High Throughput Identification of Antihypertensive Peptides from Fish Proteome Datasets
Abstract
:1. Introduction
2. Results
2.1. Construction of a Local AHTP Database
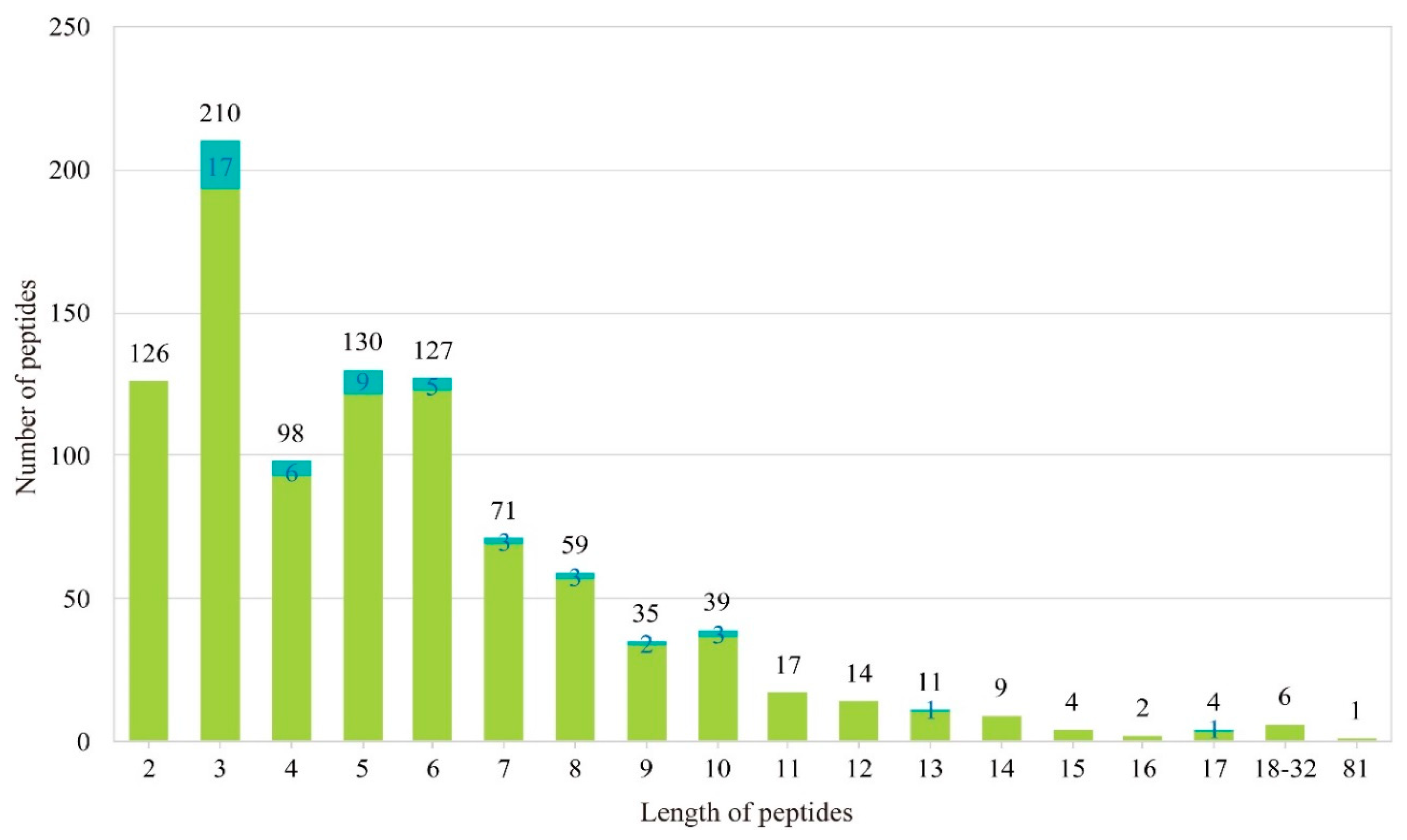
2.2. Identification of AHTPs with High Activity
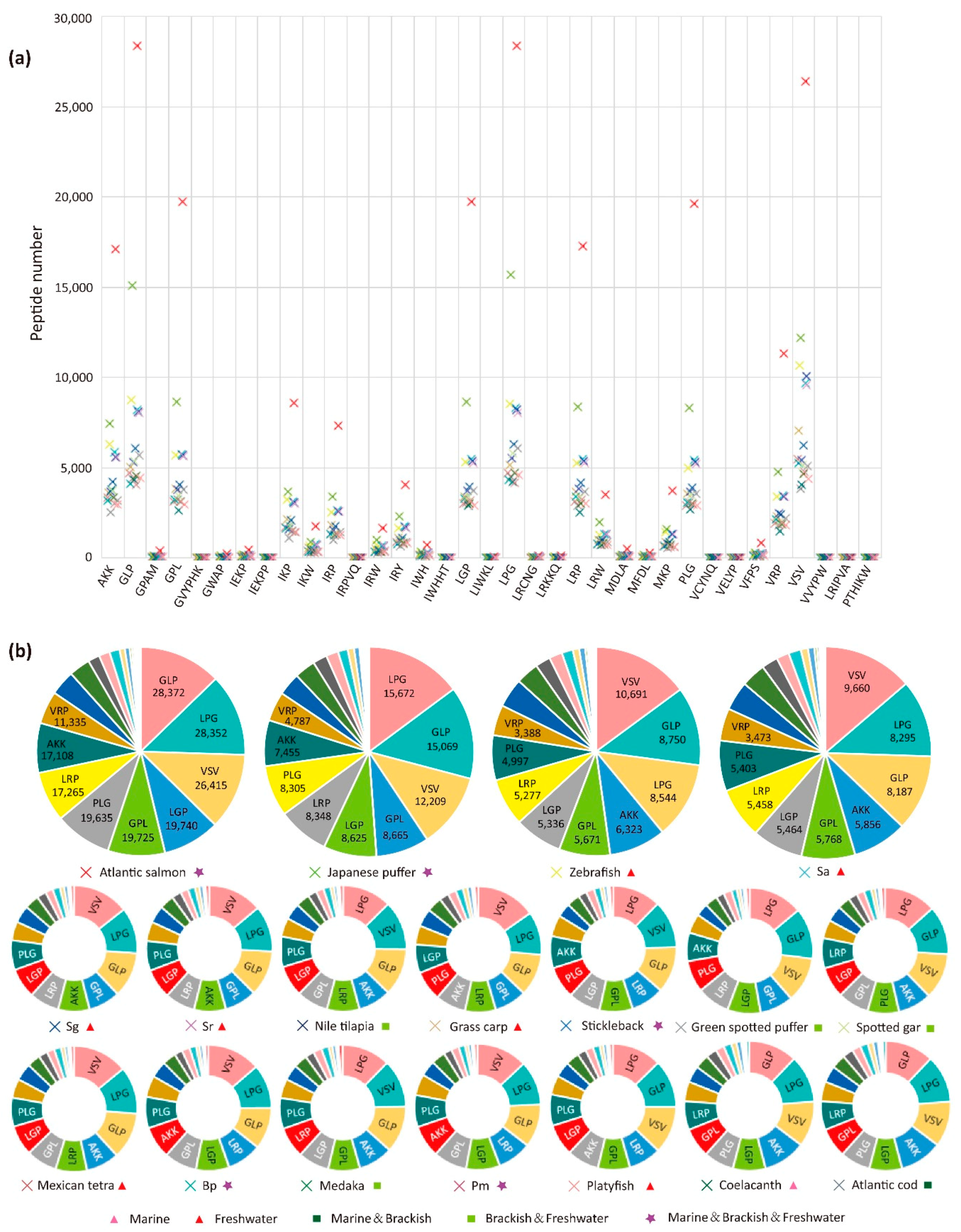
2.3. Analysis of Mapped Proteins
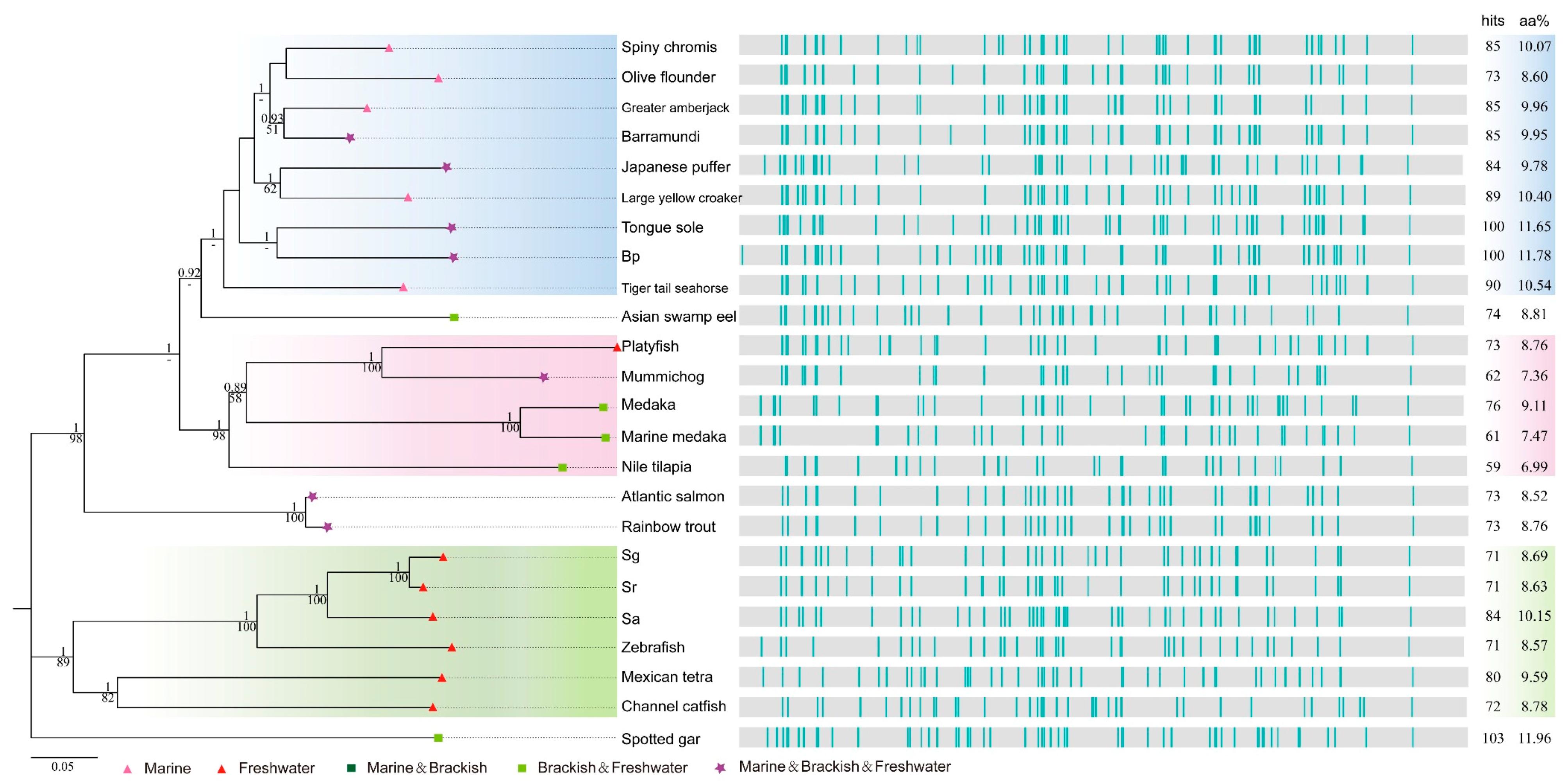
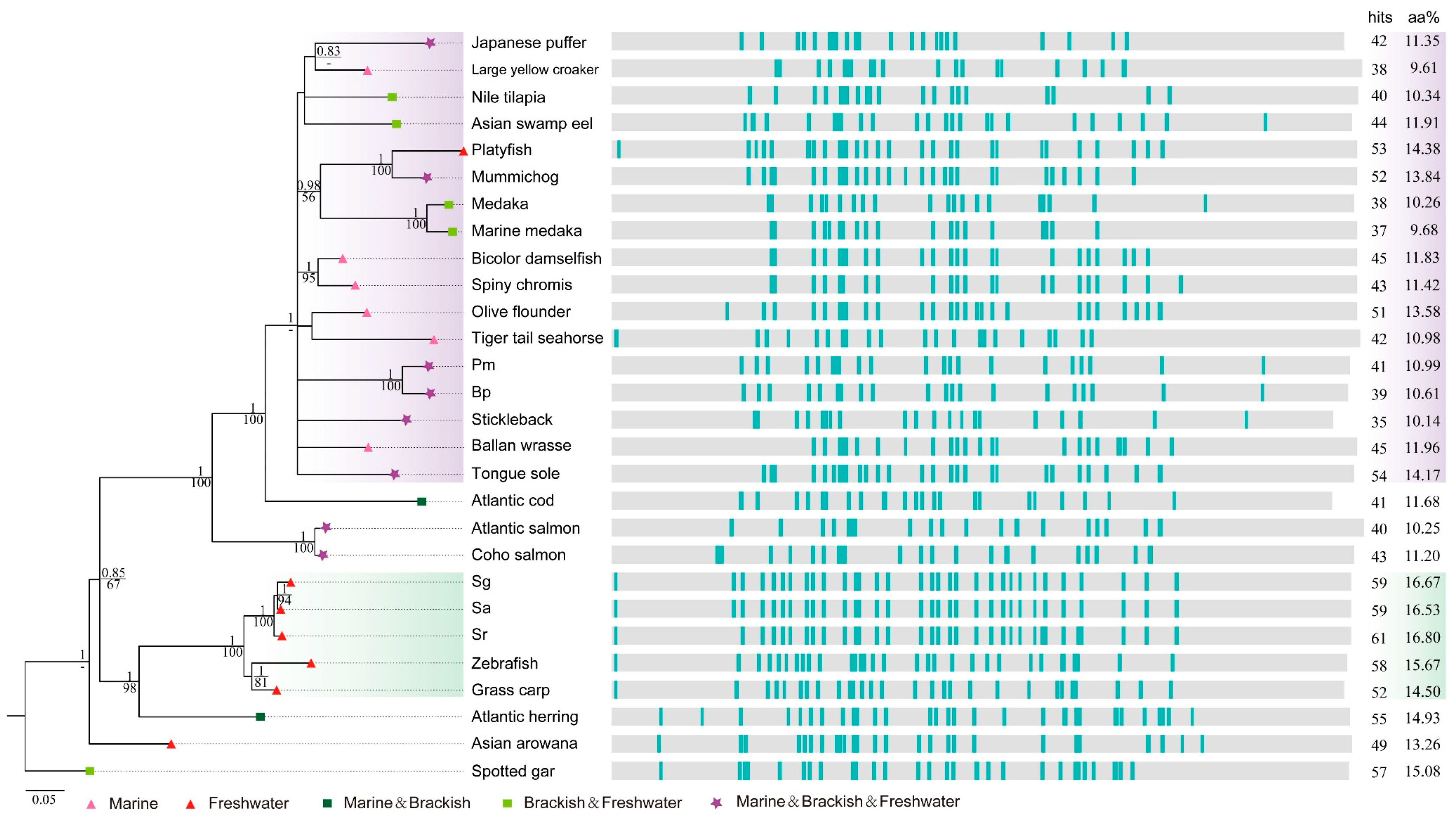
2.4. Deep Investigations into Two Collagen Subunits
3. Discussion
3.1. High-Throughput Identification of AHTPs
3.2. Potential Utility of Collagens
3.3. Prospective Development of Fish-Derived Antihypertensive Agents
4. Materials and Methods
4.1. Data Collection
4.2. Screening for AHTPs
4.3. Construction of Phylogenetic Trees
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lurie, Y. Hypertension is the biggest cost center being faced by healthcare today. Hypertension 2018. [Google Scholar]
- Chockalingam, A.; Campbell, N.R.; Fodor, J.G. Worldwide epidemic of hypertension. Can. J. Cardiol. 2006, 22, 553–555. [Google Scholar] [CrossRef] [Green Version]
- SPRINT Research Group. A randomized trial of intensive versus standard blood-pressure control. N. Engl. J. Med. 2015, 373, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- The SPRINT Study Research Group. The design and rationale of a multi-center clinical trial comparing two strategies for control of systolic blood pressure: The Systolic Blood Pressure Intervention Trial (SPRINT). Clin. Trials Lond. Engl. 2014, 11, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzidis, R.G.; Elisaf, M.S. Treatment of hypertension in chronic kidney disease. Curr. Hypertens. Rep. 2018, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- George, A.J.; Thomas, W.G.; Hannan, R.D. The renin–angiotensin system and cancer: Old dog, new tricks. Nat. Rev. Cancer 2010, 10, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, W.; Udenigwe, C.C. Revisiting the mechanisms of ACE inhibitory peptides from food proteins. Food Bioact. Evid. Health Benefits Underst. Mech. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Brook, R.D.; Appel, L.J.; Rubenfire, M.; Ogedegbe, G.; Bisognano, J.D.; Elliott, W.J.; Fuchs, F.D.; Hughes, J.W.; Lackland, D.T.; Staffileno, B.A.; et al. Beyond medications and diet: Alternative approaches to lowering blood pressure. Hypertension 2013, 61, 1360–1383. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.H.; Bartelt, D.C.; Greene, L.J. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry 1970, 9, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Majumder, K.; Wu, J. Molecular targets of antihypertensive peptides: understanding the mechanisms of action based on the pathophysiology of hypertension. Int. J. Mol. Sci. 2015, 16, 256–283. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Antihypertensive peptides of animal origin: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Plant. Biotechnol. Food Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, B.; Veryser, L.; Verbeke, F.; Wynendaele, E.; De Spiegeleer, B. Fish hydrolysates: A regulatory perspective of bioactive peptides. Protein Pept. Lett. 2016, 23, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B.M.; Oh, D.H.; Lee, B.H. Bioactive peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Nasri, M. Protein hydrolysates and biopeptides: Production, biological activities, and applications in foods and health Benefits. A review. In Advances in Food and Nutrition Research; Toldrá, F., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 109–159. ISBN 1043-4526. [Google Scholar]
