Evaluation of Prebiotic Potential of Three Marine Algae Oligosaccharides from Enzymatic Hydrolysis
Abstract
1. Introduction
2. Results
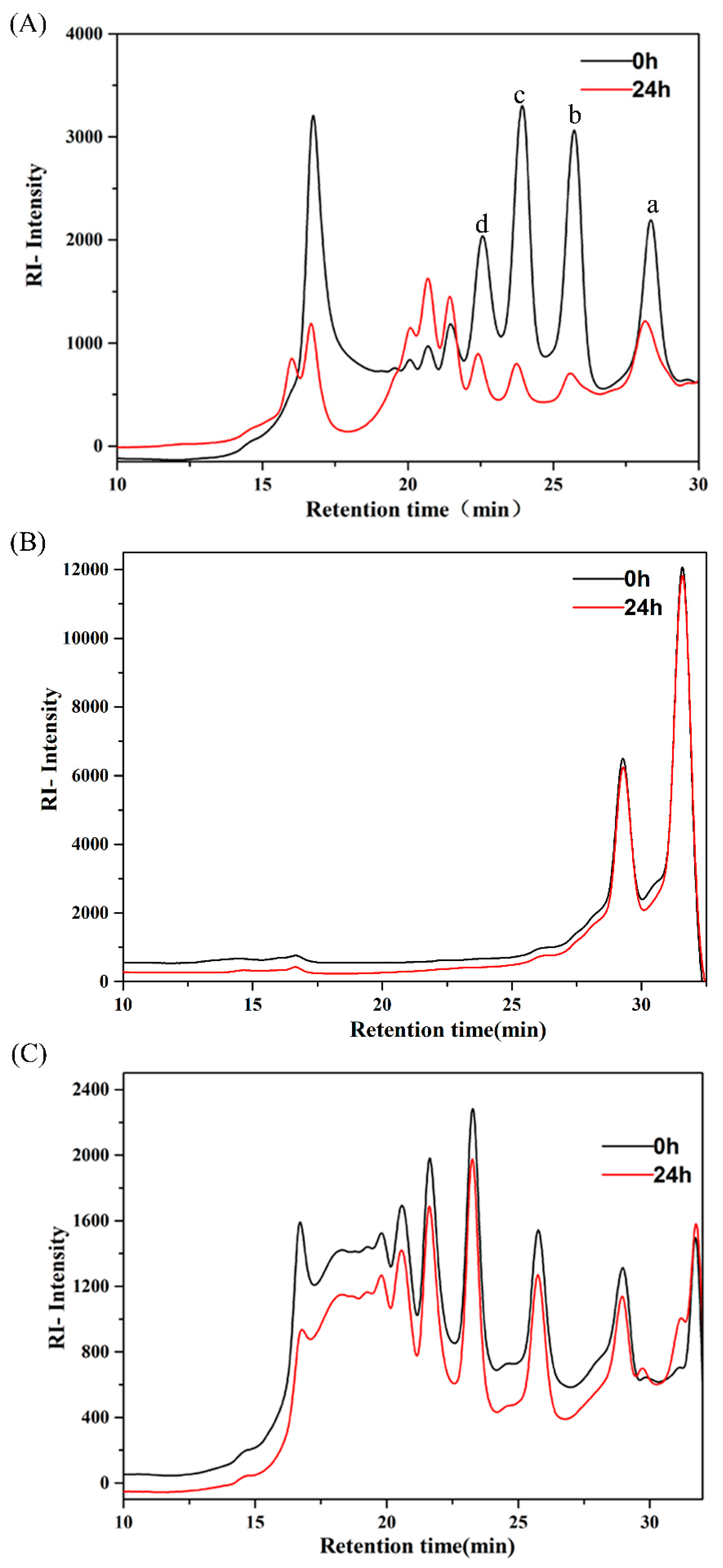
2.1. Utilization of AlgO, AO, and KCO
2.2. SCFA Production from the Fermentation
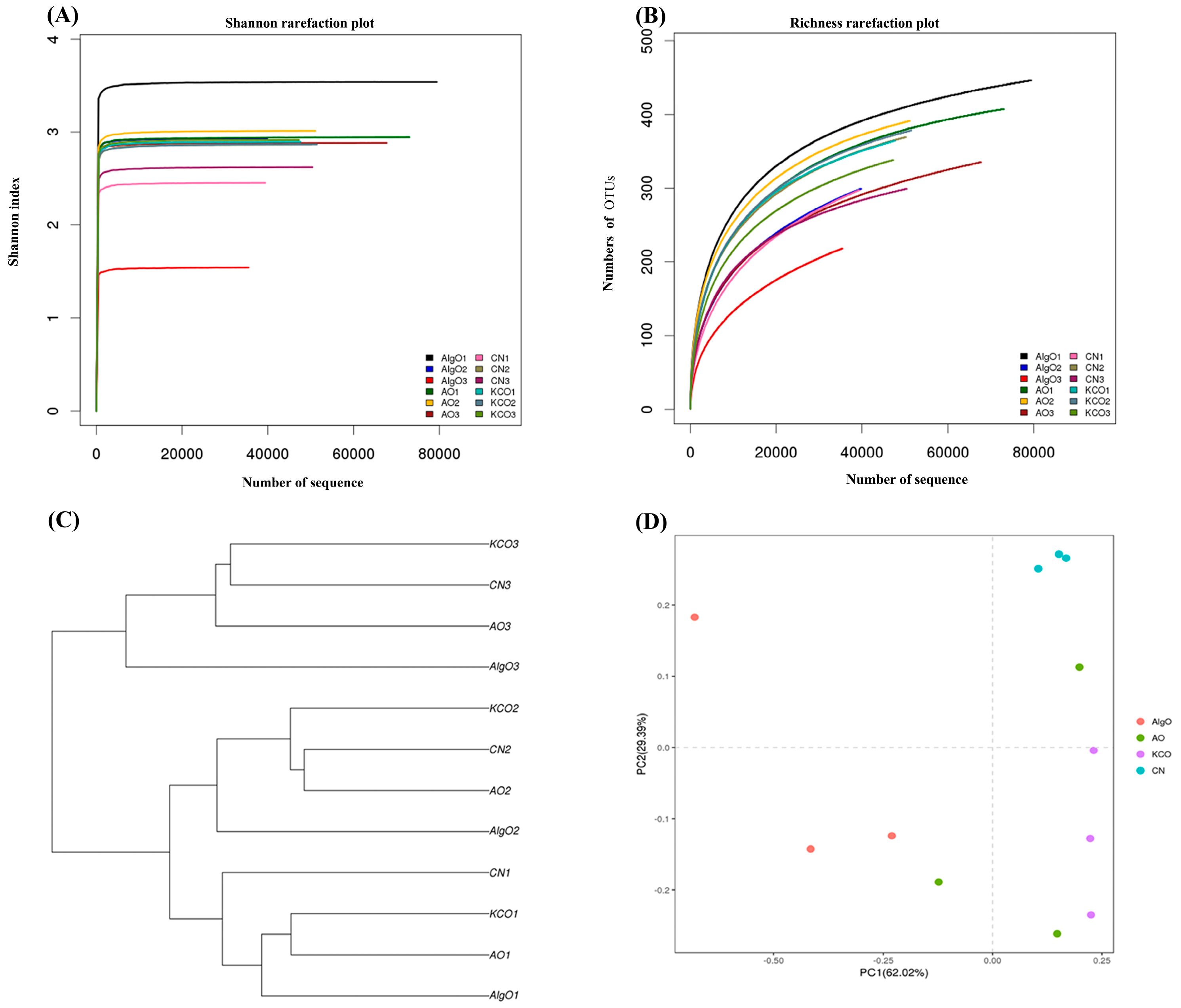
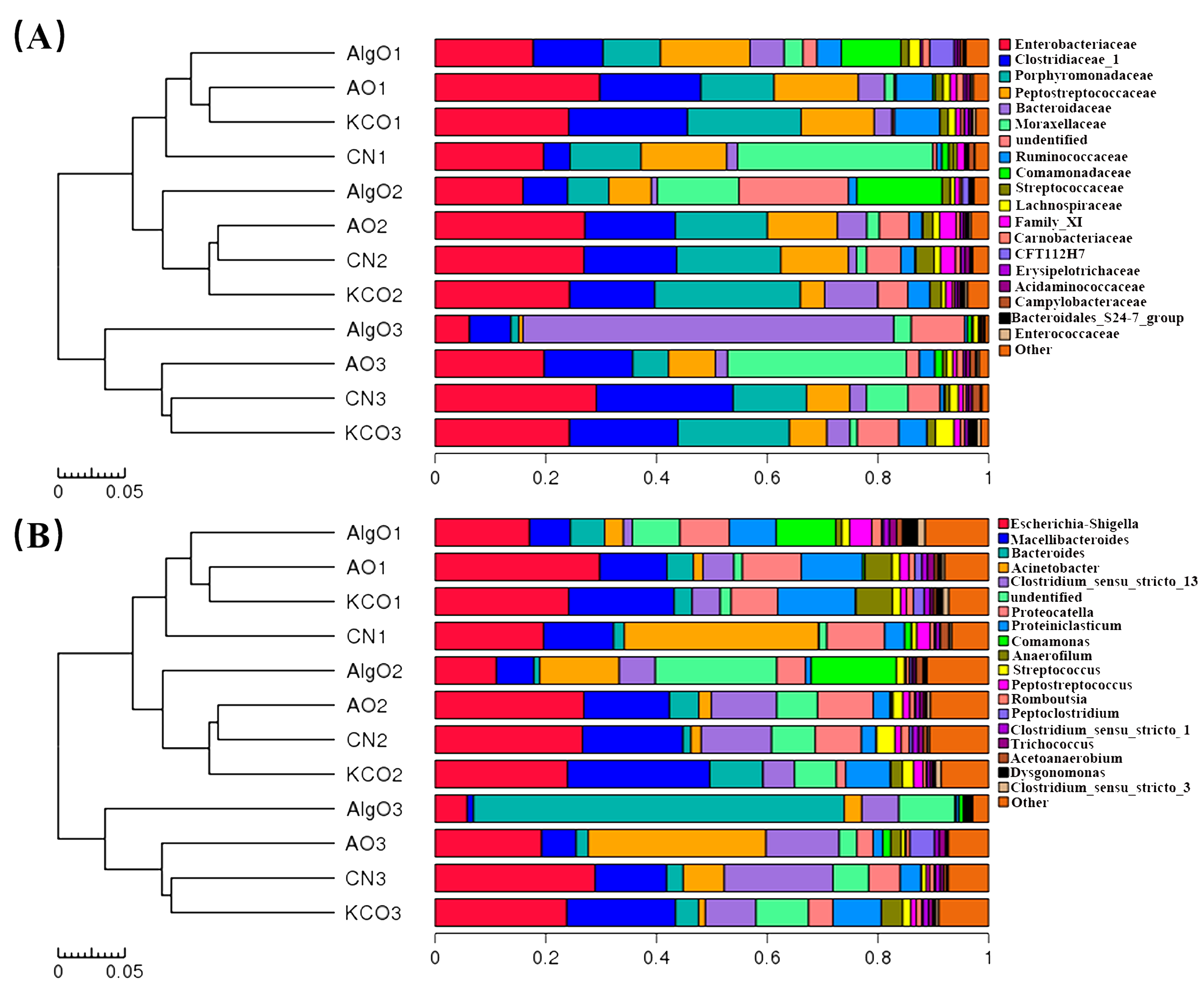
2.3. Change of the Fecal Microbiota Structure after the Fermentation of Oligosaccharides
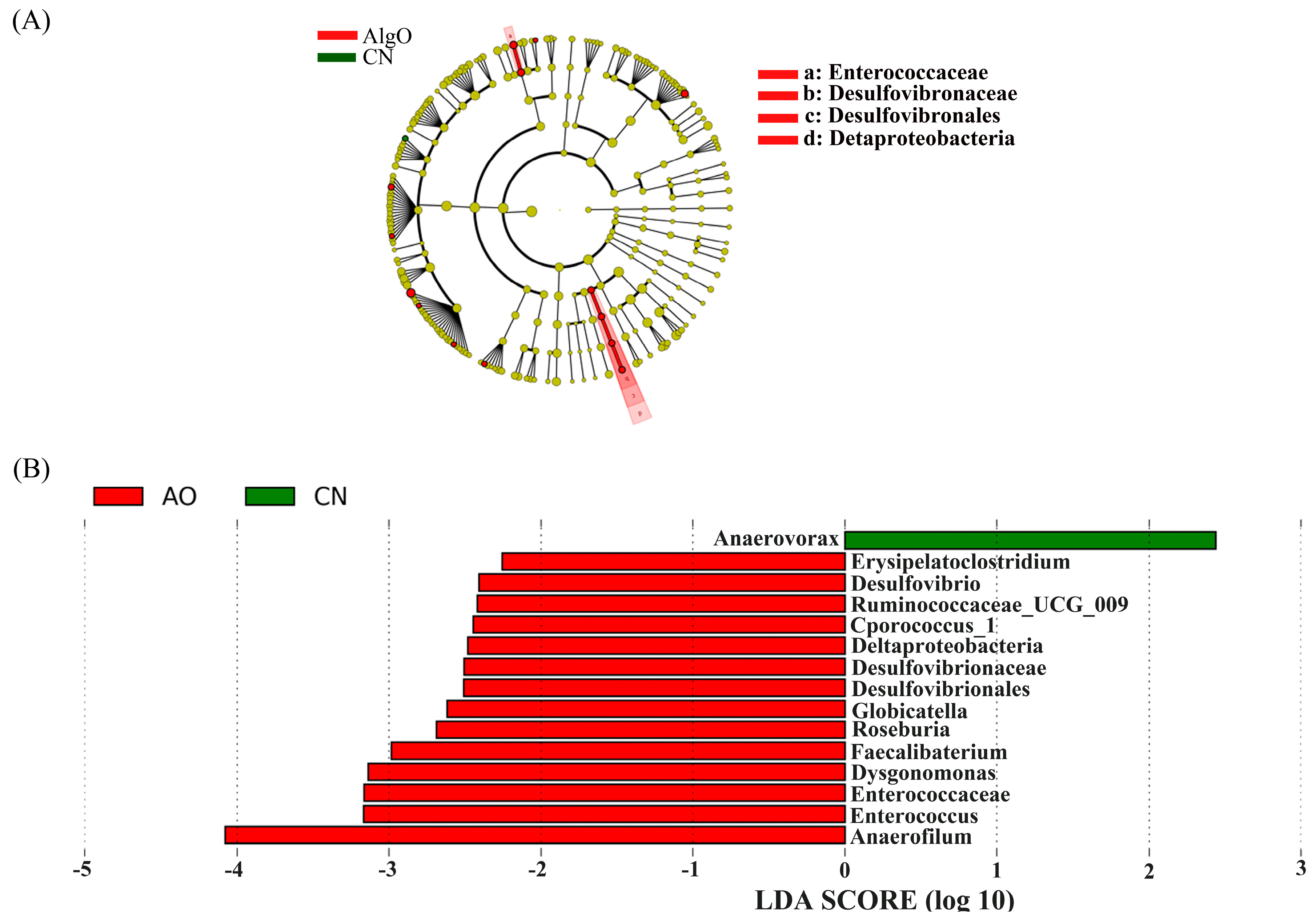
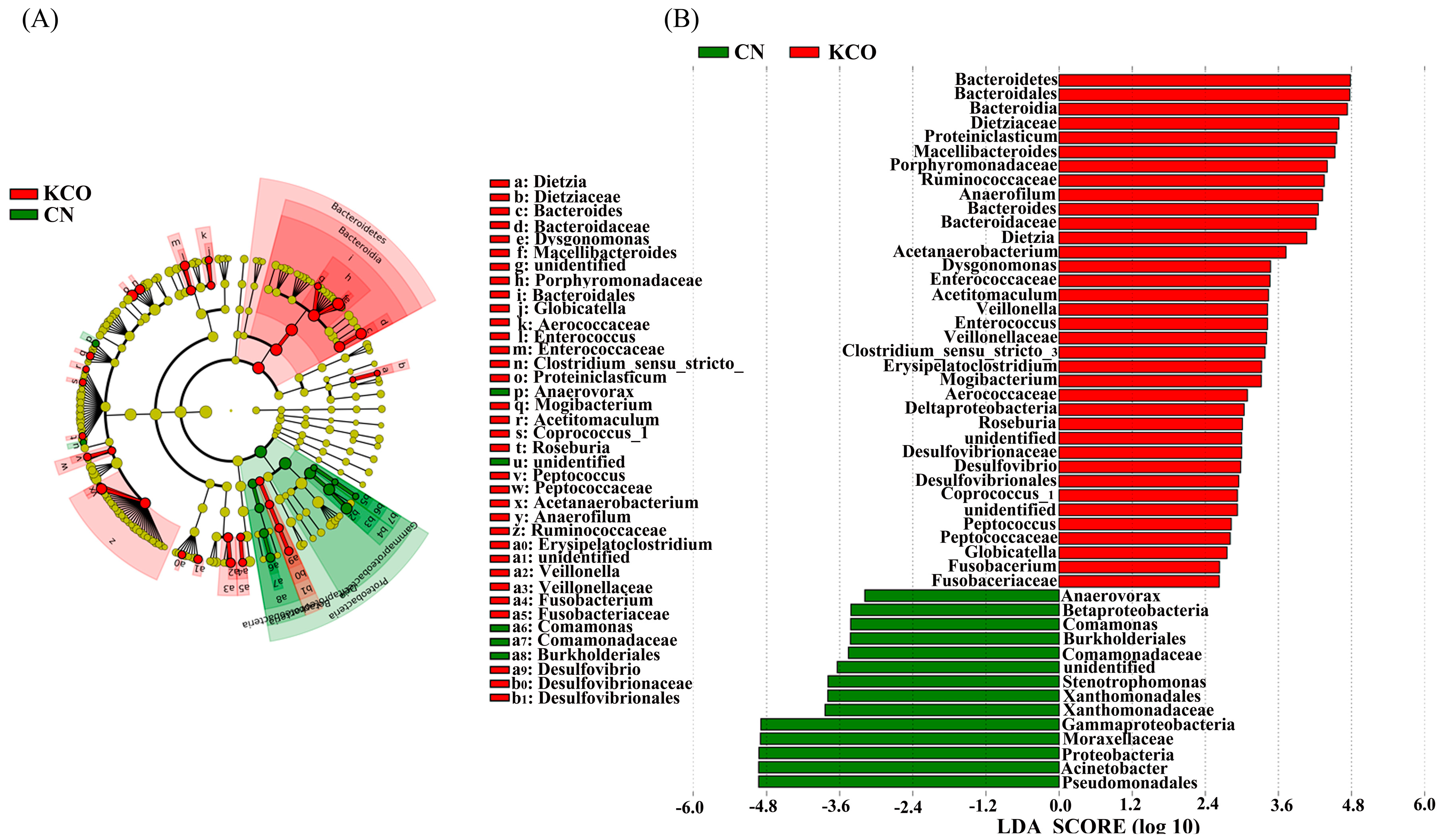
2.4. Key Phylotypes of Fecal Microbiota Modulated by the Oligosaccharides
2.5. Link Between Microbiota Composition, SCFAs, and Three Oligosaccharides
3. Discussion
3.1. Utilization and Influence of AlgO on Fermentation Pattern
3.2. Utilization and Influence of AO on Fermentation Pattern
3.3. Utilization and Influence of KCO on Fermentation Pattern
4. Materials and Methods
4.1. Materials
4.2. Preparation of AlgO, AO, and KCO
4.3. Fecal Samples and In Vitro Fermentation
4.4. Analysis of Oligosaccharides and SCFAs
4.5. DNA Extraction and High-Throughput Sequencing
4.6. Bioinformatics Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mussatto, S.I.; Mancilha, I.M. Non-digestible oligosaccharides: A review. Carbohyd. Polym. 2007, 68, 587–597. [Google Scholar] [CrossRef]
- Roberfroid, M.; Slavin, J. Nondigestible oligosaccharides. Crit. Rev. Food Sci. 2000, 40, 461–480. [Google Scholar] [CrossRef]
- Conlon, M.A.; Bird, A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Hamaker, B.R.; Tuncil, Y.E. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J. Mol. Biol. 2014, 426, 3838–3850. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Goyal, A. Functional oligosaccharides: Production, properties and applications. World J. Microbiol. Biotechnol. 2011, 27, 1119–1128. [Google Scholar] [CrossRef]
