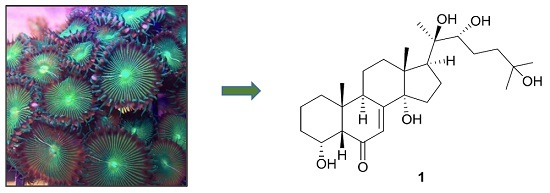
Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki
Abstract
:1. Introduction
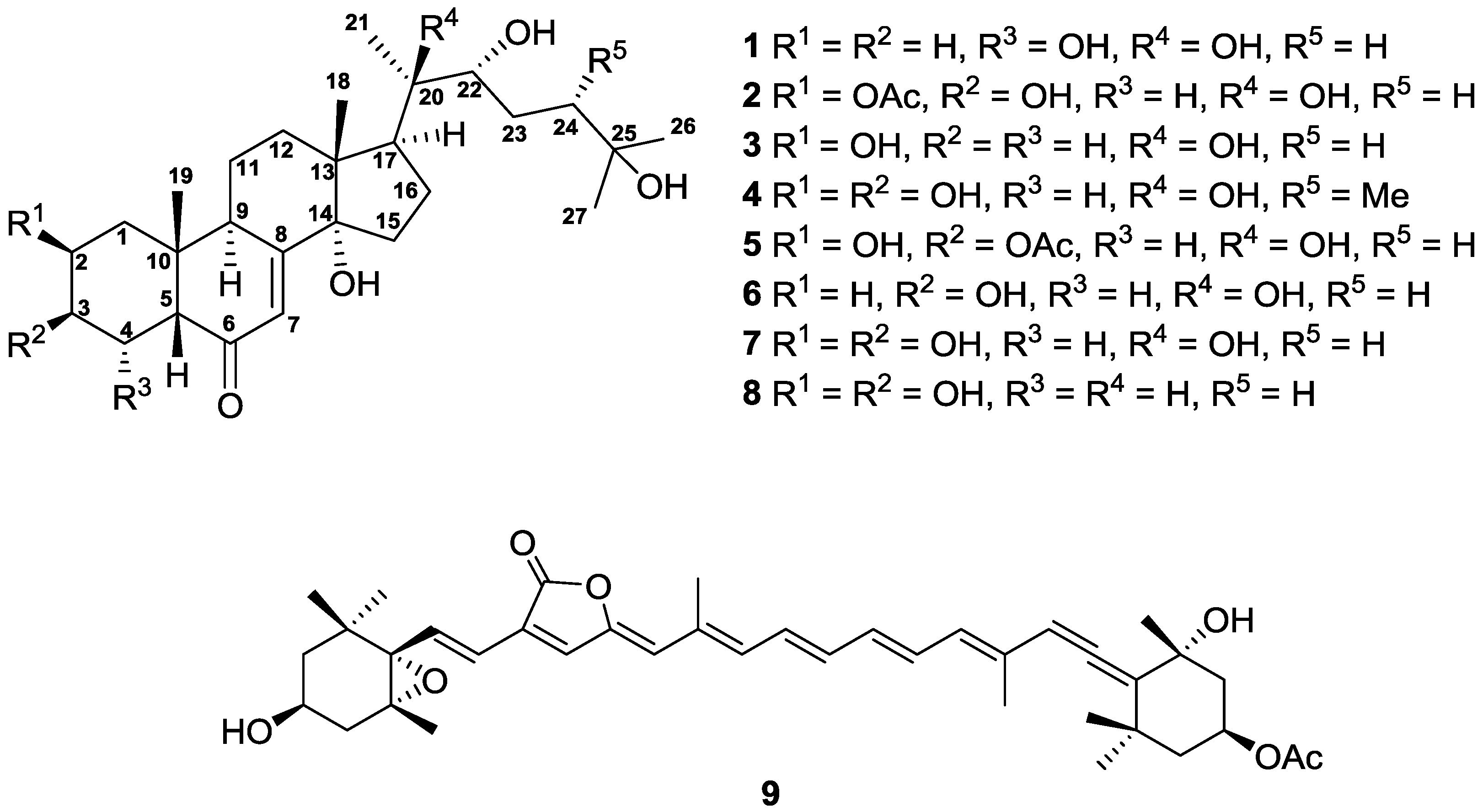
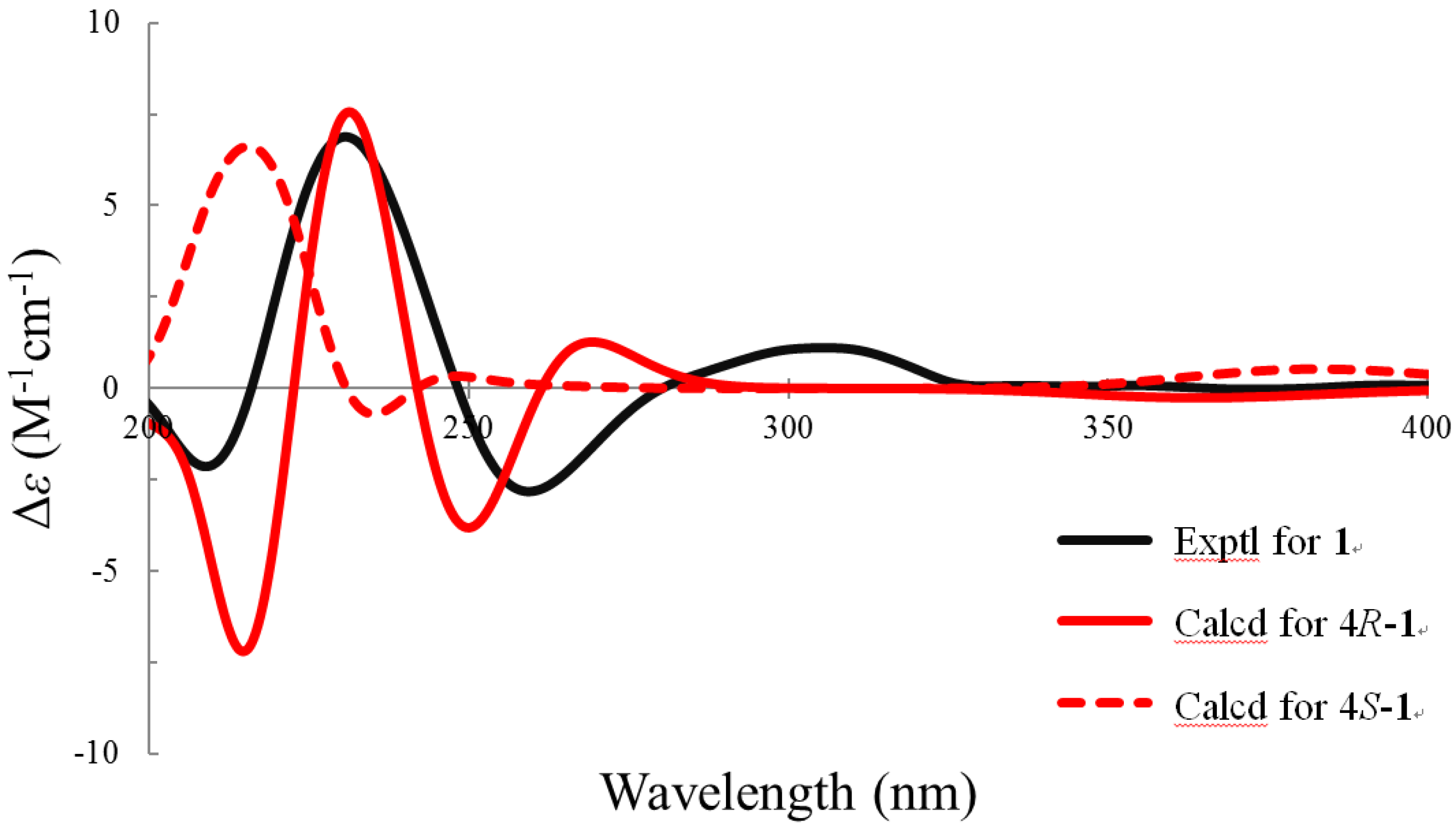
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Species Identification
3.4. Extraction and Isolation
3.5. ECD Calculations
3.6. Anti-DENV Activity Assay
3.7. Evaluation of Anti-DENV RdRp and Protease Activity
3.8. Cytotoxicity Assay
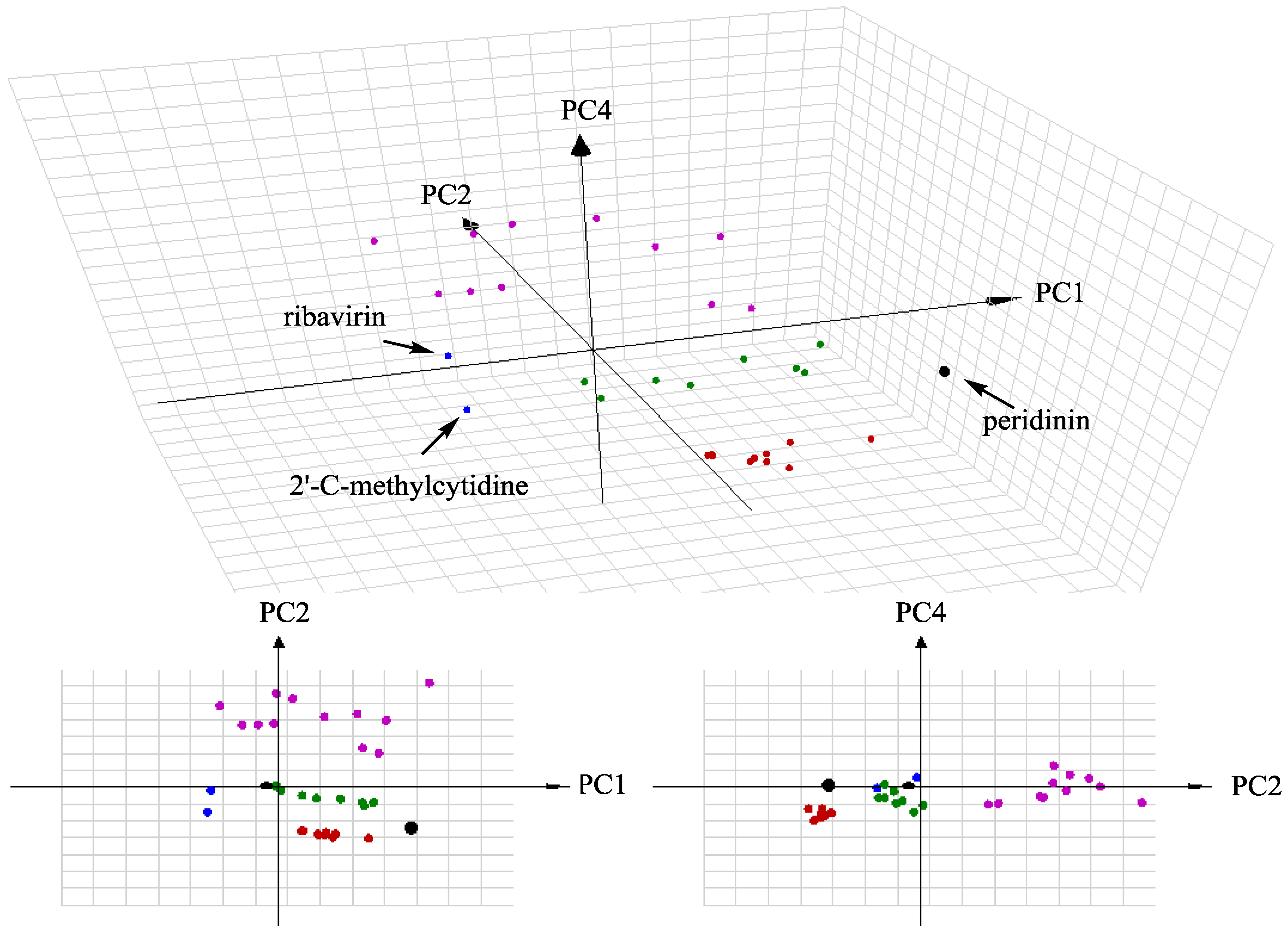
3.9. ChemGPS–NP Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Inuzuka, T.; Uemura, D.; Arimoto, H. The conformational features of palytoxin in aqueous solution. Tetrahedron 2008, 64, 7718–7723. [Google Scholar] [CrossRef]
- Takano, S.; Uemura, D.; Hirata, Y. Isolation and structure of a new amino acid, palythine, from the zoanthid Palythoa tuberculosa. Tetrahedron Lett. 1978, 26, 2299–2300. [Google Scholar] [CrossRef]
- Takano, S.; Uemura, D.; Hirata, Y. Isolation and structure of two new amino acids, palythinol and palythenr, from the zoanthid Palythoa tuberculosa. Tetrahedron Lett. 1978, 49, 4909–4912. [Google Scholar] [CrossRef]