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015, 43, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive peptides from muscle sources: Meat and fish. Nutrients 2011, 3, 765–791. [Google Scholar] [CrossRef] [PubMed]
- Jensen, I.J.; Mæhre, K.H. Preclinical and clinical studies on antioxidative, antihypertensive and cardioprotective effect of marine proteins and peptides—A review. Mar. Drugs 2016, 14, 211. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.; Hayes, M.; Carney, B. Angiotensin-I-converting enzyme and prolyl endopeptidase inhibitory peptides from natural sources with a focus on marine processing by-products. Food Chem. 2011, 129, 235–244. [Google Scholar] [CrossRef]
- Kim, M.R.; Kim, J.W.; Park, J.B.; Hong, Y.K.; Ku, S.K.; Choi, J.S. Anti-obesity effects of yellow catfish protein hydrolysate on mice fed a 45% kcal high-fat diet. Int. J. Mol. Med. 2017, 40, 784–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikoo, M.; Benjakul, S.; Xu, X. Antioxidant and cryoprotective effects of Amur sturgeon skin gelatin hydrolysate in unwashed fish mince. Food Chem. 2015, 181, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.C.; Harnedy, P.A.; O’Keeffe, M.B.; FitzGerald, R.J. Bioactive peptides from Atlantic salmon (Salmo salar) with angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory, and antioxidant activities. Food Chem. 2017, 218, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Elavarasan, K.; Shamasundar, B.A.; Badii, F.; Howell, N. Angiotensin I-converting enzyme (ACE) inhibitory activity and structural properties of oven- and freeze-dried protein hydrolysate from fresh water fish (Cirrhinus mrigala). Food Chem. 2016, 206, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.J.; Je, J.Y.; Kim, S.K. Antihypertensive effect of angiotensin I converting enzyme-inhibitory peptide from hydrolysates of bigeye tuna dark muscle, Thunnus obesus. J. Agric. Food Chem. 2007, 55, 8398–8403. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Zhong, Q.; Wu, Y.; Xia, W. Purification and characterization of a novel angiotensin-I converting enzyme (ACE) inhibitory peptide derived from enzymatic hydrolysate of grass carp protein. Peptides 2012, 33, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; FitzGerald, R.J. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Shuttleworth, C.A. Type VIII collagen. Int. J. Biochem. Cell. Biol. 1997, 29, 1145–1148. [Google Scholar] [CrossRef]
- Gordon, M.K.; Hahn, R.A. Collagens. Cell. Tissue Res. 2009, 339, 247. [Google Scholar] [CrossRef] [PubMed]
- Win, T.S.; Schaduangrat, N.; Prachayasittikul, V.; Nantasenamat, C.; Shoombuatong, W. PAAP: A web server for predicting antihypertensive activity of peptides. Future Med. Chem. 2018, 10, 1749–1767. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [PubMed]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, 45012. [Google Scholar] [CrossRef] [PubMed]
- Bleakley, S.; Hayes, M.; O’ Shea, N.; Gallagher, E.; Lafarga, T. Predicted release and analysis of novel ACE-I, renin, and DPP-IV inhibitory peptides from common oat (Avena sativa) protein hydrolysates using in silico analysis. Foods 2017, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Chaudhary, K.; Singh Chauhan, J.; Nagpal, G.; Kumar, R.; Sharma, M.; Raghava, G.P. An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci. Rep. 2015, 5, 12512. [Google Scholar] [CrossRef] [PubMed]
- Zhipeng, Y.; Yang, C.; Wenzhu, Z.; Jianrong, L.; Jingbo, L.; Feng, C. Identification and molecular docking study of novel angiotensin-converting enzyme inhibitory peptides from Salmo salar using in silico methods. J. Sci. Food Agric. 2018, 98, 3907–3914. [Google Scholar]
