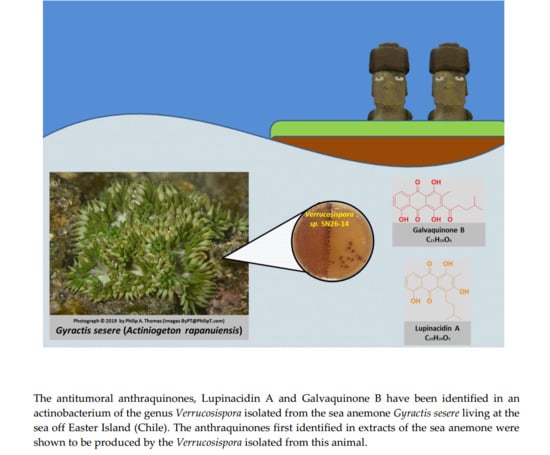
Antitumor Anthraquinones from an Easter Island Sea Anemone: Animal or Bacterial Origin?
Abstract
:1. Introduction
2. Results and Discussion
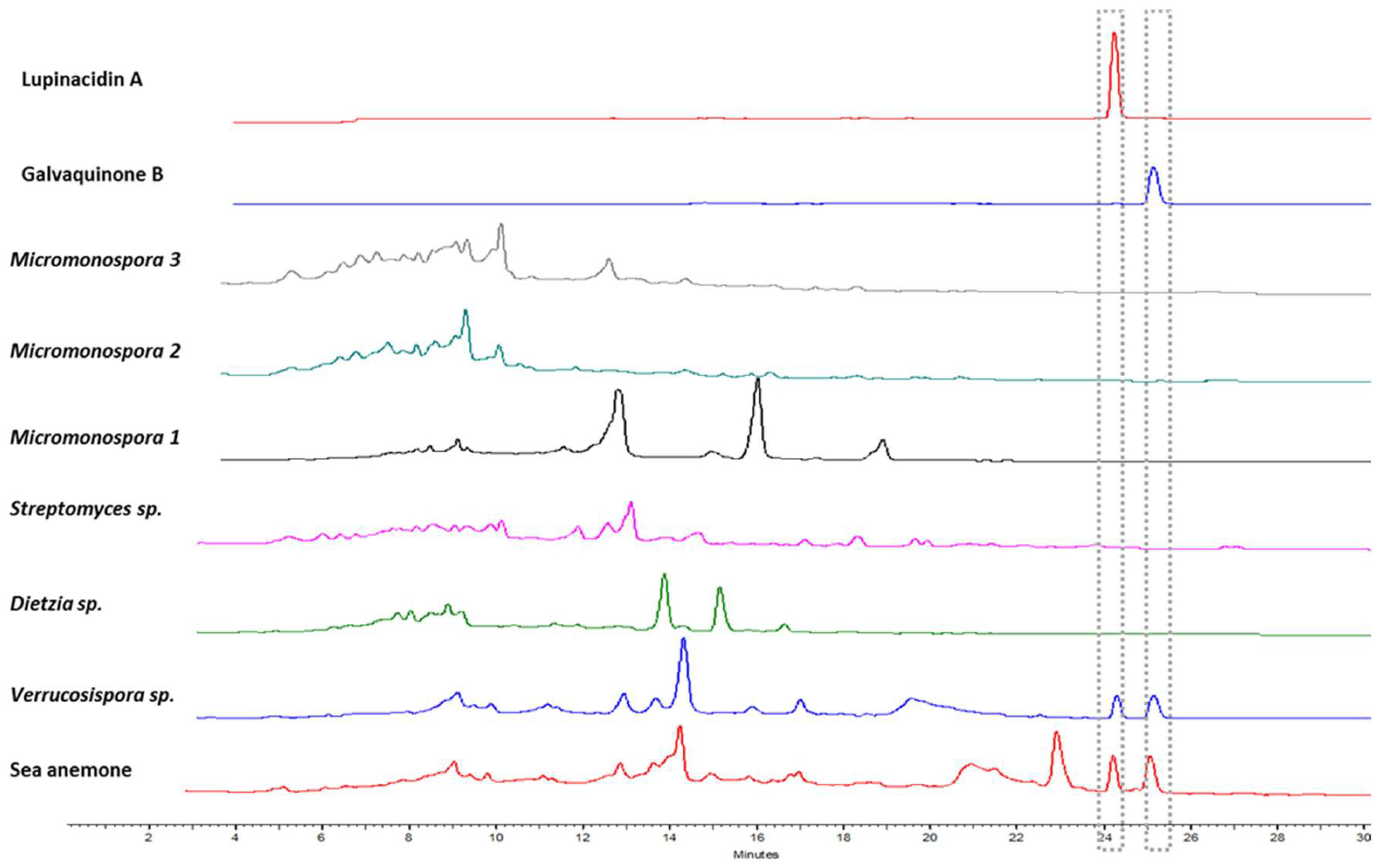
2.1. Sea Anemone Dereplication
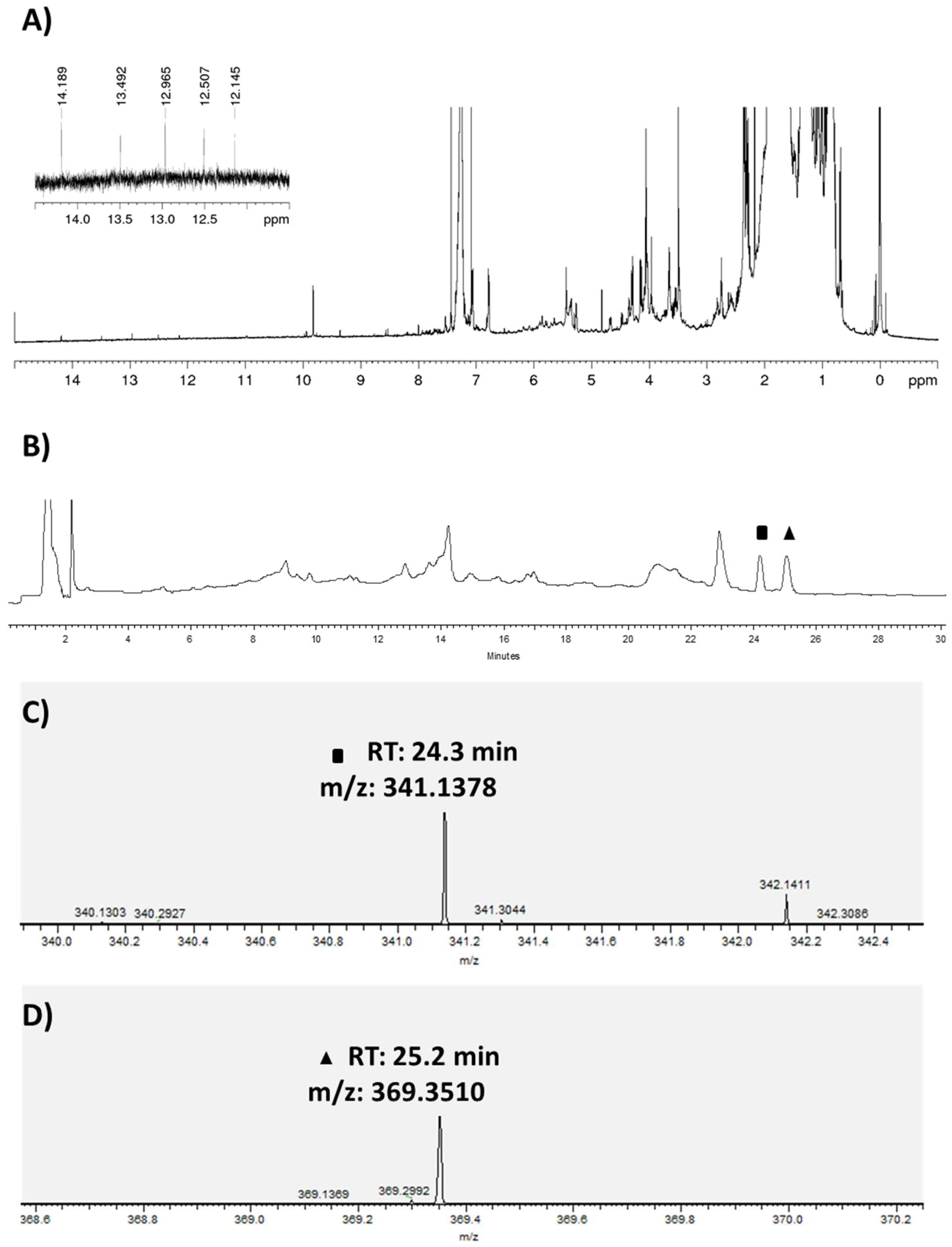
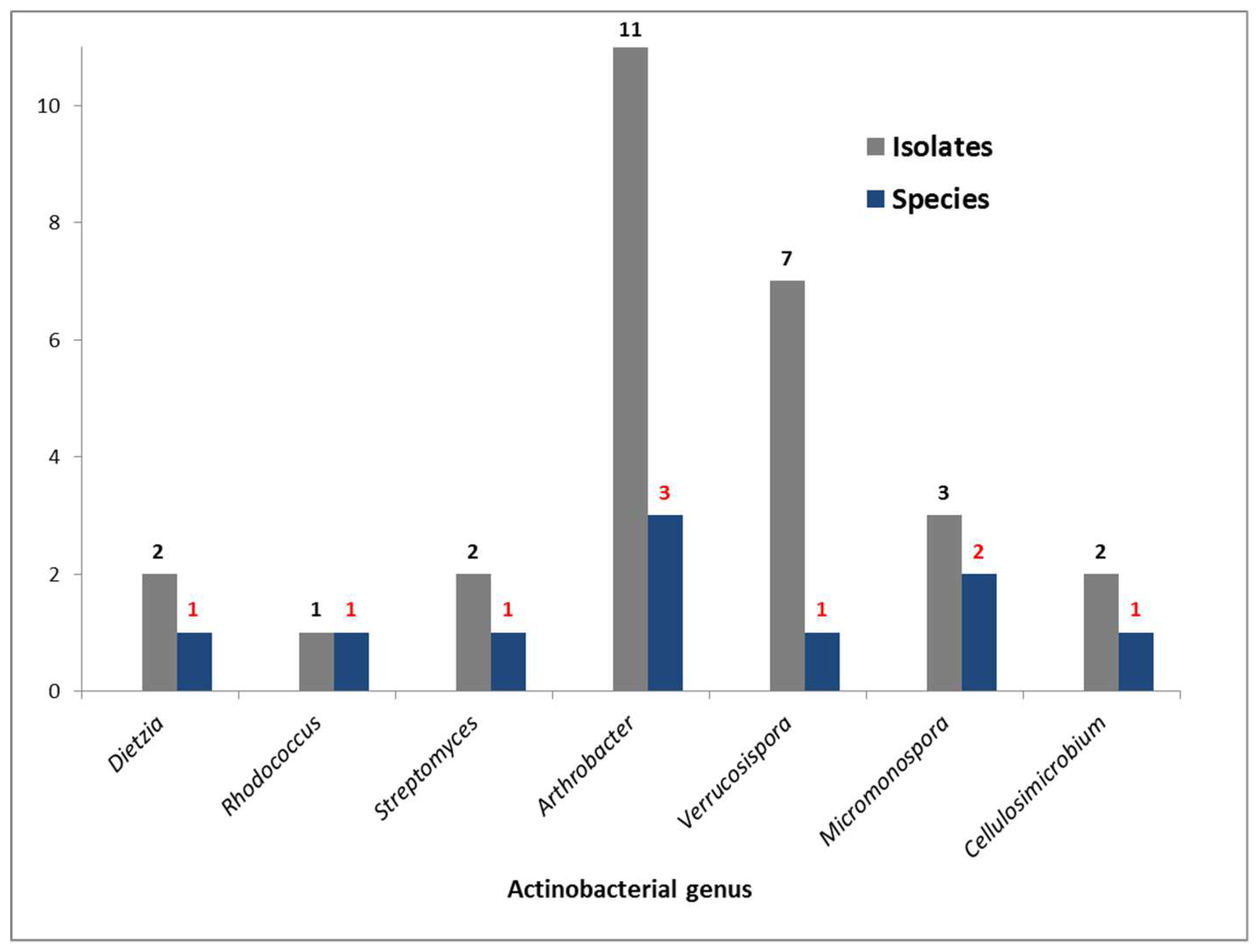
2.2. Bacterial Metabolites and Harbored Bacteria
2.3. Bacterial Growth
2.4. Actinobacterial Producer
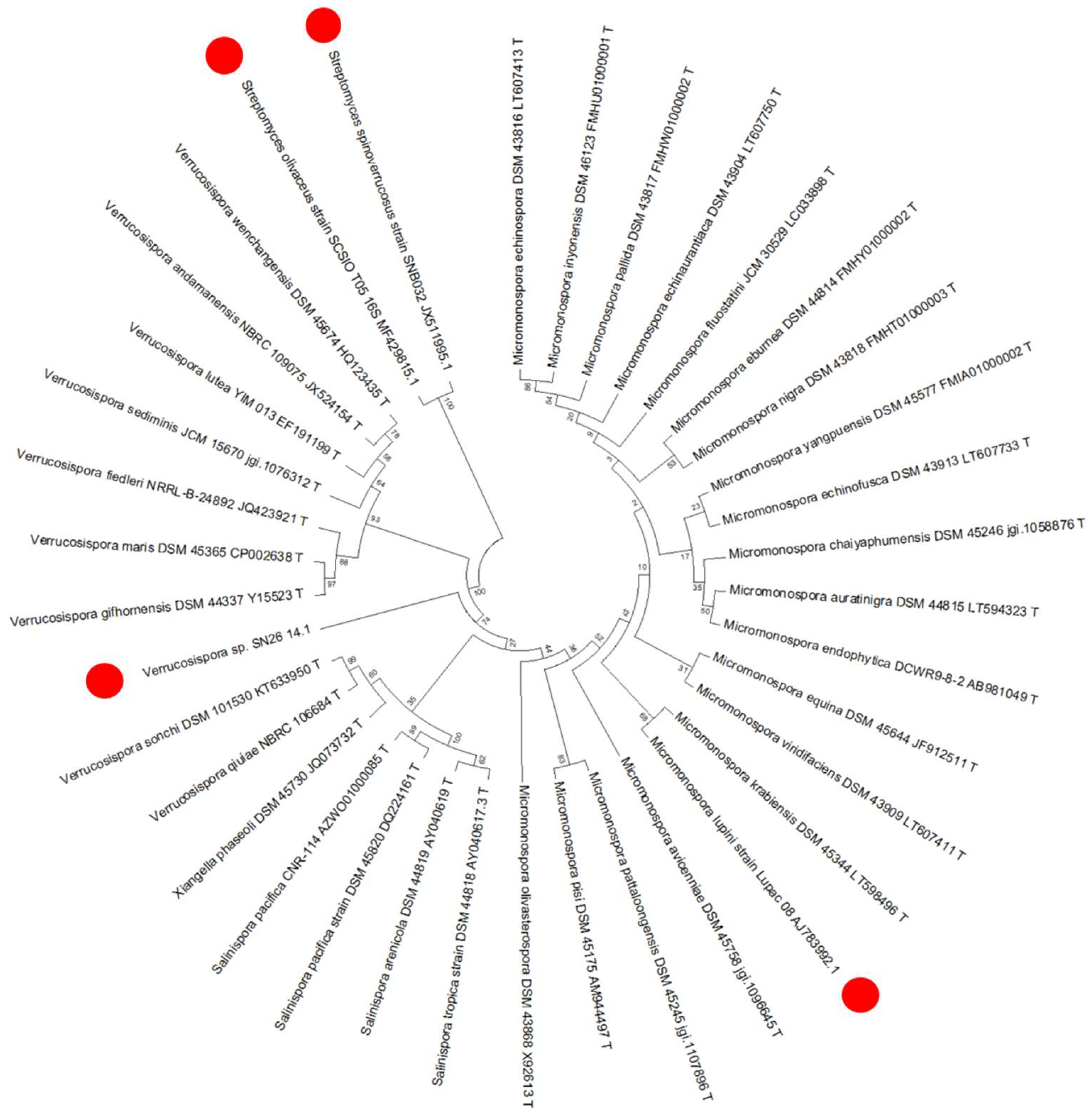
2.5. Phylogeny of the Producer
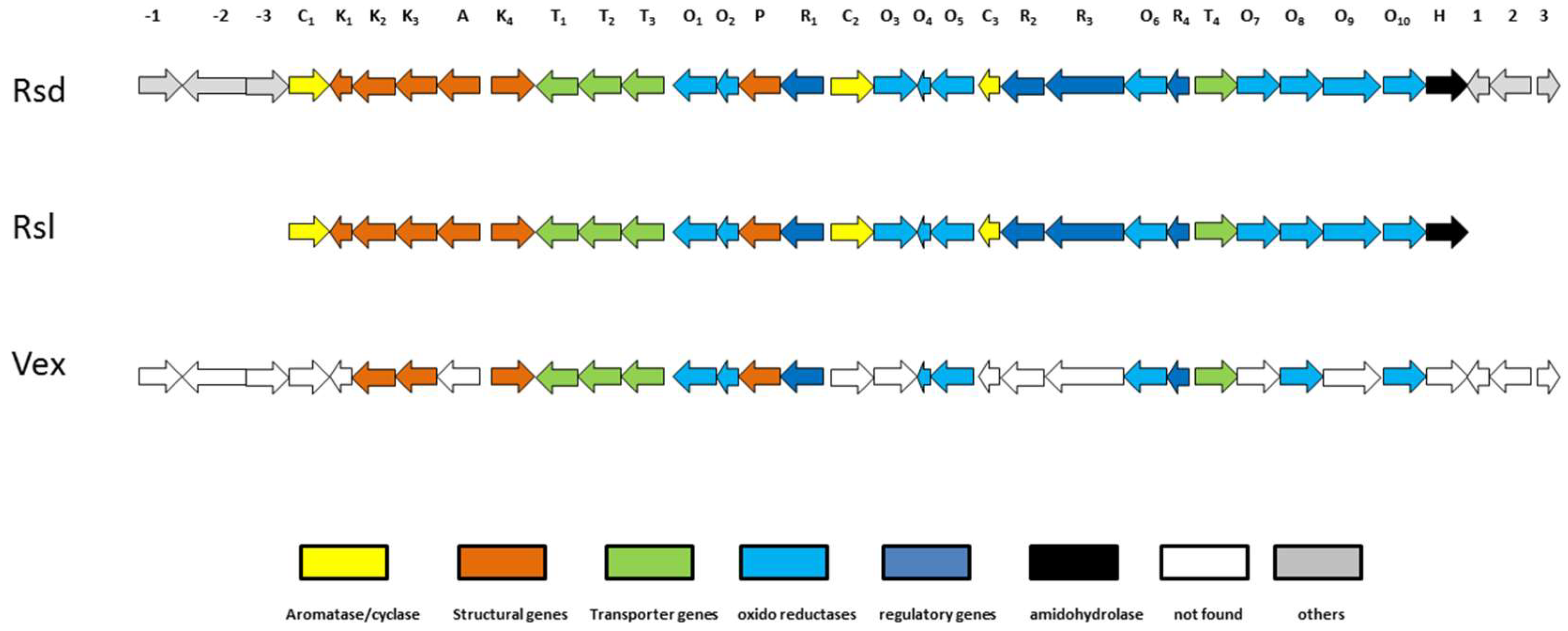
2.6. Biosynthesis
2.7. Antibiotic Activity Test
3. Materials and Methods
3.1. Sample Collection
3.2. Sea Anemone Dereplication
3.3. Bacterial Isolation
3.4. Molecular Characterization and Phylogenetic Analysis
3.5. Bacterial Growth for Secondary Metabolites Production
3.6. Chemical Extraction, Purification and Structure Elucidation
3.7. Genome Sequencing
3.8. Genome Assembly
3.9. Secondary Metabolites Gene Clusters Search
3.10. Antibiotic Activity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dransfield, J.; Flenley, J.R.; King, S.M.; Harkness, D.D.; Rapu, S. A recently extinct palm from Easter Island. Nature 1984, 312, 750. [Google Scholar] [CrossRef]