- Chung, I.K.; Beardall, J.; Mehta, S.; Sahoo, D.; Stojkovic, S. Using marine macroalgae for carbon sequestration: A critical appraisal. J. Appl. Phycol. 2011, 23, 877–886. [Google Scholar] [CrossRef]
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Iji, P.A.; Kadam, M.M. Prebiotic properties of algae and algae-supplemented products. In Functional Ingredients from Algae for Foods and Nutraceutical; Dominguez, H., Ed.; Woodhead Publishing: Cambridge, UK, 2013; pp. 658–664. [Google Scholar]
- Sardari, R.R.R.; Karlsson, E.N. Marine poly- and oligosaccharides as prebiotics. J. Agric. Food Chem. 2018, 66, 11544–11549. [Google Scholar] [CrossRef]
- Tseng, C.K. Algal biotechnology industries and research activities in China. J. Appl. Phycol. 2001, 13, 375–380. [Google Scholar] [CrossRef]
- Jonathan, M.C.; van den Borne, J.J.G.C.; Wiechen, P.; Silva, C.S.; Schols, H.A.; Gruppen, H. In vitro fermentation of 12 dietary fibres by faecal inoculum from pigs and humans. Food Chem. 2012, 133, 889–897. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Smyth, L.; Yadav, A.; Vocadlo, D.J.; Boraston, A.B. Analysis of keystone enzyme in agar hydrolysis provides insight into the degradation (of a polysaccharide from) red seaweeds. J. Biol. Chem. 2012, 287, 13985–13995. [Google Scholar] [CrossRef]
- Shang, Q.; Jiang, H.; Cai, C.; Hao, J.; Li, G.; Yu, G. Gut microbiota fermentation of marine polysaccharides and its effects on intestinal ecology: An overview. Carbohyd. Polym. 2018, 179, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, F.; Hu, B.; Li, J.; Yu, W. In vivo prebiotic properties of alginate oligosaccharides prepared through enzymatic hydrolysis of alginate. Nutr. Res. 2006, 26, 597–603. [Google Scholar] [CrossRef]
