Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis
Abstract
1. Introduction
2. Results
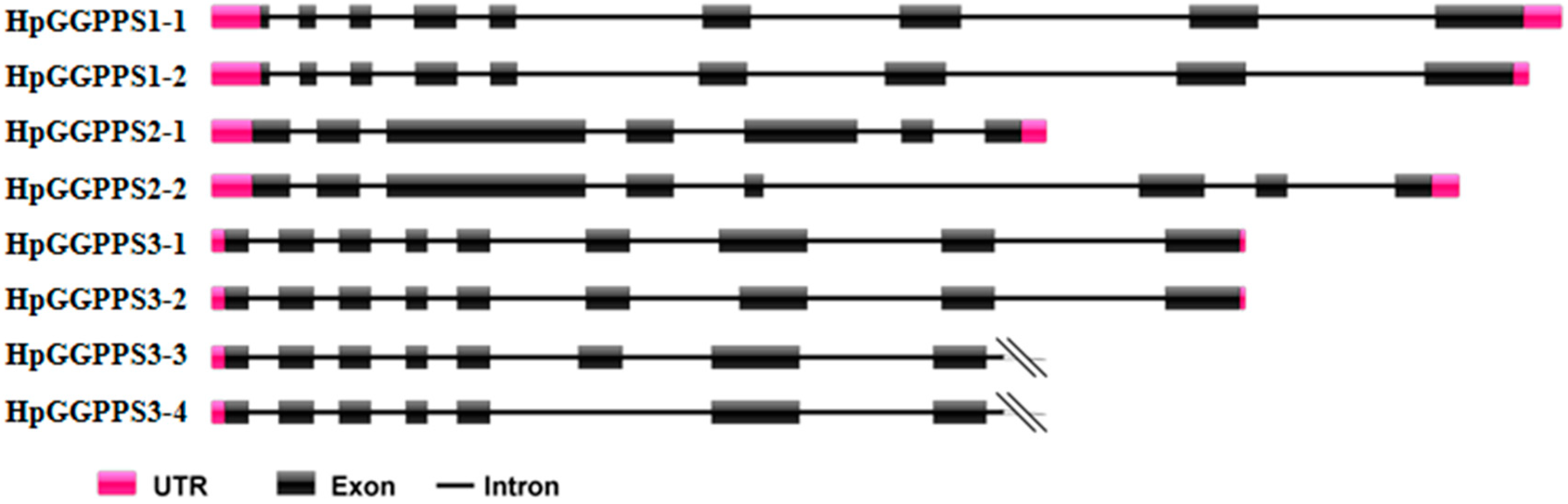
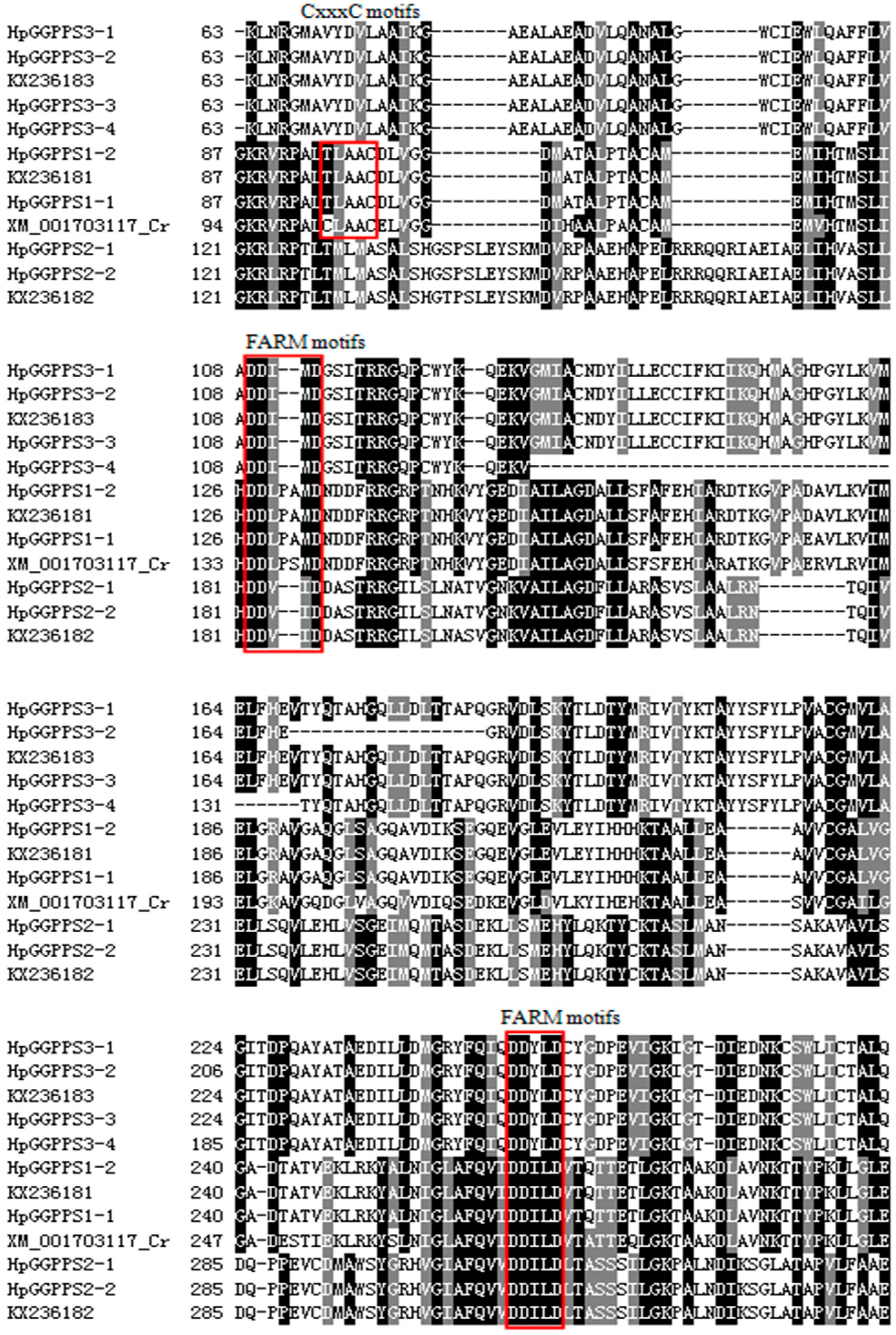
2.1. Cloning and Sequence Characterization of HpGGPPS Genes