- Boyko, C.B. The endemic marine invertebrates of Easter Island: How many species and for how long? In Easter Island: Scientific Exploration into the World’s Environmental Problems in Microcosm; Loret, J., Tanacredi, J.T., Eds.; Springer: Boston, MA, USA, 2003; pp. 155–175. [Google Scholar]
- Skottsberg, C. The Natural History of Juan Fernández and Easter Island; Almqvist & Wiksells Boktryckeri: Uppsala, Sweden, 1920. [Google Scholar]
- Kohn, A.J.; Lloyd, M.C. Marine polychaete annelids of Easter Island. Int. Rev. Hydrobiol. 1973, 58, 691–712. [Google Scholar] [CrossRef]
- Sehgal, S.N. Rapamune® (RAPA, rapamycin, sirolimus): Mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin. Biochem. 1998, 31, 335–340. [Google Scholar] [CrossRef]
- Vezina, C.; Kudelski, A.; Sehgal, S. Rapamycin (AY-22, 989), a new antifungal antibiotic. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.R. Conservation hotspots of biodiversity and endemism for Indo-Pacific coral reef fishes. Aquat. Conserv. 2008, 18, 541–556. [Google Scholar] [CrossRef]
- Horton, T.; Kroh, A.; Ahyong, S.; Bailly, N.; Boyko, C.B.; Brandão, S.N.; Costello, M.J.; Gofas, S.; Hernandez, F.; Holovachov, O.; et al. World Register of Marine Species (WoRMS); WoRMS Editorial Board: Ostend, Belgium, 2018. [Google Scholar]
