nd | 17 | CB2-7 | nd | ad |
| 6 | CB1-9 | nd | ad | 18 | CB2-8 | Vibrio sp. | ad |
| 7 | CB1-10 | Vibrio sp. | ad | 19 | CB2-9 | Vibrio sp. | ad |
| 8 | CB1-11 | Vibrio sp. | ad | 20 | CB2-10 | nd | ad |
| 9 | CB1-12 | nd | nd | 21 | CB2-11 | Vibrio sp. | + |
| 10 | CB1-13 | nd | nd | 22 | CB2-12 | Vibrio sp. | ad |
| 11 | CB1-14 | nd | + | 23 | CB2-13 | nd | nd |
| 12 | CB2-1 | Vibrio sp. | nd |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makarieva, T.; Shubina, L.; Kurilenko, V.; Isaeva, M.; Chernysheva, N.; Popov, R.; Bystritskaya, E.; Dmitrenok, P.; Stonik, V. Marine Bacterium Vibrio sp. CB1-14 Produces Guanidine Alkaloid 6-epi-Monanchorin, Previously Isolated from Marine Polychaete and Sponges. Mar. Drugs 2019, 17, 213. https://doi.org/10.3390/md17040213
Makarieva T, Shubina L, Kurilenko V, Isaeva M, Chernysheva N, Popov R, Bystritskaya E, Dmitrenok P, Stonik V. Marine Bacterium Vibrio sp. CB1-14 Produces Guanidine Alkaloid 6-epi-Monanchorin, Previously Isolated from Marine Polychaete and Sponges. Marine Drugs. 2019; 17(4):213. https://doi.org/10.3390/md17040213
Chicago/Turabian StyleMakarieva, Tatyana, Larisa Shubina, Valeria Kurilenko, Marina Isaeva, Nadezhda Chernysheva, Roman Popov, Evgeniya Bystritskaya, Pavel Dmitrenok, and Valentin Stonik. 2019. "Marine Bacterium Vibrio sp. CB1-14 Produces Guanidine Alkaloid 6-epi-Monanchorin, Previously Isolated from Marine Polychaete and Sponges" Marine Drugs 17, no. 4: 213. https://doi.org/10.3390/md17040213
APA StyleMakarieva, T., Shubina, L., Kurilenko, V., Isaeva, M., Chernysheva, N., Popov, R., Bystritskaya, E., Dmitrenok, P., & Stonik, V. (2019). Marine Bacterium Vibrio sp. CB1-14 Produces Guanidine Alkaloid 6-epi-Monanchorin, Previously Isolated from Marine Polychaete and Sponges. Marine Drugs, 17(4), 213. https://doi.org/10.3390/md17040213