- Ngo, D.H.; Ryu, B.; Kim, S.K. Active peptides from skate (Okamejei kenojei) skin gelatin diminish angiotensin-I converting enzyme activity and intracellular free radical-mediated oxidation. Food Chem. 2014, 143, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure−activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Ondetti, M.; Rubin, B.; Cushman, D. Design of specific inhibitors of angiotensin-converting enzyme: New class of orally active antihypertensive agents. Science 1977, 196, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Bhat, I.; Karunasagar, I. Antihypertensive activity of fish protein hydrolysates and its peptides. Crit. Rev. Food Sci. Nutr. 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Felician, F.F.; Xia, C.; Qi, W.; Xu, H. Collagen from marine biological sources and medical applications. Chem. Biodivers. 2018, 15, e1700557. [Google Scholar] [CrossRef] [PubMed]
- Boonmaleerat, K.; Wanachewin, O.; Phitak, T.; Pothacharoen, P.; Kongtawelert, P. Fish collagen hydrolysates modulate cartilage metabolism. Cell. Biochem. Biophys. 2018, 76, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Woo, Y.M.; Shen, Z.; Yao, H.; Cai, Y.; Lin, M.C.; Poon, W.S. Enhanced expression of Vastatin inhibits angiogenesis and prolongs survival in murine orthotopic glioblastoma model. BMC Cancer 2017, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.U.B.; Willumsen, N.; Sand, J.M.B.; Larsen, L.; Karsdal, M.A.; Leeming, D.J. Type VIII collagen is elevated in diseases associated with angiogenesis and vascular remodeling. Clin. Biochem. 2016, 49, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, T.C.; Lin, B.H.; Wang, Y.W.; Johnson, S.L.; Yu, J. Collagen IX is required for the integrity of collagen II fibrils and the regulation of vascular plexus formation in Zebrafish caudal fins. Dev. Biol. 2009, 332, 360–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdillahi, S.M.; Maaß, T.; Kasetty, G.; Strömstedt, A.A.; Baumgarten, M.; Tati, R.; Nordin, S.L.; Walse, B.; Wagener, R.; Schmidtchen, A.; et al. Collagen VI contains multiple host defense peptides with potent in vivo activity. J. Immunol. 