- Brownlee, I.A.; Allen, A.; Pearson, J.P.; Dettmar, P.W.; Havler, M.E.; Atherton, M.R.; Onsøyen, E. Alginate as a source of dietary fiber. Crit. Rev. Food Sci. 2005, 45, 497–510. [Google Scholar] [CrossRef]
- Hu, B.; Gong, Q.; Wang, Y.; Ma, Y.; Li, J.; Yu, W. Prebiotic effects of neoagaro-oligosaccharides prepared by enzymatic hydrolysis of agarose. Anaerobe 2006, 12, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Relleve, L.; Abad, L. Characterization and antioxidant properties of alcoholic extracts from gamma irradiated κ-carrageenan. Radiat. Phys. Chem. 2015, 112, 40–48. [Google Scholar] [CrossRef]
- Mou, H.; Jiang, X.; Guan, H. A κ-carrageenan derived oligosaccharide prepared by enzymatic degradation containing anti-tumor activity. J. Appl. Phycol. 2003, 15, 297–303. [Google Scholar]
- Yuan, H.; Song, J.; Li, X.; Li, N.; Dai, J. Immunomodulation and antitumor activity of κ-carrageenan oligosaccharides. Cancer Lett. 2006, 243, 228–234. [Google Scholar] [CrossRef]
- Pang, X.; Hua, X.; Yang, Q.; Ding, D.; Che, C.; Cui, L.; Jia, W.; Bucheli, P.; Zhao, L. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 2007, 1, 156–162. [Google Scholar] [CrossRef]
- Tøndervik, A.; Sletta, H.; Klinkenberg, G.; Emanuel, C.; Powell, L.C.; Pritchard, M.F.; Khan, S.; Craine, K.M.; Onsøyen, E.; Rye, P.D.; et al. Alginate oligosaccharides inhibit fungal cell growth and potentiate the activity of antifungals against Candida and Aspergillus spp. PLoS ONE 2014, 9, e112518. [Google Scholar] [CrossRef]
- Xu, X.; Wu, X.; Wang, Q.; Cai, N.; Zhang, H.; Jiang, Z.; Wan, M.; Oda, T. Immunomodulatory effects of alginate oligosaccharides on murine macrophage RAW264.7 cells and their structure-activity relationships. J. Agric. Food Chem. 2014, 62, 3168–3176. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Endo, T.; Nakakita, R.; Murata, K.; Yonemoto, Y.; Okayama, K. Effect of depolymerized alginates on the growth of bifidobacteria. Biosci. Biotechnol. Biochem. 1992, 56, 355–356. [Google Scholar] [CrossRef]
