Metabolomic Investigations on Nesterenkonia flava Revealed Significant Differences between Marine and Terrestrial Actinomycetes
Abstract
:1. Introduction
2. Results and Discussion
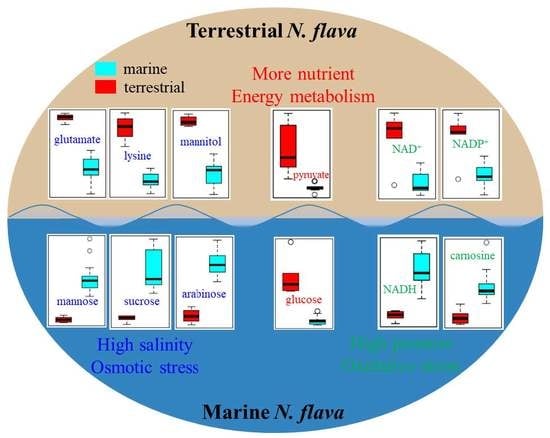
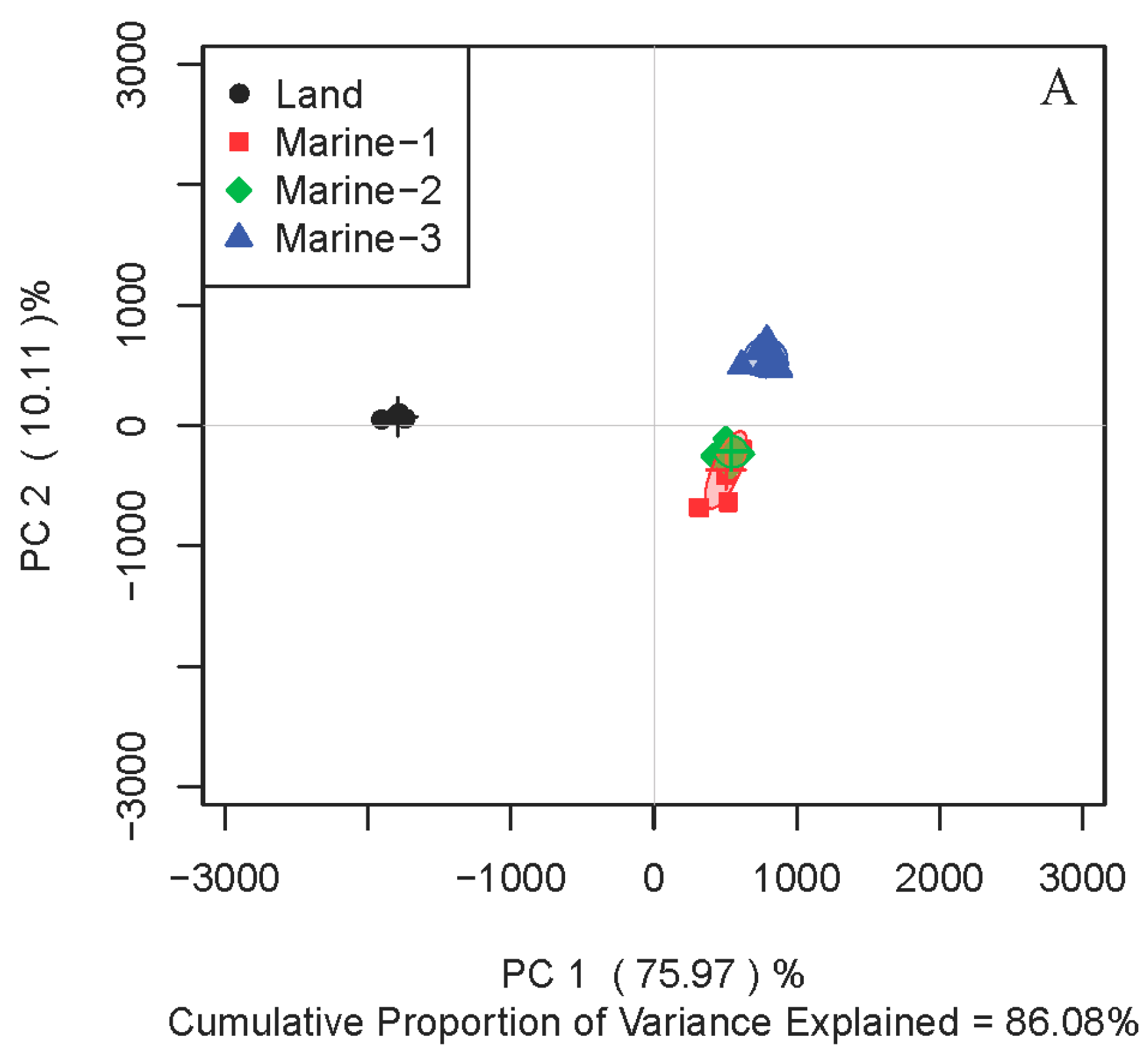
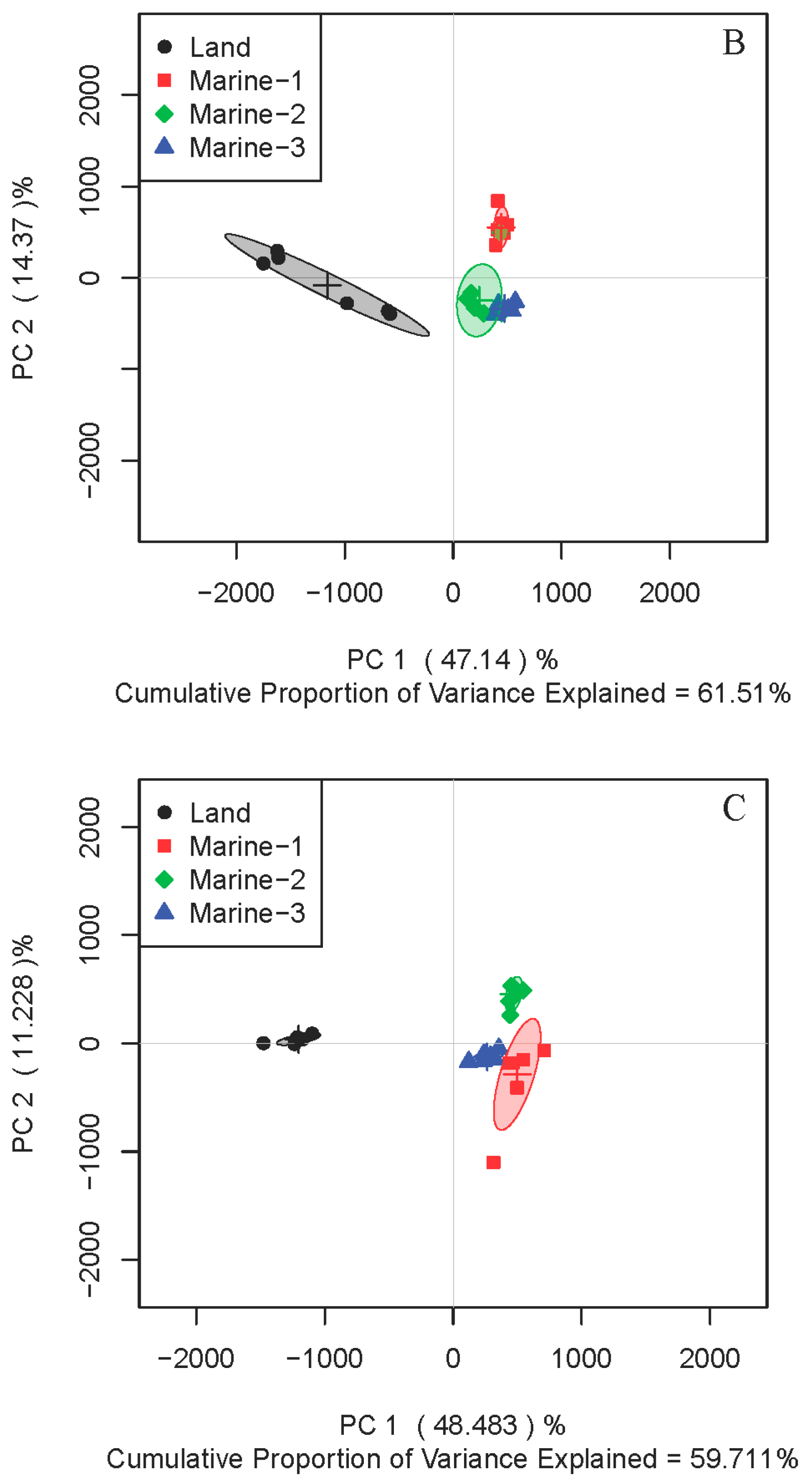
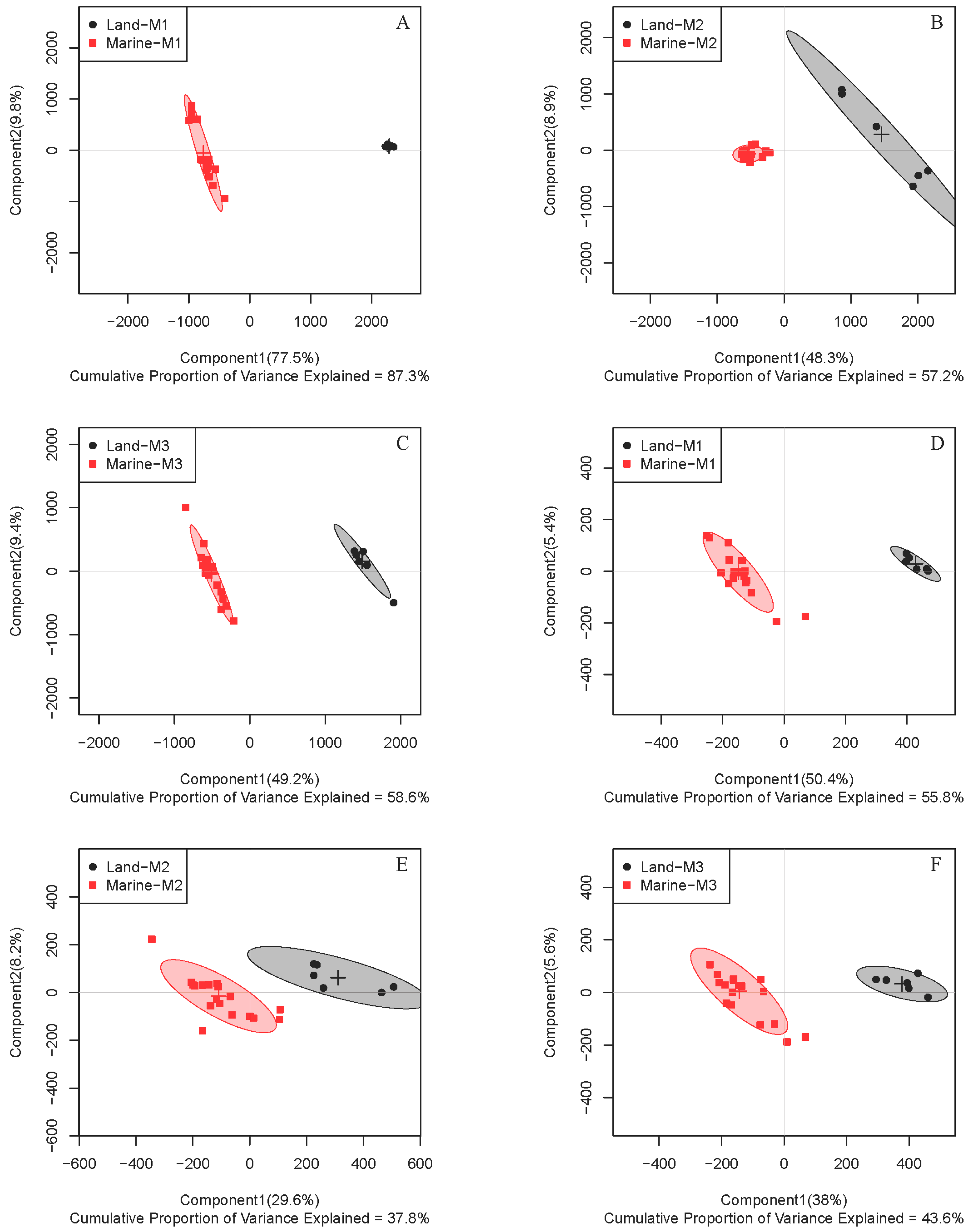
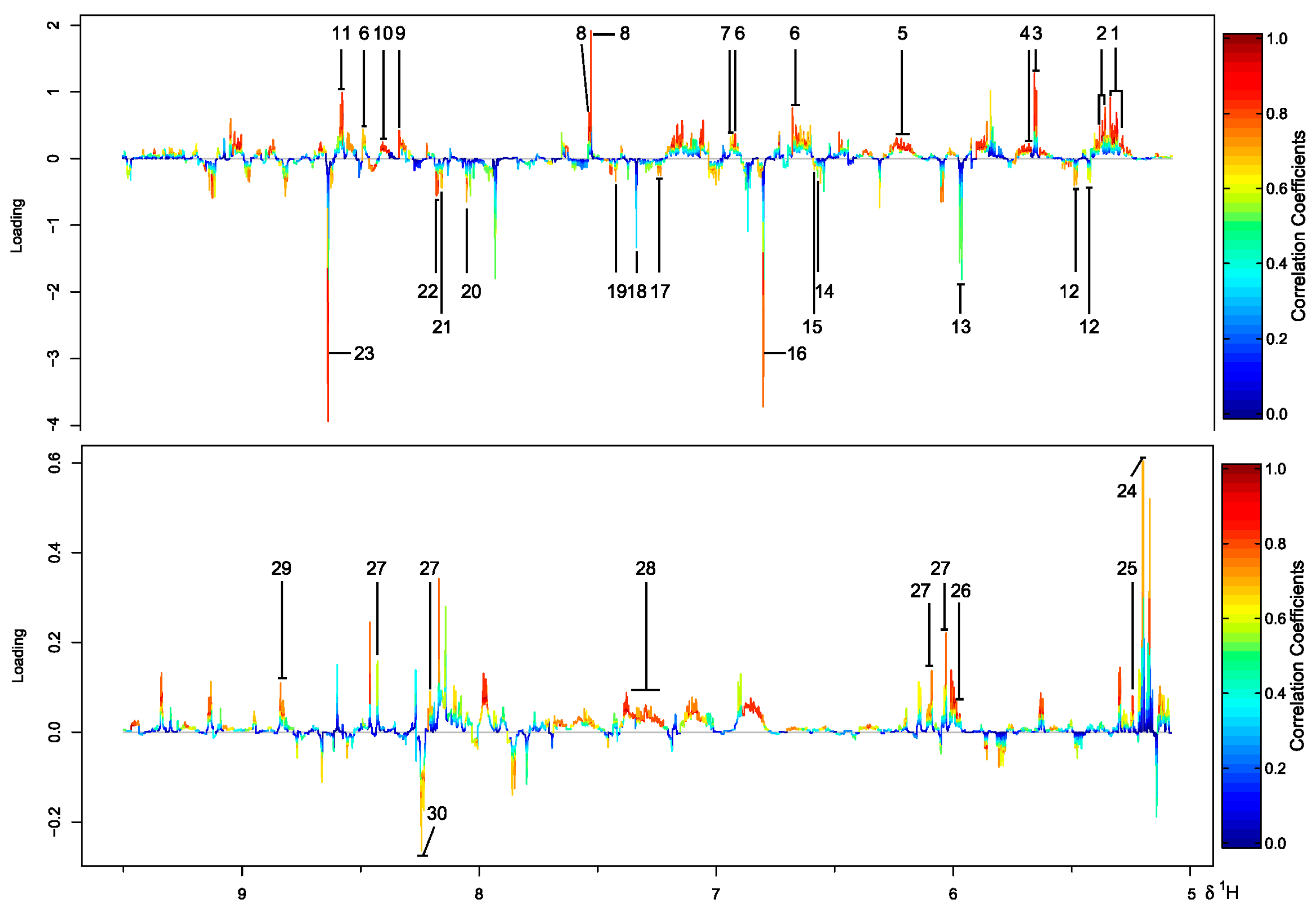
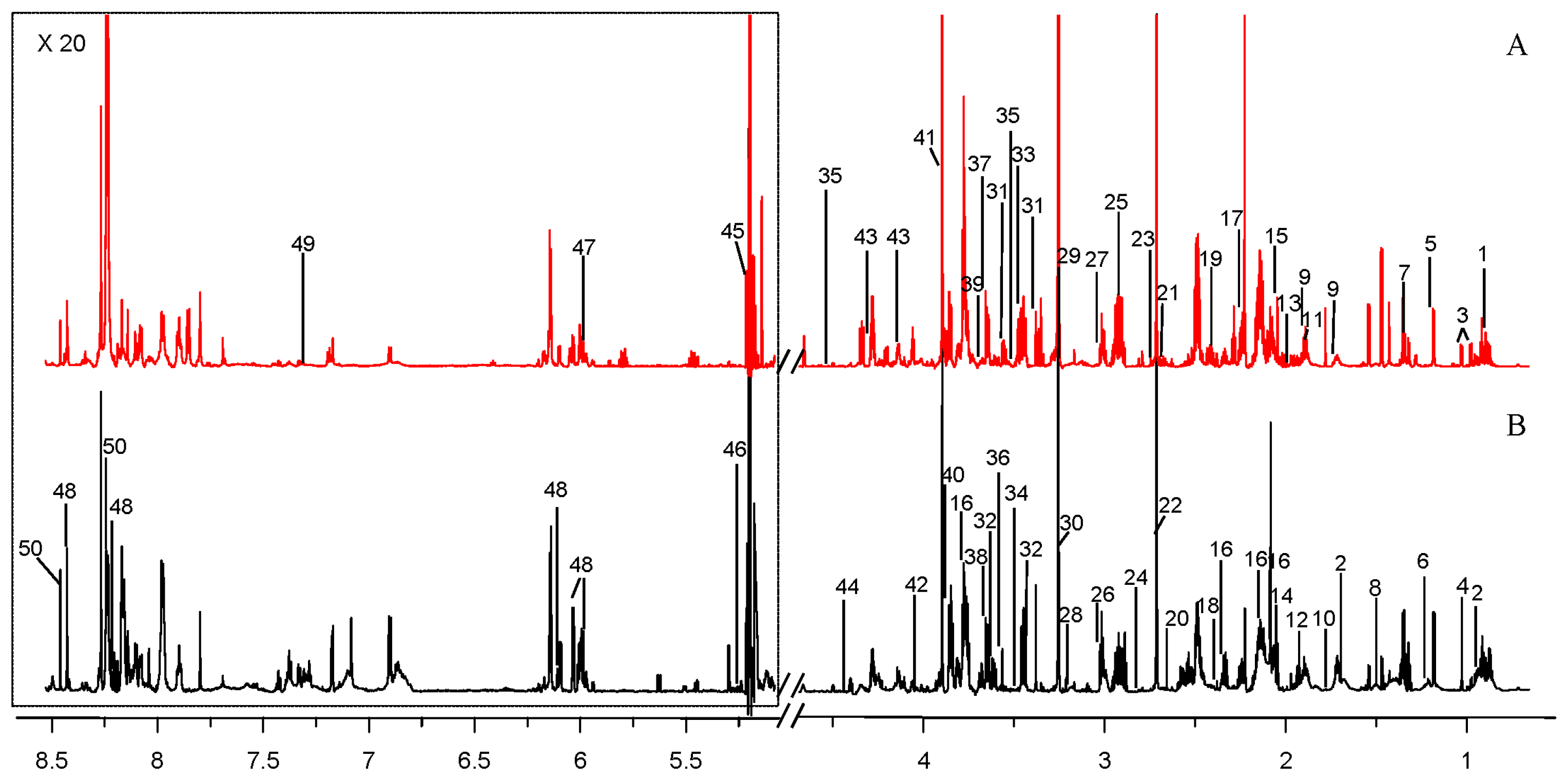
2.1. Comparison of Metabolite Profiles Between Marine and Terrestrial N. flava
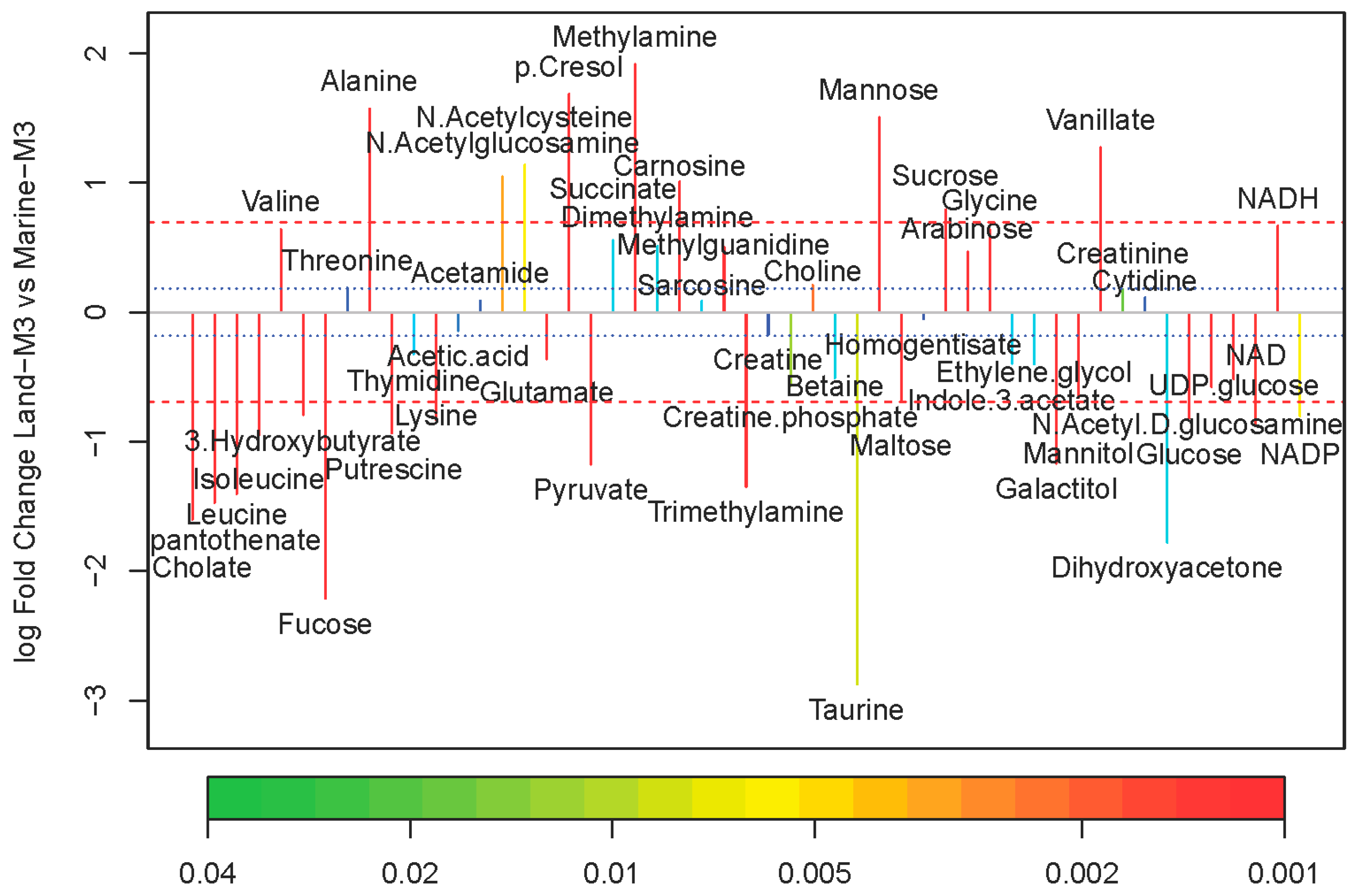
2.2. Comparison of Metabolite Concentrations Between Marine and Terrestrial N. flava
2.3. Inter-Metabolite Correlations for Marine and Terrestrial N. flava
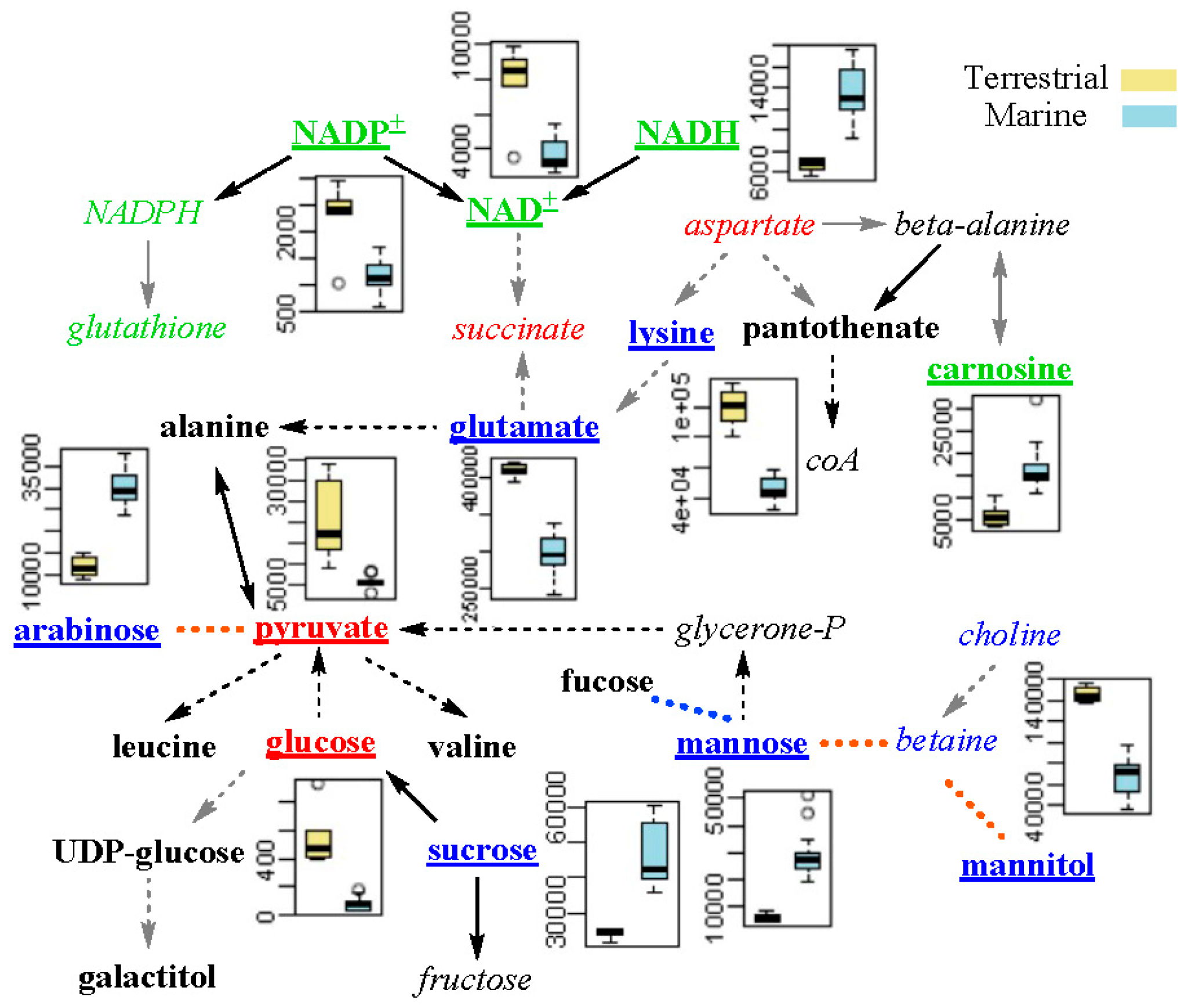
2.4. Differential Metabolic Network Between Marine and Terrestrial N. flava
3. Materials and Methods