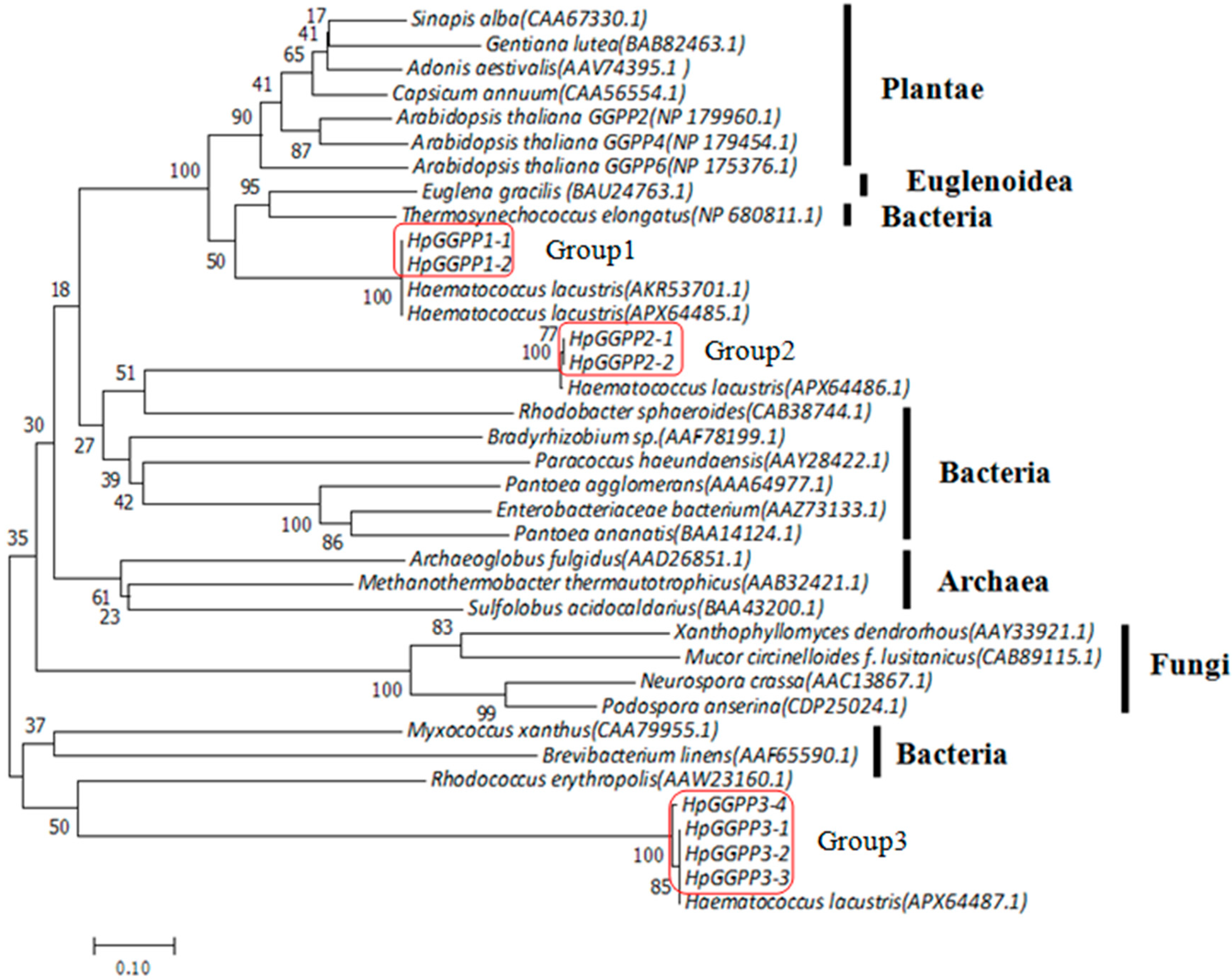
2.2. Molecular Evolution of HpGGPPS Genes
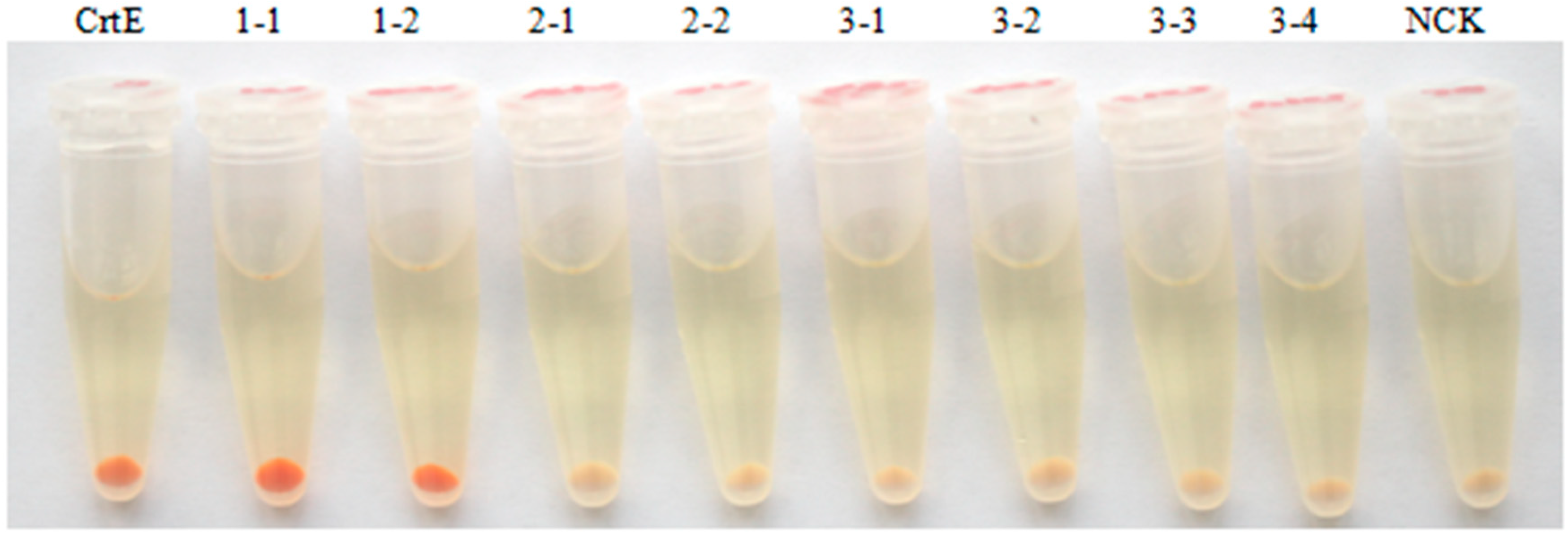
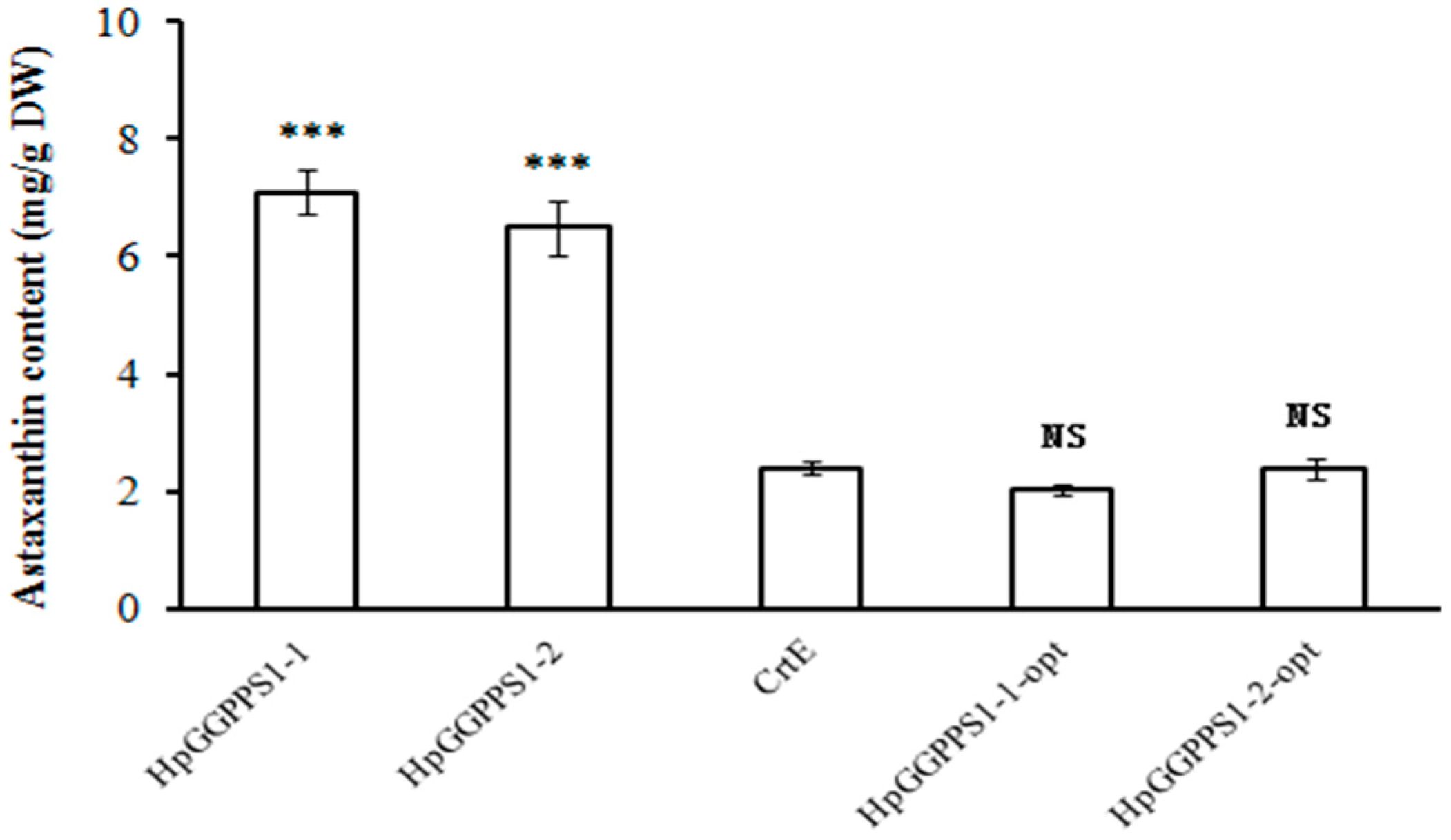
2.3. Functional Identification of HpGGPPS Genes in Escherichia coli
3. Discussion
4. Materials and Methods