- Urda, C.; Fernández, R.; Pérez, M.; Rodríguez, J.; Jiménez, C.; Cuevas, C. Protoxenicins A and B, cytotoxic long-chain acylated xenicanes from the soft coral Protodendron repens. J. Nat. Prod. 2017, 80, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, F.; Kudo, N.; Tomachi, Y.; Nakata, A.; Takemoto, M.; Ito, A.; Tabei, H.; Arai, D.; de Voogd, N.; Yoshida, M.; et al. Halistanol sulfates I and J, new SIRT1–3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria. J. Antibiot. 2017, 71, 273. [Google Scholar] [CrossRef] [PubMed]
- Aminin, D.; Menchinskaya, E.; Pisliagin, E.; Silchenko, A.; Avilov, S.; Kalinin, V. Anticancer activity of sea cucumber triterpene glycosides. Mar. Drugs 2015, 13, 1202–1223. [Google Scholar] [CrossRef] [PubMed]
- Hay, M.E. Marine chemical ecology: What’s known and what’s next? J. Exp. Mar. Biol. Ecol. 1996, 200, 103–134. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160–258. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar] [CrossRef] [PubMed]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Unson, M.D.; Holland, N.D.; Faulkner, D.J. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 1994, 119, 1–11. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Obraztsova, A.Y.; Davidson, S.K.; Faulkner, D.J.; Haygood, M.G. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-Proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 2000, 136, 969–977. [Google Scholar] [CrossRef]
