20-Nor-Isopimarane Epimers Produced by Aspergillus wentii SD-310, a Fungal Strain Obtained from Deep Sea Sediment
Abstract
1. Introduction
2. Results and Discussion
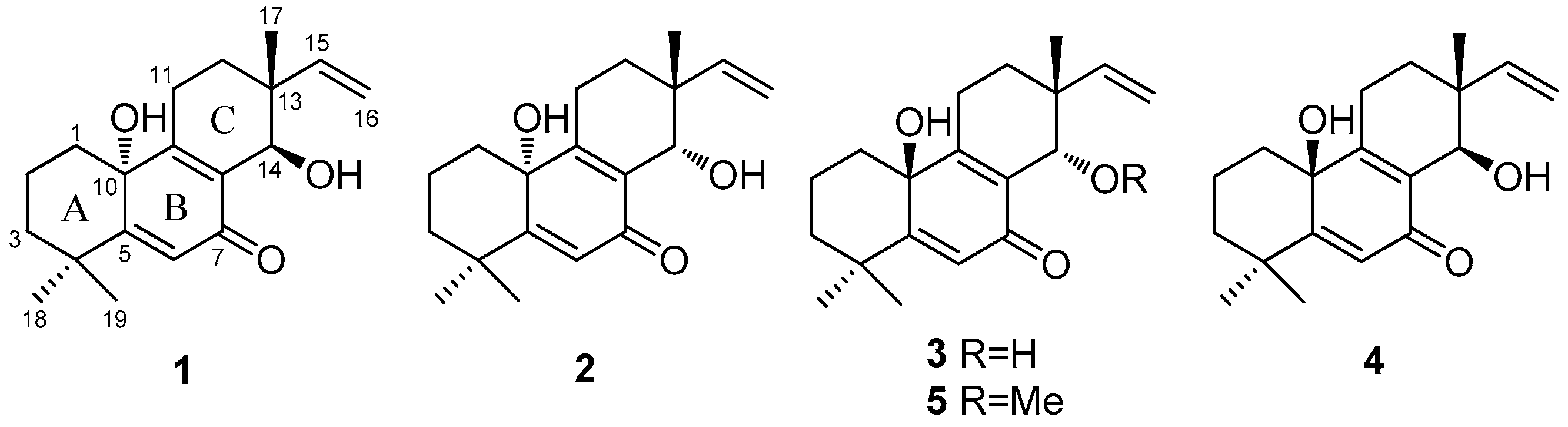
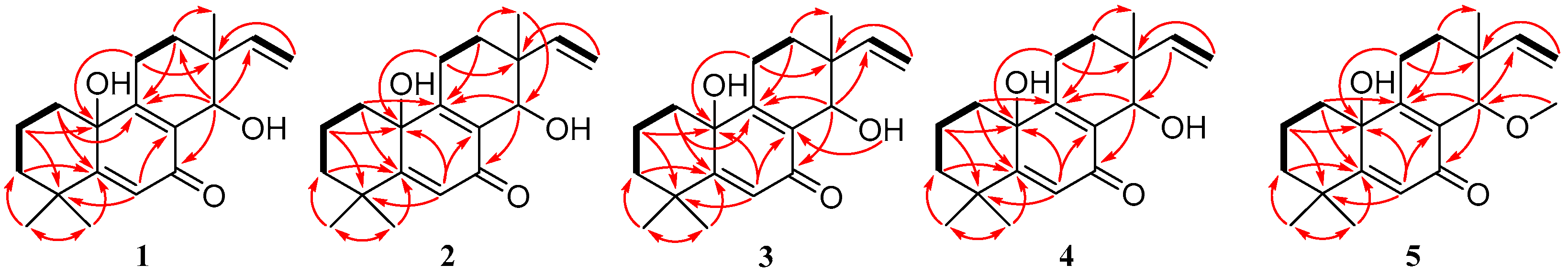
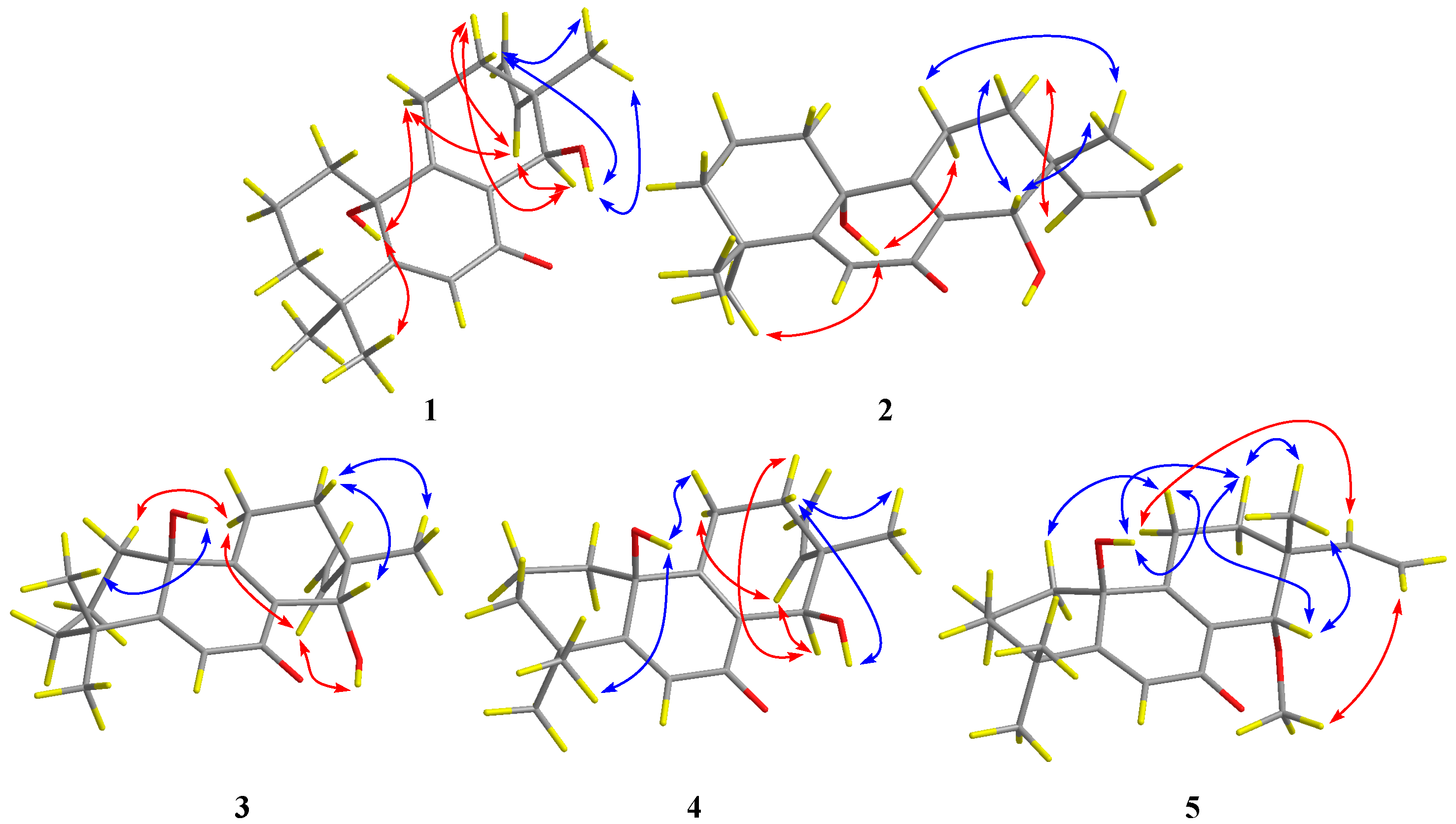
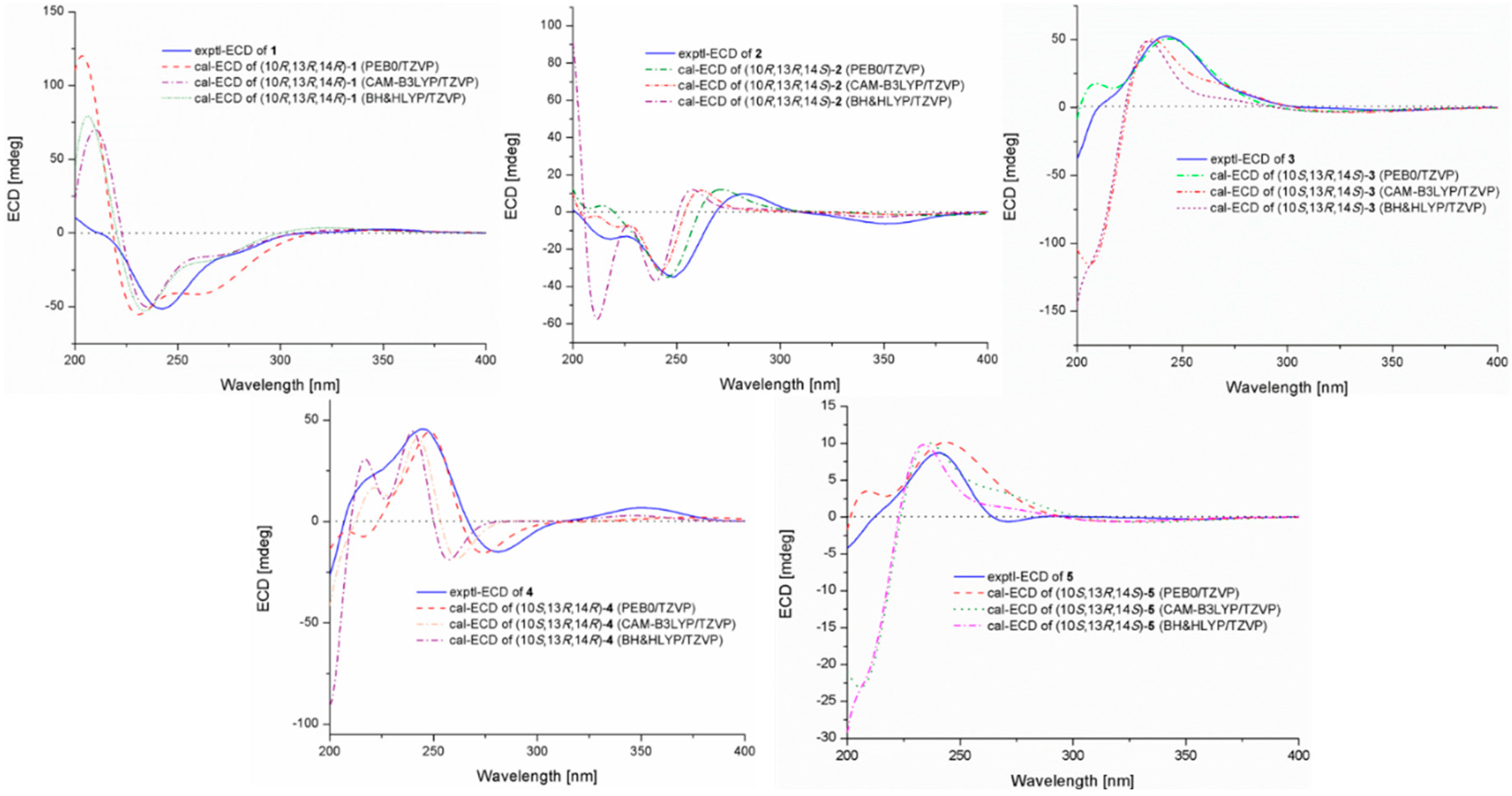
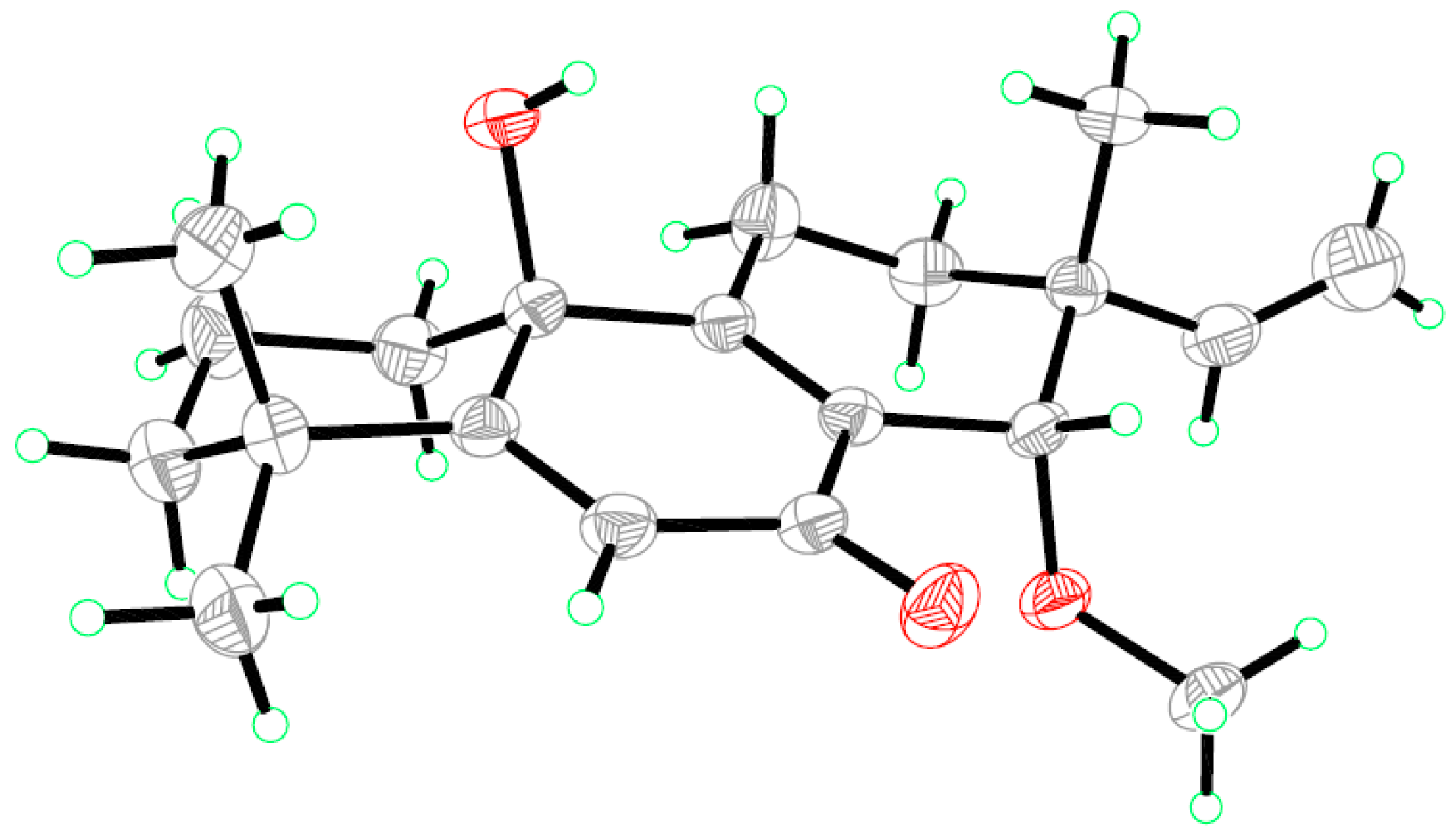
2.1. Structure Elucidation of the New Compounds
2.2. Biological Activities of the Isolated Compounds
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Isolation
3.5. Antimicrobial and Brine Shrimp Lethality Assays
3.6. X-ray Crystallographic Analysis
3.7. Computational Section
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References and Note
- Choma, A.; Wiater, A.; Komaniecka, I.; Paduch, R.; Pleszczyńska, M.; Szczodrak, J. Chemical characterization of a water insoluble (1→3)-α-d-glucan from an alkaline extract of Aspergillus wentii. Carbohydr. Polym. 2013, 91, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.-R.; Miao, F.