4.1. Microalga Culture, Treatment, and Sample Collection
4.2. Discovery, Isolation, and Sequencing of HpGGPPS Genes
4.3. Sequences Analysis and Molecular Evolution of HpGGPPS Genes
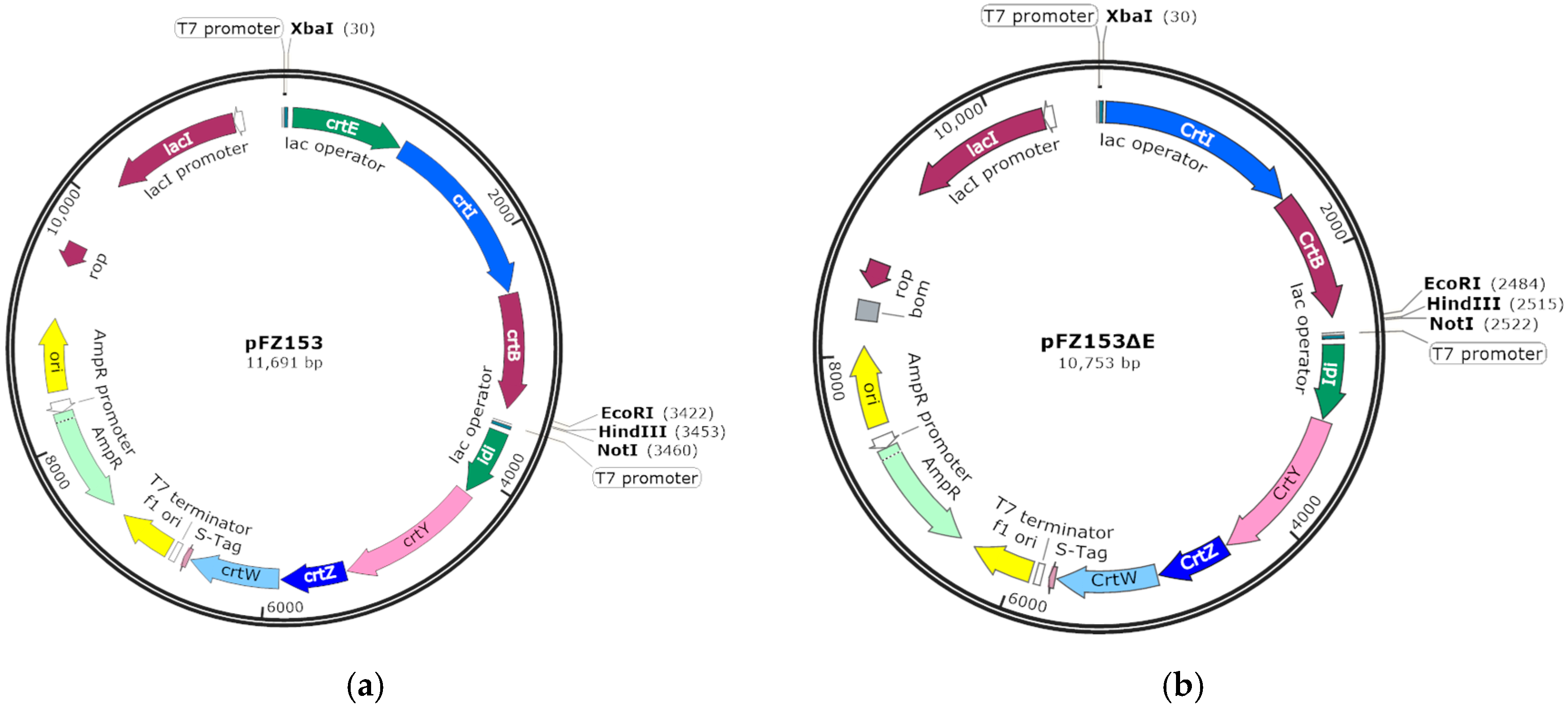
4.4. Plasmids Construction and Heterologous Expression
4.5. Pigment Extraction from E. coli and HPLC Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grand View Research. Astaxanthin Market Analysis by Source (Natural [Yeast, Krill/Shrimp, Microalgae] And Synthetic), by Product (Dried Biomass/Powder, Oil, Soft gels, Liquid), by Application, and Segment Forecasts, 2018–2025. Available online: https://www.grandviewresearch.com/industry-analysis/global-astaxanthin-market (accessed on 20 October 2017).