- Jonathan, M.C.; Souza da Silva, C.; Bosch, G.; Schols, H.A.; Gruppen, H. In vivo degradation of alginate in pigs in the presence and in the absence of resistant starch. Food Chem. 2015, 172, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Jonathan, M.C.; Bosch, G.; Schols, H.A.; Gruppen, H. Separation and identification of individual alginate oligosaccharides in the feces of alginate-fed pigs. J. Agric. Food Chem. 2013, 61, 553–560. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Acad. Natl. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Canani, R.B.; Di Costanzo, M.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- McNabney, S.; Henagan, T. Short chain fatty acids in the colon and peripheral tissues: A focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients 2017, 9, 1348. [Google Scholar] [CrossRef]
- Tian, L.; Bruggeman, G.; van den Berg, M.; Borewicz, K.; Scheurink, A.J.W.; Bruininx, E.; Vos, P.; Smidt, H.; Schols, H.A.; Gruppen, H. Effects of pectin on fermentation characteristics, carbohydrate utilization, and microbial community composition in the gastrointestinal tract of weaning pigs. Mol. Nutr. Food Res. 2017, 61, 1600186. [Google Scholar] [CrossRef]
- Salvador, V.; Cherbut, C.; Barry, J.; Bertrand, D. Sugar composition production during of dietary fibre and short-chain fatty acid in vitro fermentation by human bacteria. Brit. J. Nutr. 1993, 70, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.M.; Lu, P.Z.; Zhang, H. Bacterial communities in manures of piglets and adult pigs bred with different feeds revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biot. 2014, 98, 2657–2665. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Keller-Costa, T.; Jousset, A.; van Overbeek, L.; van Elsas, J.D.; Costa, R. The freshwater sponge Ephydatia fluviatilis harbours diverse Pseudomonas species (Gammaproteobacteria, Pseudomonadales) with broad-spectrum antimicrobial activity. PLoS ONE 2014, 9, e88429. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Miura, T.; Okamura, Y.; Aki, T.; Matsumura, Y.; Tajima, T.; Kato, J.; Nakashimada, Y. Dysgonomonas alginatilytica sp. nov., an alginate-degrading bacterium isolated from a microbial consortium. Int. J. Syst. Evol. Microbiol. 2015, 65, 3570–3575. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Miura, T.; Kawata, S.; Yamaguchi, T.; Okamura, Y.; Aki, T.; Matsumura, Y.; Tajima, T.; Kato, J.; Nishio, N.; Nakashimada, Y. Bacterial community structure and predicted alginate metabolic pathway in an alginate-degrading bacterial consortium. J. Biosci. Bioeng. 2016, 121, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Møller, K. Culture-independent analysis of gut bacteria: The pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, G.; Zhu, L.; Yin, Y.; Zhao, X.; Xiang, C.; Yu, G.; Wang, X. Isolation and characterization of an agaro-oligosaccharide (AO)-hydrolyzing bacterium from the gut microflora of Chinese individuals. PLoS ONE 2014, 9, e91106. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Nelson, E.; Chu, P.Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; EI-Osta, A.; Mackay, C.R.; Kaye, D.M. High fibre diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart Failure in DOCA-salt hypertensive mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, Q.; Fu, X.; Liu, R.H. In vitro fermentation of mulberry fruit polysaccharides by human fecal inocula and impact on microbiota. Food Funct. 