-P.; Zhang, J.; Wang, G.; Yin, X.-L.; Ji, N.-Y. Three new xanthone derivatives from an algicolous isolate of Aspergillus wentii. Magn. Reson. Chem. 2013, 51, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.-M.; Xu, G.-M.; Li, C.-S.; Wang, B.-G. Antioxidant metabolites from marine alga-derived fungus Aspergillus wentii EN-48. Phytochem. Lett. 2014, 7, 120–123. [Google Scholar] [CrossRef]
- Dorner, J.W.; Cole, R.J.; Springer, J.P.; Cox, R.H.; Cutler, H.; Wicklow, D.T. Isolation and identification of two new biologically active norditerpene dilactones from Aspergillus wentii. Phytochemistry 1980, 19, 1157–1161. [Google Scholar] [CrossRef]
- Sun, H.-F.; Li, X.-M.; Meng, L.; Cui, C.-M.; Gao, S.-S.; Li, C.-S.; Huang, C.-G.; Wang, B.-G. Asperolides A−C, tetranorlabdane diterpenoids from the marine alga-derived endophytic fungus Aspergillus wentii EN-48. J. Nat. Prod. 2012, 75, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.-P.; Liang, X.-R.; Liu, X.-H.; Ji, N.-Y. Aspewentins A−C, norditerpenes from a cryptic pathway in an algicolous strain of Aspergillus wentii. J. Nat. Prod. 2014, 77, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-D.; Li, X.-M.; Xu, G.-M.; Zhang, P.; Wang, B.-G. Antimicrobial phenolic bisabolanes and related derivatives from Penicillium aculeatum SD-321, a deep sea sediment-derived fungus. J. Nat. Prod. 2015, 78, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-H.; Wang, C.-Y.; Mándi, A.; Li, X.-M.; Hu, X.-Y.; Kassack, M.-U.; Kurtán, T.; Wang, B.-G. Three diketopiperazine alkaloids with spirocyclic skeletons and one bisthiodiketopiperazine derivative from the mangrove-derived endophytic fungus Penicillium brocae MA-231. Org. Lett. 2016, 18, 5304–5307. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-L.; Li, X.-M.; Liu, H.; Meng, L.-H.; Wang, B.-G. Two new diphenylketones and a new xanthone from Talaromyces islandicus EN-501, an endophytic fungus derived from the marine red alga Laurencia okamurai. Mar. Drugs 2016, 14, 223. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-L.; Xu, R.; Li, X.