- Higuera-ciapara, I.; Felix-valenzuela, L.; Goycoolea, F.M. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Krause, W.; Henrich, K.; Paust, J.; Ernst, H. Preparation of Astaxanthin. Available online: https://www.google.com/patents/US5654488 (accessed on 5 August 1997).
- Koller, M.; Muhr, A.; Braunegg, G. Microalgae as versatile cellular factories for valued products. Algal Res. 2014, 6, 52–63. [Google Scholar] [CrossRef]
- Lorenz, R.T.; Cysewski, G.R. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 2000, 18, 160–167. [Google Scholar] [CrossRef]
- McGarvey, D.J.; Croteau, R. Terpenoid metabolism. Plant Cell 1995, 7, 1015–1026. [Google Scholar] [PubMed]
- Shah, M.M.R.; Yuanmei, L.; Cheng, J.J.; Maurycy, D. Astaxanthin-producing green microalga Haematococcus pluvialis: From single cell to high value commercial products. Front. Plant Sci. 2016, 7, 531. [Google Scholar] [CrossRef]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 295, 517–524. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, W.; Zhang, X.; Fan, X.; Xu, D.; Ye, N.; Yang, Q. Isolation and expression analyses of methyl-d-erythritol 4-phosphate (MEP) pathway genes from Haematococcus pluvialis. J. Appl. Phycol. 2015, 28, 1–10. [Google Scholar] [CrossRef]
- Sun, Z.; Cunningham, F.X., Jr.; Gantt, E. Differential expression of two isopentenyl pyrophosphate isomerases and enhanced carotenoid accumulation in a unicellular chlorophyte. Proc. Natl. Acad. Sci. USA 1998, 95, 11482–11488. [Google Scholar] [CrossRef]
- Hoeffler, J.F.; Hemmerlin, A.; Grosdemange-Billiard, C.; Bach, T.J.; Rohmer, M. Isoprenoid biosynthesis in higher plants and in Escherichia coli: On the branching in the methylerythritol phosphate pathway and the independent biosynthesis of isopentenyl diphosphate and dimethylallyl diphosphate. Biochem. J. 2002, 366, 573–583. [Google Scholar] [CrossRef]
- Gwak, Y.; Hwang, Y.S.; Wang, B.; Kim, M.; Jeong, J.; Lee, C.G.; Jin, E. Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photo oxidative stress in Haematococcus pluvialis. J. Exp. Bot. 2014, 65, 4317–4334. [Google Scholar] [CrossRef]
- Britton, G. Biosynthesis of carotenoids. In Carotenoids in Photosynthesis; Young, A., Britton, G., Eds.; Springer: London, UK, 1993; pp. 96–126. [Google Scholar]
- Mende, K.; Homann, V.; Tudzynski, B. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: Isolation and expression. Mol. Gen. Genet. 1997, 255, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Kainou, T.; Kawamura, K.; Tanaka, K.; Matsuda, H.; Kawamukai, M. Identification of the GGPS1 genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim. Biophys. Acta. 1999, 1437, 333–340. [Google Scholar] [CrossRef]
- Coman, D.; Altenhoff, A.; Zoller, S.; Gruissem, W.; Vranová, E. Distinct evolutionary strategies in the GGPPS family from plants. Front. Plant Sci. 2014, 5, 230. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Gonzalez-Garay, M.L. Introduction to isoform sequencing using pacific biosciences technology (Iso-Seq). In Transcriptomics and Gene Regulation; Wu, J., Ed.; Springer: Dordrecht, The Netherlands, 2016; pp. 141–160. [Google Scholar]
- Abdelghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Reddy, A.S. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef]