- Feng, Y.; Khokhar, S.; Davis, R.A. Crinoids: Ancient organisms, modern chemistry. Nat. Prod. Rep. 2017, 34, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Khokhar, S.; Pierens, G.K.; Hooper, J.N.A.; Ekins, M.G.; Feng, Y.; Davis, R.A. Rhodocomatulin-type anthraquinones from the australian marine invertebrates Clathria hirsuta and Comatula rotalaria. J. Nat. Prod. 2016, 79, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Tietze, L.F.; Gericke, K.M.; Schuberth, I. Synthesis of highly functionalized anthraquinones and evaluation of their antitumor activity. Eur. J. Org. Chem. 2007, 2007, 4563–4577. [Google Scholar] [CrossRef]
- Yang, K.-L.; Wei, M.-Y.; Shao, C.-L.; Fu, X.-M.; Guo, Z.-Y.; Xu, R.-F.; Zheng, C.-J.; She, Z.-G.; Lin, Y.-C.; Wang, C.-Y. Antibacterial anthraquinone derivatives from a sea anemone-derived fungus Nigrospora sp. J. Nat. Prod. 2012, 75, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Lee, C.-H.; Kim, H.-G.; Lee, H.-S. Anthraquinones isolated from Cassia tora (Leguminosae) Seed Show an Antifungal Property against Phytopathogenic Fungi. J. Agric. Food Chem. 2004, 52, 6096–6100. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Martinez, E.D.; MacMillan, J.B. Anthraquinones from a marine-derived Streptomyces spinoverrucosus. J. Nat. Prod. 2012, 75, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Trujillo, M.E.; Martínez-Molina, E.; Yanase, S.; Miyanaga, S.; Obata, T.; Sakurai, H.; Saiki, I.; Fujita, T.; Furumai, T. Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg. Med. Chem. Lett. 2007, 17, 3702–3705. [Google Scholar] [CrossRef] [PubMed]
- Haddon, A.C.; Shackleton, A.M. Description of some new species of Actiniaria from Torres Straits. R. Dublin Soc. 1893, 8. [Google Scholar]
- Carlgren, O. Actiniaria und zoantharia von Juan Fernandez und der Osterinsel. In The Natural History of Juan Fernandez and Easter Island; Skottsberg, C., Ed.; Almquist & Wiksells Boktryckeri: Uppsala, Sweden, 1922; pp. 145–160. [Google Scholar]
- Zhang, C.; Sun, C.; Huang, H.; Gui, C.; Wang, L.; Li, Q.; Ju, J. Biosynthetic Baeyer–Villiger chemistry enables access to two anthracene scaffolds from a single gene cluster in deep-sea-derived Streptomyces olivaceus SCSIO T05. J. Nat. Prod. 2018, 81, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Probst, K.; Linnenbrink, A.; Arnold, M.; Paululat, T.; Zeeck, A.; Bechthold, A. Cloning and heterologous expression of three type II PKS gene clusters from Streptomyces bottropensis. ChemBioChem 2012, 13, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Fiedler, H.-P. A guide to successful bioprospecting: Informed by actinobacterial systematics. J. Microb. 2010, 98, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Patin, N.V.; Duncan, K.R.; Dorrestein, P.C.; Jensen, P.R. Competitive strategies differentiate closely related species of marine Actinobacteria. ISME J. 2015, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Waglechner, N.; Wright, G.D. Antibiotic resistance–mediated isolation of scaffold-specific natural product producers. Nat. Protoc. 2014, 9, 1469. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Staufenberger, T.; Thiel, V.; Wiese, J.; Imhoff, J.F. Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol. Ecol. 2008, 64, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Morgan, P.; Weightman, A.J.; Fry, J.C. Cultivation-dependent and independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microb. 2003, 69, 3223. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microb. 2008, 74, 2461. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.B.; Jensen, P.R.; Fenical, W. Cytotoxic and antimicrobial napyradiomycins from two marine-derived Streptomyces strains. Eur. J. Org. Chem. 2013, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 May 2018).