-M.; Yang, S.-Q.; Meng, L.-H.; Wang, B.-G. Simpterpenoid A, a meroterpenoid with a highly functionalized cyclohexadiene moiety featuring gem-propane-1,2-dione and methylformate groups, from the mangrove-derived Penicillium simplicissimum MA-332. Org. Lett. 2018, 20, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Q.; Li, X.-M.; Li, X.; Chi, L.-P.; Wang, B.-G. Two new diketomorpholine derivatives and a new highly conjugated ergostane-type steroid from the marine algal-derived endophytic fungus Aspergillus alabamensis EN-547. Mar. Drugs 2018, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-D.; Li, X.-M.; Li, X.; Xu, G.-M.; Liu, Y.; Wang, B.-G. Aspewentins D−H, 20-nor-isopimarane derivatives from the deep sea sediment-derived fungus Aspergillus wentii SD-310. J. Nat. Prod. 2016, 79, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.-M.; Li, X.-D.; Xu, G.-M.; Liu, Y.; Wang, B.-G. 20-nor-isopimarane cycloethers from the deep-sea sediment-derived fungus Aspergillus wentii SD-310. RSC Adv. 2016, 6, 75981–75987. [Google Scholar] [CrossRef]
- Li, X.; Li, X.-D.; Li, X.-M.; Xu, G.-M.; Liu, Y.; Wang, B.-G. Wentinoids A–F, six new isopimarane diterpenoids from Aspergillus wentii SD-310, a deep-sea sediment derived fungus. RSC Adv. 2017, 4, 4387–4394. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Pierce, C.-G.; Uppuluri, P.; Tristan, A.-R.; Wormley, F.-L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.-L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Zhang, P.; Li, X.-M.; Li, C.-S.; Cui, C.-M.; Wang, B.-G. Cyclohexadepsipeptides of the isaridin class from the marine-derived fungus Beauveria felina EN-135. J. Nat. Prod. 2014, 77, 1164–1169. [Google Scholar] [CrossRef] [PubMed]
- Crystallographic data of compound 5 have been deposited in the Cambridge Crystallographic Data Centre as CCDC 1835494. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/data/request/cif (or from the CCDC, 12 Union Road, Cambridge CB21EZ, U.K.; fax: + 44-1223-336-033; e-mail: Deposit@ccdc.cam.ac.uk).