- Luo, Q.; Bian, C.; Tao, M.; Huang, Y.; Zheng, Y.; Lv, Y.; Xu, J. Genome and transcriptome sequencing of the astaxanthin-producing green microalga, Haematococcus pluvialis. Genome Biol. Evol. 2019, 11, 166–173. [Google Scholar] [CrossRef]
- Wayama, M.; Ota, S.; Matsuura, H.; Nango, N.; Hirata, A.; Kawano, S. Three-dimensional ultra structural study of oil and astaxanthin accumulation during encystment in the green alga Haematococcus pluvialis. PLoS ONE 2013, 8, e53618. [Google Scholar] [CrossRef]
- Capelli, B.; Bagchi, D.; Cysewski, G.R. Synthetic astaxanthin is significantly inferior to algal-based astaxanthin as an antioxidant and may not be suitable as a human nutraceutical supplement. Nutrafoods 2013, 12, 145–152. [Google Scholar] [CrossRef]
- Linden, H. Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim. Biophys. Acta Gene Struct. Expr. 1999, 1446, 203–212. [Google Scholar] [CrossRef]
- Lotan, T.; Hirschberg, J. Cloning and expression in Escherichia coli of the gene coding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett. 1995, 364, 125–128. [Google Scholar]
- Kajiwara, S.; Kakizono, T.; Saito, T.; Saito, T.; Kondo, K.; Ohtani, T.; Nishio, N.; Misawa, N. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol. Biol. 1995, 29, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.; Chen, F.; Sandmann, G. Stress-related differential expression of multiple β-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J. Biotech. 2006, 122, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhu, X.; Wu, T.; Wang, W.; Zhao, D.; Bi, C.; Zhang, X. Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol. Biofuels 2018, 11, 278–287. [Google Scholar] [CrossRef]
- Henke, N.A.; Wendisch, V.F. Improved astaxanthin production with Corynebcterium glutamicum by application of a membrane fusion protein. Mar. Drugs 2019, 17, 621. [Google Scholar] [CrossRef]
- Wu, Y.; Yan, P.; Liu, X.; Wang, Z.; Tang, Y.J.; Chen, T.; Zhao, X. Combinatorial expression of different β-carotene hydroxylases and ketolases in Escherichia coli for increased astaxanthin production. J. Ind. Microbiol. Biotechnol. 2019, 46, 1505–1516. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Coman, D.; Beck, G.; Barja, M.; Colinas, M.; Graf, A.; Gruissem, W. Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 2016, 209, 252–264. [Google Scholar] [CrossRef]
- Liang, P.H.; Ko, T.P.; Wang, A.H.J. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef]
- Tholl, D.; Kish, C.M.; Orlova, I.; Sherman, D.; Gershenzon, J.; Pichersky, E.; Dudareva, N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyldiphosphate synthases. Plant Cell 2004, 16, 977–992. [Google Scholar] [CrossRef]
- Beck, G.; Coman, D.; Herren, E.; Ruiz-Sola, M.Á.; Rodríguez-Concepción, M.; Gruissem, W.; Vranová, E. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 2013, 82, 393–416. [Google Scholar] [CrossRef]
- Hsieh, F.L.; Chang, T.H.; Ko, T.P.; Wang, A.H.J. Structure and mechanism of an Arabidopsis medium/long-chain-length prenylpyrophosphate synthase. Plant Physiol. 2011, 155, 1079–1090. [Google Scholar] [CrossRef]