2018, 201, 1007. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Liang, C.H.; Wu, H.T.; Pang, H.Y.; Chen, C.; Wang, G.H.; Chan, L.P. Antioxidant and anti-inflammatory capacities of collagen peptides from milkfish (Chanos chanos) scales. J. Food Sci. Technol. 2018, 55, 2310–2317. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, A.; Koyama, Y.; Ishihara, S.; Kobayashi, S.; Tometsuka, C.; Kusubata, M.; Kuwaba, K.; Hayashida, O.; Hattori, S.; Katagiri, K. Collagen-derived peptides modulate CD4(+) T-cell differentiation and suppress allergic responses in mice. Immun. Inflamm. Dis. 2018, 6, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jin, S.; Li, X.; Li, X.; Hu, X.; Chen, Y.; Huang, F.; Yang, Z.; Yu, F.; Ding, G. Physicochemical properties and biocompatibility evaluation of collagen from the skin of giant croaker (Nibea japonica). Mar. Drugs 2018, 16, 222. [Google Scholar] [CrossRef] [PubMed]
- Jie, L.; MingYan, Y.; JunHong, L.; Song, Q. Research progress of fish collagen. J. Food Saf. Qual. 2015, 6, 3941–3946. [Google Scholar]
- Puwastien, P.; Judprasong, K.; Kettwan, E.; Vasanachitt, K.; Nakngamanong, Y.; Bhattacharjee, L. Proximate composition of raw and cooked Thai freshwater and marine fish. J. Food Compos. Anal. 1999, 12, 9–16. [Google Scholar] [CrossRef]
- Shaviklo, A.R. Development of fish protein powder as an ingredient for food applications: A review. J. Food Sci. Technol. 2015, 52, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinforma. Oxf. Engl. 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 4 October 2016).