- Sheldrick, G.M. SADABS, Software for Empirical Absorption Correction; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXTL, Structure Determination Software Programs; Bruker Analytical X-ray System Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97 and SHELXS-97, Program for X-ray Crystal Structure Solution and Refinement; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1α | 1.14, m | 1.07, m | 1.04, m | 0.97, m | 1.10, m |
| 1β | 2.17, d (4.8) | 2.15, m | 2.18, m | 2.08, m | 2.20, m |
| 2α | 1.45, m | 1.45, m | 1.46, m | 1.42, m | 1.46, m |
| 2β | 1.98, m | 1.99, m | 1.99, m | 1.97, m | 2.00, m |
| 3α | 1.26, m | 1.24, m | 1.22, m | 1.24, m | 1.28, m |
| 3β | 1.62, m | 1.60, m | 1.64, m | 1.60, m | 1.63, m |
| 6 | 5.95, s | 5.94, s | 5.96, s | 5.93, s | 5.99, s |
| 11α | 2.31, m | 2.20, m | 2.37, m | 2.13, m | 2.40, m |
| 11β | 2.40, m | 2.66, m | 2.39, m | 2.55, m | 2.42, m |
| 12α | 1.37, m | 1.38, m | 1.26, m | 1.40, m | 1.33, m |
| 12β | 1.69, m | 1.81, m | 1.83, m | 1.70, m | 1.82, m |
| 14 | 4.15, d (4.8) | 4.09, br s | 4.11, d (4.7) | 4.20, br s | 3.82, s |
| 15 | 5.75, dd | 6.05, dd | 6.04, dd | 5.61, dd | 6.04, dd |
| (17.6, 10.9) | (17.7, 10.9) | (17.7, 10.9) | (17.7, 11.1) | (17.5, 11.1) | |
| 16a | 4.91, br d | 4.95, br d | 4.95, br d | 4.84, br d | 5.02, br d |
| (10.9) | (10.9) | (10.9) | (17.7) | (11.1) | |
| 16b | 4.97, br d | 4.98, br d | 4.98, br d | 4.87, br d | 5.04, br d |
| (17.6) | (17.7) | (17.7) | (11.1) | (17.5) | |
| 17 | 0.99, s | 0.75, s | 0.80, s | 0.99, s | 0.79, s |
| 18 | 1.34, s | 1.34, s | 1.35, s | 1.33, s | 1.36, s |
| 19 | 1.13, s | 1.11, s | 1.12, s | 1.10, s | 1.13, s |
| 10-OH | 5.24, s | 5.42, br s | 5.29, br s | 5.44, br s | 5.34, br s |
| 14-OH | 4.67, d | 4.57, br s | 4.55, d | 4.78, br s | |
| (4.8) | (4.7) | ||||
| 14-OMe | – | – | – | – | 3.23, s |
| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 38.9, CH2 | 38.5, CH2 | 38.9, CH2 | 38.5, CH2 | 39.1, CH2 |
| 2 | 17.9, CH2 | 17.9, CH2 | 17.9, CH2 | 17.8, CH2 | 18.0, CH2 |
| 3 | 42.4, CH2 | 42.2, CH2 | 42.4, CH2 | 42.2, CH2 | 42.3, CH2 |
| 4 | 38.1, qC | 38.0, qC | 38.1, qC | 38.0, qC | 38.2, qC |
| 5 | 169.2, qC | 168.9, qC | 169.2, qC | 168.9, qC | 169.5, qC |
| 6 | 122.9, CH | 122.6, CH | 122.8, CH | 122.5, CH | 122.6, CH |
| 7 | 185.6, qC | 185.2, qC | 185.4, qC | 185.1, qC | 185.4, qC |