- Lao, Y.M.; Jin, H.; Zhou, J.; Zhang, H.J.; Zhu, X.S.; Cai, Z.H. A novel hydrolytic activity of tri-functional geranylgeranyl pyrophosphate synthase in Haematococcus pluvialis. Plant Cell Physiol. 2018, 59, 2536–2548. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, H.; Su, P.; Chen, T.; Guo, J.; Gao, W.; Huang, L.Q. Molecular cloning and functional characterization of multiple geranylgeranyl pyrophosphate synthase (ApGGPPS) from Andrographis paniculata. Plant Cell Rep. 2019, 38, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Kane, J.F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 1995, 6, 494–500. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, Y.; Wu, G.; Li, G.; Sun, H.; Deng, S.; Zhang, X. Transcriptome analysis in Haematococcus pluvialis: Astaxanthin induction by salicylic acid (SA) and jasmonic acid (JA). PLoS ONE 2015, 10, e0140609. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Z.; Tao, M.; Li, J.; Hu, Z. Effects of selenite on green microalga Haematococcus pluvialis: Bioaccumulation of selenium and enhancement of astaxanthin production. Aquat. Toxicol. 2017, 183, 21–27. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ma, T.; Zhou, Y.; Li, X.; Zhu, F.; Cheng, Y.; Liu, Y.; Liu, T. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli. Biotechnol. J. 2016, 11, 228–237. [Google Scholar] [CrossRef]
| Primer ID | Sequences (5’–3’) | Used for |
|---|---|---|
| GGPP1F1 | CACTGCCTATCCCCGTTTCCAATC | Cloning of HpGGPPS1-1 and HpGGPPS1-2 |
| GGPP1R1 | GCACCTGCTGACCCGCTCTG | |
| GGPP2F1 | GGCGACGCGGGCAAATCAGT | Cloning of HpGGPPS2-1 and HpGGPPS2-2 |
| GGPP2R1 | AAGCGCCAGGGAATACCAAACATA | |
| GGPP3F1 | GCTCTCTTCGCACTTCTTGG | Cloning of HpGGPPS3-1, HpGGPPS3-2, HpGGPPS3-3 and HpGGPPS3-4 |
| GGPP3R1 | TGATGCCTAGACAGCTCACTT | |
| fGGPP1F2 | GTATCTGAATTCAAAAAATGATCCGAGCGATGCACA † | Subcloning of HpGGPPS1-1 |
| GGPP1R2 | CATAGAAAGCTTTCAGTTCTTGCGGTATCCTA | |
| GGPP1F3 | GTATCTGAATTCAAAAAATGATCCGAGCGATGCACA | Subcloning of HpGGPPS1-2 |
| GGPP1R3 | CATAGAAAGCTTTCAGTTCTTGCGGTACCCT | |
| GGPP2F2 | GTATCTGAATTCAAAAAATGAGGGGCCTAGCGGGCAA | Subcloning of HpGGPPS2-1 and HpGGPPS2-2 |
| GGPP2R2 | CATAGAGCGGCCGCCTATTTCTTTCTGCTCAGGACTC | |
| GGPP3F2 | GTATCTGAATTCAAAAAATGGTATCGGATGTGATGCAAG | Subcloning of HpGGPPS3-1 |
| GGPP3R2 | CATAGAAAGCTTTCACTTGCAGCGCTTGTAAATC | |
| GGPP3F3 | GTATCTGAATTCAAAAAATGGTATCGGATGTGATGCAAG | Subcloning of HpGGPPS3-2, HpGGPPS3-3, and HpGGPPS3-4 |
| GGPP3R3 | CATAGAAAGCTTTCACTTGCAGCGCTTGTAAATC | |
| CrtIB-F2 | GTATCTTCTAGAGTAAGGATCCTAACATGAAACCGACCACGGTGA | Subcloning of CrtIB |
| CrtIB-R2 | CATAGAGAATTCATGTCGACAAGTTACAGCGGACGTTGCCAC | |
| CrtE-F1 | GTATCTGAATTCATACCATGACCGTGTGTGCGAA | Subcloning of CrtE |
| CrtE-R1 | ATAGAAAGCTTTCCTTTACGACACCGCTGCCA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, D.; Liu, W.; Li, A.; Wang, C.; Hu, Z. Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis. Mar. Drugs 2019, 17, 696. https://doi.org/10.3390/md17120696
Huang D, Liu W, Li A, Wang C, Hu Z. Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis. Marine Drugs. 2019; 17(12):696. https://doi.org/10.3390/md17120696
Chicago/Turabian StyleHuang, Danqiong, Wenfu Liu, Anguo Li, Chaogang Wang, and Zhangli Hu. 2019. "Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis" Marine Drugs 17, no. 12: 696. https://doi.org/10.3390/md17120696
APA StyleHuang, D., Liu, W., Li, A., Wang, C., & Hu, Z. (2019). Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis. Marine Drugs, 17(12), 696. https://doi.org/10.3390/md17120696