- Bolger, A.M.; Usadel, B.; Lohse, M. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Tesler, G.; Vyahhi, N.; Saveliev, V. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bondi, A., Jr.; Spaulding, E.H.; Smith, D.E.; Dietz, C. A routine method for the rapid determination of susceptibility to penicillin and other antibiotics. Am. J. Med. Sci. 1947, 213, 221–225. [Google Scholar] [CrossRef] [PubMed]
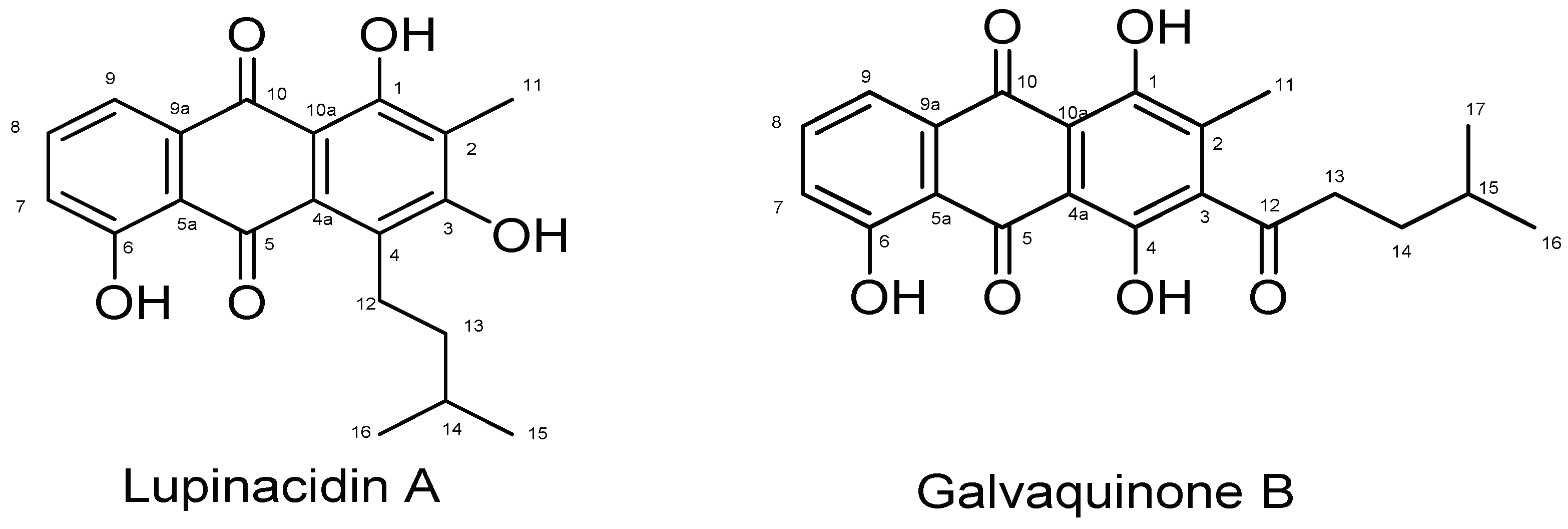
| Lupinacidin A | Galvaquinone B | |||||||
|---|---|---|---|---|---|---|---|---|
| Position | δC | δH Mult (J in Hz) | HMBC | COSY | δC | δH Mult (J in Hz) | HMBC | COSY |
| 1 | 162.5 | 157.5 | ||||||
| 2 | 117.5 | 137.1 | ||||||
| 3 | 159.6 | 141.1 | ||||||
| 4 | 130.4 | 153.8 | ||||||
| 4a | 127.9 | 116.2 | ||||||
| 5 | 190.2 | 190.7 | ||||||
| 5a | 117.1 | 116.2 | ||||||
| 6 | 162.6 | 162.7 | ||||||
| 7 | 124.3 | 7.26, d (8.2) | 5a, 6, 9 | H-8 | 124.8 | 7.32, d (8.5, 1.3) | 5a, 6, 9 | H-8 |
| 8 | 136 | 7.62, dd (8.2, 7.5) | 6, 9a | H-7, H-9 | 137.1 | 7.72, dd (8.5, 7.6) | 6, 9, 9a | H-7, H-9 |
| 9 | 118.3 | 7.79, d (7.5) | 5a, 7, 10 | H-8 | 119.7 | 7.90, d (7.6, 1.3) | 5a, 7, 10, | H-9 |
| 9a | 133 | 133.4 | ||||||
| 10 | 186.9 | 186.5 | ||||||
| 10a | 110.8 | 111.7 | ||||||
| 11 | 8.4 | 2.27, s | 1, 2, 3 | 13.2 | 2.25, s | 1, 2, 3 | ||
| 12 | 24.8 | 3.21, m (6.6) | 3, 4a, 13, 14 | H-13 | 204.9 | |||
| 13 | 37.7 | 1.46, m (6.6) | 4a, 12, 14, 15 | H-12, H-14 | 42.4 | 2.85, m | 12, 14, 15 | H-14 |
| 14 | 28.4 | 1.80, m (6.6) | 12, 13, 15, 16 | H-13, H-15, H-16 | 31.9 | 1.63, m | 14, 15, 16, 17 | H-15 |