3.1. Bacterial Material
3.2. Culture Media
3.3. Bacteria Culture
3.4. Extraction of Metabolites
3.5. 1H Nuclear Magnetic Resonance (NMR) Analysis of Samples
3.6. Data Processing, Bioinformatics, and Statistical Analyses
3.7. Identification of Intracellular Metabolites
3.8. Univariate Analysis
3.9. Correlation Network Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Marine actinobacterial metabolites: Current status and future perspectives. Microbiol. Res. 2013, 168, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Kang, K.H.; Sivakumar, K.; Li-Chan, E.C.; Oh, H.M.; Kim, S.K. Marine actinobacteria: An important source of bioactive natural products. Environ. Toxicol. Pharmacol. 2014, 38, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.; Hamann, M.T. Marine natural products and their potential applications as anti-infective agents. Lancet Infect. Dis. 2003, 3, 338–348. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.E.; Pond, C.D.; Pierce, E.; Harmer, Z.P.; Kwan, J.; Zachariah, M.M.; Harper, M.K.; Wyche, T.P.; Matainaho, T.K.; Bugni, T.S.; et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol. 2018, 14, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.A.; Rutherford, L.T.; Hodson, R.E. Evidence for indigenous Streptomyes populations in a marine environment determined with a 16S rRNA probe. Appl. Environ. Microbiol. 1995, 61, 3695–3700. [Google Scholar] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.G.; Oh, J.; Keasling, A.; Colson, K.L.; Hamann, M.T. The utility of metabolomics in natural product and biomarker characterization. Biochim. Biophys. Acta 2014, 1840, 