- Diop, M.; Leung-Tack, D.; Braekman, J.-C.; Kornprobst, J.-M. Composition en Stérols de Quatre Zoanthaires du Genre Palythoa de la Presqu-île du Cap-Vert. Biochem. Syst. Ecol. 1986, 14, 151–154. [Google Scholar] [CrossRef]
- Fedorov, S.N.; Stonik, V.A.; Elyakov, G.B. Identification of ecdysteroids of hexactinic corals. Chem. Nat. Compd. 1988, 24, 517–518. [Google Scholar] [CrossRef]
- Han, C.; Qi, J.; Shi, X.; Sakagami, Y.; Shibata, T.; Uchida, K.; Ojika, M. Prostaglandins from a zoanthid paclitaxel-like neurite-degenerating and microtubule-stabilizating activities. Biosci. Biotechnol. Biochem. 2006, 70, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.G.; Maia, A.I.; Wilke, D.V.; Silveira, E.R.; Braz-Filho, R.; La Clair, J.J.; Costa-Lotufo, L.V.; Pessoa, O.D. Palyosulfonoceramides A and B: Unique sulfonylated ceramides from the Brazilian zoanthids Palythoa caribaeorum and Protopalythoa variabilis. Mar. Drugs 2012, 10, 2846–2860. [Google Scholar] [CrossRef] [PubMed]