- Wallace, S. Perl Graphics Programming: Creating SVG, SWF (Flash), JPEG and PNG Files with Perl; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2002; ISBN 1-4493-5830-6. [Google Scholar]


| Species | Common Name | The Top Hit | Hit Number | The Top Score | Hit Number |
|---|---|---|---|---|---|
| Latimeria chalumnae | Coelacanth | Col4a5 | 89 | Col8a1 | 39 |
| Lepisosteus oculatus | Spotted gar | Col4a5 | 102 | Col8a1b | 57 |
| Danio rerio | Zebrafish | Elastin b | 85 | Birc2 | 1 |
| Ctenopharyngodon idella | Grass carp | Titin | 107 | Col8a1 | 52 |
| Sinocyclocheilus anshuiensis | Sa | Titin | 95 | Unknown 1 | 9 |
| Sinocyclocheilus rhinocerous | Sr | Titin | 98 | Col8a1 | 61 |
| Sinocyclocheilus grahami | Sg | Col4a6 | 68 | Col8a1 | 59 |
| Astyanax mexicanus | Mexican tetra | Col4a5 | 79 | Col8a2 | 46 |
| Salmo salar | Atlantic salmon | Xylanase | 122 | Xylanase | 122 |
| Gadus morhua | Atlantic cod | Titin | 84 | Col8a1 | 29 |
| Oryzias latipes | Medaka | Col4a6 | 68 | Col25a1 | 24 |
| Xiphophorus maculatus | Platyfish | Titin | 75 | Col8a1a | 53 |
| Gasterosteus aculeatus | Stickleback | Titin | 80 | Col8a1 | 47 |
| Takifugu rubripes | Japanese puffer | Col4a5 | 86 | Col8a1a | 42 |
| Tetraodon nigroviridis | Green spotted puffer | Col4a5 | 82 | Col14a1 | 5 |
| Oreochromis niloticus | Nile tilapia | Col4a6 | 68 | Col25a1 | 33 |
| Boleophthalmus pectinirostris | Bp | Titin | 108 | Epsin | 89 |
| Periophthalmus magnuspinnatus | Pm | Col4a5 | 53 | Unknown 2 | 5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Y.; Lv, Y.; Zhang, L.; Yang, J.; Shi, Q. High Throughput Identification of Antihypertensive Peptides from Fish Proteome Datasets. Mar. Drugs 2018, 16, 365. https://doi.org/10.3390/md16100365
Yi Y, Lv Y, Zhang L, Yang J, Shi Q. High Throughput Identification of Antihypertensive Peptides from Fish Proteome Datasets. Marine Drugs. 2018; 16(10):365. https://doi.org/10.3390/md16100365
Chicago/Turabian StyleYi, Yunhai, Yunyun Lv, Lijun Zhang, Jian Yang, and Qiong Shi. 2018. "High Throughput Identification of Antihypertensive Peptides from Fish Proteome Datasets" Marine Drugs 16, no. 10: 365. https://doi.org/10.3390/md16100365
APA StyleYi, Y., Lv, Y., Zhang, L., Yang, J., & Shi, Q. (2018). High Throughput Identification of Antihypertensive Peptides from Fish Proteome Datasets. Marine Drugs, 16(10), 365. https://doi.org/10.3390/md16100365