3460–3474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña, A.; Valens, M.; Santos, F.; Buczolits, S.; Antón, J.; Kämpfer, P.; Busse, H.J.; Amann, R.; Rosselló-Mora, R. Intraspecific comparative analysis of the species Salinibacter ruber. Extremophiles 2005, 9, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Dastager, S.G.; Krishnamurthi, S.; Rameshkumar, N.; Dharne, M. The Family Micrococcaceae. In The Prokaryotes–Actinobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 455–498. [Google Scholar]
- Ramawat, K.G. Halophiles, Biodiversity and Sustainable Exploitation. In Sustainable Development and Biodiversity; Maheshwari, D.K., Saraf, M., Eds.; Springer: Cham, Switzerland, 2015; Volume 6, p. 12. [Google Scholar]
- Beales, N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: A review. Compr. Rev. Food Sci. Food Saf. 2004, 3, 1–20. [Google Scholar] [CrossRef]
- Xie, Y.; Chou, L.S.; Cutler, A.; Weimer, B. DNA macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 2004, 70, 6738–6747. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Rodrigues, L.N.; de Almeida Brito, W.; Parente, A.F.A.; Weber, S.S.; Bailão, A.M.; Casaletti, L.; Borges, C.L.; de Almeida Soares, C.M. Osmotic stress adaptation of Paracoccidioides lutzii, Pb01, monitored by proteomics. Fungal Genet. Biol. 2016, 95, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Musa, H.; Kasim, F.H.; Nagoor Gunny, A.A.; Gopinath, S.C.B. Salt-adapted moulds and yeasts: Potentials in industrial and environmental biotechnology. Process Biochem. 2018, 69, 33–44. [Google Scholar] [CrossRef]
- Kimura, K.; Morimatsu, K.; Inaoka, T.; Yamamoto, K. Injury and recovery of Escherichia coli ATCC25922 cells treated by high hydrostatic pressure at 400–600 MPa. J. Biosci. Bioeng. 