- Elbagory, A.M.; Meyer, M.; Ali, A.H.; Ameer, F.; Parker-Nance, S.; Benito, M.T.; Doyagüez, E.G.; Jimeno, M.L.; Hussein, A.A. New polyhydroxylated sterols from Palythoa tuberculosa and their apoptotic activity in cancer cells. Steroids 2015, 101, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-B.; Lee, J.-C.; Lo, I.-W.; Chen, S.-R.; Hu, H.-C.; Wu, Y.-H.; Wu, Y.-C.; Chang, F.-R. Ecdysones from Zoanthus spp. with inhibitory activity against dengue virus 2. Bioorg. Med. Chem. Lett. 2016, 26, 2344–2348. [Google Scholar] [CrossRef] [PubMed]
- Suksamrarn, A.; Pattanaprateep, P. Selective acetylation of 20-hydroxyecdysone partial synthesis of some minor ecdysteroids and analogues. Tetrahedron 1995, 51, 10633–10650. [Google Scholar] [CrossRef]
- Suksamrarn, A.; Charoensuk, S.; Yingyongnarongkul, B. Synthesis and biological activity of 3-deoxyecdysteroid analogues. Tetrahedron 1996, 52, 10673–10684. [Google Scholar] [CrossRef]
- Zhu, N.; Kikuzaki, H.; Vastano, B.C.; Nakatani, N.; Karwe, M.V.; Rosen, R.T.; Ho, C.-T. Ecdysteroids of quinoa seeds (Chenopodium quinoa Willd.). J. Agric. Food Chem. 2001, 49, 2576–2578. [Google Scholar] [CrossRef] [PubMed]