| 8 | 131.6, qC | 130.7, qC | 131.4, qC | 131.3, qC | 130.0, qC |
| 9 | 161.4, qC | 159.6, qC | 160.7, qC | 160.2, qC | 161.6, qC |
| 10 | 70.7, qC | 69.9, qC | 70.6, qC | 70.0, qC | 70.8, qC |
| 11 | 21.9, CH2 | 21.3, CH2 | 21.5, CH2 | 22.0, CH2 | 21.3, CH2 |
| 12 | 28.6, CH2 | 25.9, CH2 | 26.7, CH2 | 27.6, CH2 | 26.5, CH2 |
| 13 | 39.1, qC | 38.9, qC | 38.9, qC | 39.4, qC | 38.8, qC |
| 14 | 66.2, CH | 66.3, CH | 66.7, CH | 65.1, CH | 76.1, CH |
| 15 | 144.5, CH | 147.2, CH | 146.9, CH | 144.0, CH | 146.7, CH |
| 16 | 113.0, CH2 | 111.7, CH2 | 111.8, CH2 | 112.8, CH2 | 112.1, CH2 |
| 17 | 22.9, CH3 | 20.6, CH3 | 21.2, CH3 | 25.0, CH3 | 20.4, CH3 |
| 18 | 27.7, CH3 | 27.6, CH3 | 27.7, CH3 | 27.7, CH3 | 27.7, CH3 |
| 19 | 31.2, CH3 | 31.2, CH3 | 31.2, CH3 | 31.2, CH3 | 31.2, CH3 |
| 14-OMe | – | – | – | – | 59.4, CH3 |
| Strains | 1 | 2 | 3 | 4 | 5 | Positive Control |
|---|---|---|---|---|---|---|
| Escherichia colib | 32 | – | – | – | 4.0 | |
| Aeromonas hydrophiliab | – | – | 16 | – | – | 4.0 |
| Edwardsiella tardab | 8.0 | 8.0 | – | 32 | – | 4.0 |
| Pseudomonas aeruginosab | 32 | – | – | – | – | 4.0 |
| Vibrio anguillarumb | – | – | 32 | – | 32 | 0.5 |
| Vibrio harveyib | 8.0 | 8.0 | – | 32 | 32 | 4.0 |
| Vibrio parahaemolyticusb | 8.0 | 8.0 | – | – | – | 1.0 |
| Fusarium graminearumc | – | – | – | – | 4.0 | 4.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-D.; Li, X.; Li, X.-M.; Xu, G.-M.; Liu, Y.; Wang, B.-G. 20-Nor-Isopimarane Epimers Produced by Aspergillus wentii SD-310, a Fungal Strain Obtained from Deep Sea Sediment. Mar. Drugs 2018, 16, 440. https://doi.org/10.3390/md16110440
Li X-D, Li X, Li X-M, Xu G-M, Liu Y, Wang B-G. 20-Nor-Isopimarane Epimers Produced by Aspergillus wentii SD-310, a Fungal Strain Obtained from Deep Sea Sediment. Marine Drugs. 2018; 16(11):440. https://doi.org/10.3390/md16110440
Chicago/Turabian StyleLi, Xiao-Dong, Xin Li, Xiao-Ming Li, Gang-Ming Xu, Yang Liu, and Bin-Gui Wang. 2018. "20-Nor-Isopimarane Epimers Produced by Aspergillus wentii SD-310, a Fungal Strain Obtained from Deep Sea Sediment" Marine Drugs 16, no. 11: 440. https://doi.org/10.3390/md16110440
APA StyleLi, X.-D., Li, X., Li, X.-M., Xu, G.-M., Liu, Y., & Wang, B.-G. (2018). 20-Nor-Isopimarane Epimers Produced by Aspergillus wentii SD-310, a Fungal Strain Obtained from Deep Sea Sediment. Marine Drugs, 16(11), 440. https://doi.org/10.3390/md16110440