2017, 123, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress alters the physiological responses and improves the biofuel potential of green microalgae Acutodesmus dimorphus. Bioresour. Technol. 2017, 244, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Kwon, H.Y.; Kwon, O.B.; Kang, J.H. Hydrogen peroxide-mediated Cu, Zn-superoxide dismutase fragmentation: Protection by carnosine, homocarnosine and anserine. Biochim. Biophys. Acta 1999, 1472, 651–657. [Google Scholar] [CrossRef]
- Sindhi, V.; Gupta, V.; Sharma, K.; Bhatnagar, S.; Kumari, R.; Dhaka, N. Potential applications of antioxidants—A review. J. Pharm. Res. 2013, 7, 828–835. [Google Scholar] [CrossRef]
- Bianchi, A.; Van Wambeke, F.; Garcin, J. Bacterial utilization of glucose in the water column from eutrophic to oligotrophic pelagic areas in the eastern North Atlantic Ocean. J. Mar. Syst. 1998, 14, 45–55. [Google Scholar] [CrossRef]
- Luo, H.Y.; Miao, L.H.; Fang, C.; Yang, P.L.; Wang, Y.R.; Shi, P.J.; Yao, B.; Fan, Y.L. Nesterenkonia flava sp. nov., isolated from papermill effluent. Int. J. Syst. Evol. Microbiol. 2008, 58, 1927–1930. [Google Scholar] [CrossRef] [PubMed]
- Dai, X. Diversity of Cultivated Bacteria in the Eastern Pacific Nodule Area and the Taxonomic Analysis of Three Novel Marine Bacteria. Master’s Thesis, Ocean University of China, Qingdao, China, May 2015. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing on JSTOR. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, C.-L.; Xia, J.-M.; Wang, J.-S.; Lin, D.-H.; Yang, X.-W. Metabolomic Investigations on Nesterenkonia flava Revealed Significant Differences between Marine and Terrestrial Actinomycetes. Mar. Drugs 2018, 16, 356. https://doi.org/10.3390/md16100356
Xie C-L, Xia J-M, Wang J-S, Lin D-H, Yang X-W. Metabolomic Investigations on Nesterenkonia flava Revealed Significant Differences between Marine and Terrestrial Actinomycetes. Marine Drugs. 2018; 16(10):356. https://doi.org/10.3390/md16100356
Chicago/Turabian StyleXie, Chun-Lan, Jin-Mei Xia, Jun-Song Wang, Dong-Hai Lin, and Xian-Wen Yang. 2018. "Metabolomic Investigations on Nesterenkonia flava Revealed Significant Differences between Marine and Terrestrial Actinomycetes" Marine Drugs 16, no. 10: 356. https://doi.org/10.3390/md16100356
APA StyleXie, C.-L., Xia, J.-M., Wang, J.-S., Lin, D.-H., & Yang, X.-W. (2018). Metabolomic Investigations on Nesterenkonia flava Revealed Significant Differences between Marine and Terrestrial Actinomycetes. Marine Drugs, 16(10), 356. https://doi.org/10.3390/md16100356