2016, 7, 4637–4643. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Ivanov, I.; Mills, D.A.; Noratto, G. Infuence of whole-wheat consumption on fecal microbial community structure of obese diabetic mice. PeerJ 2016, 4, e1702. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, S.; Matamoros, S.; Cani, P.D.; Neyrinck, A.M.; Jamar, F.; Stärkel, P.; Windey, K.; Tremaroli, V.; Bäckhed, F.; Verbeke, K.; de Timary, P.; Delzenne, N.M. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA 2014, 111, E4485–E4493. [Google Scholar] [CrossRef]
- Schnabl, B.; Brenner, D.A. Interactions between the intestinal microbiome and liver diseases. J. Gastroenterol. 2014, 146, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; Ferrante, M.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2013, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Mao, X.; Li, R.W.; Hou, E.; Wang, Y.; Xue, C.; Tang, Q. Neoagarotetraose protects mice against intense exercise-induced fatigue damage by modulating gut microbial composition and function. Mol. Nutr. Food Res. 2017, 61, 1600585. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012, 10, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Doble, M. κ-Carrageenan from marine red algae, Kappaphycus alvarezii—A functional food to prevent colon carcinogenesis. J. Funct. Foods 2015, 15, 354–364. [Google Scholar] [CrossRef]
- Candela, M.; Maccaferri, S.; Turroni, S.; Carnevali, P.; Brigidi, P. Functional intestinal microbiome, new frontiers in prebiotic design. Int. J. Food Microbiol. 2010, 140, 93–101. [Google Scholar] [CrossRef]