- Mamadalieva, N.Z.; Saatov, Z.; Kachala, V.V.; Shashkov, A.S. Phytoecdysteroids of plants of the Silene genus. 2-deoxyecdysterone-25-aceate from Silene wallichiana. Chem. Nat. Compd. 2002, 38, 179–181. [Google Scholar] [CrossRef]
- Gvazava, L.N.; Kukoladze, V.S. Phytoecdysteroids from Digitalis ciliate and D. purpurea leaves. Chem. Nat. Compd. 2010, 46, 146–147. [Google Scholar] [CrossRef]
- Mamadalieva, N.Z.; Zibareva, L.N.; Saatov, Z. Phytoecdysteroids of Silene linicola. Chem. Nat. Compd. 2002, 38, 268–271. [Google Scholar] [CrossRef]
- Shaaban, M.; Shaaban, K.A.; Ghani, M.A. Hurgadacin: A new steroid from Sinularia polydactyla. Steroids 2013, 78, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Okuzumi, K.; Hara, N.; Uekusa, H.; Fujimoto, Y. Structure elucidation of cyasterone stereoisomers isolated from Cyathula officinalis. Org. Biomol. Chem. 2005, 3, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Tseng, C.-K.; Wu, Y.-H.; Kaushik-Basu, N.; Lin, C.-K.; Chen, W.-C.; Wu, H.