| 15 | 22.5 | 1.04, d (6.6) | 13, 14, 16 | H-14 | 27.6 | 1.63, m | 14, 15, 16, 17 | H-13, H-16, H-17 |
| 16 | 22.5 | 1.04, d (6.6) | 13, 14, 15 | H-14 | 22.4 | 0.93, d, (6.2) | 14, 15, 17 | H-15 |
| 17 | 22.4 | 0.93, d, (6.2) | 14, 15, 16 | H-15 | ||||
| 1-OH | 14.18 | 1, 2, 10a, | 13.49, s | 1, 2, 10a | ||||
| 3-OH | 5.62 | 2, 4a, 3 | ||||||
| 4-OH | 12.50, s | 3, 4, 10a | ||||||
| 6-OH | 12.96 | 5a, 6, 7 | 12.14, s | 5a, 6, 8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sottorff, I.; Künzel, S.; Wiese, J.; Lipfert, M.; Preußke, N.; Sönnichsen, F.D.; Imhoff, J.F. Antitumor Anthraquinones from an Easter Island Sea Anemone: Animal or Bacterial Origin? Mar. Drugs 2019, 17, 154. https://doi.org/10.3390/md17030154
Sottorff I, Künzel S, Wiese J, Lipfert M, Preußke N, Sönnichsen FD, Imhoff JF. Antitumor Anthraquinones from an Easter Island Sea Anemone: Animal or Bacterial Origin? Marine Drugs. 2019; 17(3):154. https://doi.org/10.3390/md17030154
Chicago/Turabian StyleSottorff, Ignacio, Sven Künzel, Jutta Wiese, Matthias Lipfert, Nils Preußke, Frank D. Sönnichsen, and Johannes F. Imhoff. 2019. "Antitumor Anthraquinones from an Easter Island Sea Anemone: Animal or Bacterial Origin?" Marine Drugs 17, no. 3: 154. https://doi.org/10.3390/md17030154
APA StyleSottorff, I., Künzel, S., Wiese, J., Lipfert, M., Preußke, N., Sönnichsen, F. D., & Imhoff, J. F. (2019). Antitumor Anthraquinones from an Easter Island Sea Anemone: Animal or Bacterial Origin? Marine Drugs, 17(3), 154. https://doi.org/10.3390/md17030154