- Yu, Y. Study on Heterologous Expression and Application of κ-Carrageenase in Pichia pastoris. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2018. [Google Scholar]
- Li, M. The Degradation and Utilization Study of Agarose, κ-Carrageenan and Their Oligosaccharides by Human Gut Microbiota. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2014. [Google Scholar]
- Bhardwaj, S.B. Gut flora and its modification as a therapy. Rev. Med. Microbiol. 2013, 24, 52–54. [Google Scholar] [CrossRef]
- Ohkusa, T.; Koido, S. Intestinal microbiota and ulcerative colitis. J. Infect. Chemother. 2015, 21, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Jin, T.; Yu, Y.; Yang, M.; Mou, H.; Li, L. Extracellular expression of a novel β-agarase from Microbulbifer sp. Q7, isolated from the gut of sea cucumber. AMB Express 2017, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yu, Y.; Yang, S.; Shi, X.; Mou, H.; Li, L. Expression and characterization of a new polyG-specific alginate lyase from marine bacterium Microbulbifer sp. Q7. Front. Microbiol. 2018, 9, 02894. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yang, M.; Yang, J.; Su, Q.; Mou, H. Composition and characteristics of continuous enzymatic hydrolysis products from Kappaphycus striatum. J. Appl. Phycol. 2017, 29, 1647–1656. [Google Scholar] [CrossRef]
- Charoensiddhi, S.; Conlon, M.A.; Vuaran, M.S.; Franco, C.M.M.; Zhang, W. Impact of extraction processes on prebiotic potential of the brown seaweed Ecklonia radiata by in vitro human gut bacteria fermentation. J. Funct. Foods 2016, 24, 221–230. [Google Scholar] [CrossRef]
- He, S.; Fan, X.; Katukuri, N.R.; Yuan, X.; Wang, F.; Guo, R.B. Enhanced methane production from microalgal biomass by anaerobic bio-pretreatment. Bioresour. Technol. 2016, 204, 145–151. [Google Scholar] [CrossRef]



| Substrates | pH | Total SCFA | Acetic Acid | Propionic Acid | Isobutyric Acid | Butyric Acid | Isovaleric Acid | Valeric Acid | Caproic Acid |
|---|---|---|---|---|---|---|---|---|---|
| AlgO | 7.54 ± 0.33 | 8.08 ± 0.60 | 3.84 ± 0.47 | 1.11 ± 0.05 | 0.63 ± 0.02 | 0.79 ± 0.03 ** | 0.65 ± 0.01 | 0.53 ± 0.003 ** | 0.54 ± 0.003 |
| AO | 7.7 ± 0.06 | 11.33 ± 2.39 | 6.74 ± 1.93 * | 1.24 ± 0.30 | 0.68 ± 0.02 ** | 0.86 ± 0.11 ** | 0.74 ± 0.02 ** | 0.53 ± 0.003 ** | 0.54 ± 0.004 |
| KCO | 7.64 ± 0.28 | 8.04 ± 0.26 | 3.87 ± 0.14 | 1.10 ± 0.06 | 0.62 ± 0.009 | 0.77 ± 0.03 ** | 0.63 ± 0.01 | 0.52 ± 0.0006 | 0.53 ± 0.001 |
| CN | 7.65 ± 0.04 | 6.90 ± 0.83 | 3.13 ± 0.68 | 0.94 ± 0.10 | 0.61 ± 0.01 | 0.54 ± 0.01 | 0.63 ± 0.02 | 0.52 ± 0.0003 | 0.53 ± 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.-L.; Yang, M.; Fu, X.-D.; Chen, M.; Su, Q.; Zhao, Y.-H.; Mou, H.-J. Evaluation of Prebiotic Potential of Three Marine Algae Oligosaccharides from Enzymatic Hydrolysis. Mar. Drugs 2019, 17, 173. https://doi.org/10.3390/md17030173
Han Z-L, Yang M, Fu X-D, Chen M, Su Q, Zhao Y-H, Mou H-J. Evaluation of Prebiotic Potential of Three Marine Algae Oligosaccharides from Enzymatic Hydrolysis. Marine Drugs. 2019; 17(3):173. https://doi.org/10.3390/md17030173
Chicago/Turabian StyleHan, Zhen-Lian, Min Yang, Xiao-Dan Fu, Meng Chen, Qian Su, Yuan-Hui Zhao, and Hai-Jin Mou. 2019. "Evaluation of Prebiotic Potential of Three Marine Algae Oligosaccharides from Enzymatic Hydrolysis" Marine Drugs 17, no. 3: 173. https://doi.org/10.3390/md17030173
APA StyleHan, Z.-L., Yang, M., Fu, X.-D., Chen, M., Su, Q., Zhao, Y.-H., & Mou, H.-J. (2019). Evaluation of Prebiotic Potential of Three Marine Algae Oligosaccharides from Enzymatic Hydrolysis. Marine Drugs, 17(3), 173. https://doi.org/10.3390/md17030173