-N. Characterization of the activity of 2′-C-methylcytidine against dengue virus replication. Antiviral Res. 2015, 116, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zandi, K.; Lim, T.-H.; Rahim, N.-A.; Shu, M.-H.; Teoh, B.-T.; Sam, S.-S.; Danlami, M.-B.; Tan, K.-K.; Abubakar, S. Extract of Scutellaria baicalensis inhibits dengue virus replication. BMC Complement. Altern. Med. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Zandi, K.; Teoh, B.-T.; Sam, S.-S.; Wong, P.-F.; Mustafa, M.R.; Abubakar, S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011, 8. [Google Scholar] [CrossRef] [PubMed]
- Brandão, G.C.; Kroon, E.G.; Souza, D.E.R.; Filho, J.D.S.; Oliveira, A.B. Chemistry and Antiviral Activity of Arrabidaea pulchra (Bignoniaceae). Molecules 2013, 18, 9919–9932. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.; Gottfries, J.; Muresan, S.; Backlund, A. ChemGPS–NP: Tuned for navigation in biologically relevant chemical space. J. Nat. Prod. 2007, 70, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Rosén, J.; Lövgren, A.; Kogej, T.; Muresan, S.; Gottfries, J.; Backlund, A. ChemGPS–NPWeb: Chemical space navigation online. J. Comput. Aided Mol. Des. 2009, 23, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.C.; Kofron, J.L.; Traphagen, L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002, 45, 4350–4358. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-B.; Chien, Y.-T.; Lee, J.-C.; Tseng, C.-K.; Wang, H.-C.; Lo, I.-W.; Wu, Y.-H.; Wang, S.-Y.; Wu, Y.-C.; Chang, F.-R. Limonoids from the seeds of Swietenia macrophylla with inhibitory activity against dengue virus 2. J. Nat. Prod. 2014, 77, 2367–2374. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-H.; Lin, C.-K.; Chen, Y.-L.; Hsu, C.-Y.; Wu, H.-N.; Tseng, C.-K.; Lee, J.-C. Synthesis, antiproliferative and anti-dengue virus evaluations of 2-aroyl-3-arylquinoline derivatives. Eur. J. Med. Chem. 2014, 79, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B–NS3 protease–helicase as a target for antiviral drug development. Antiviral Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
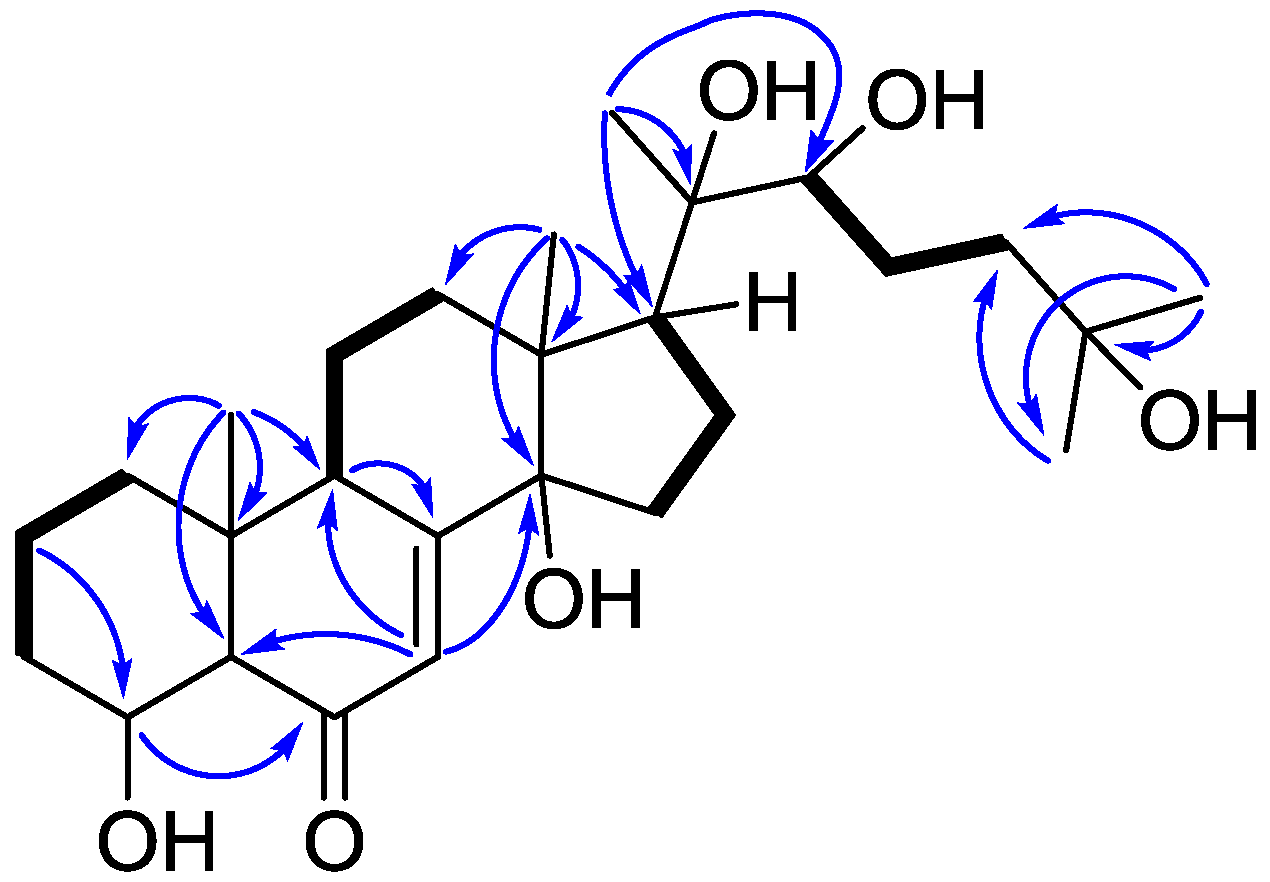
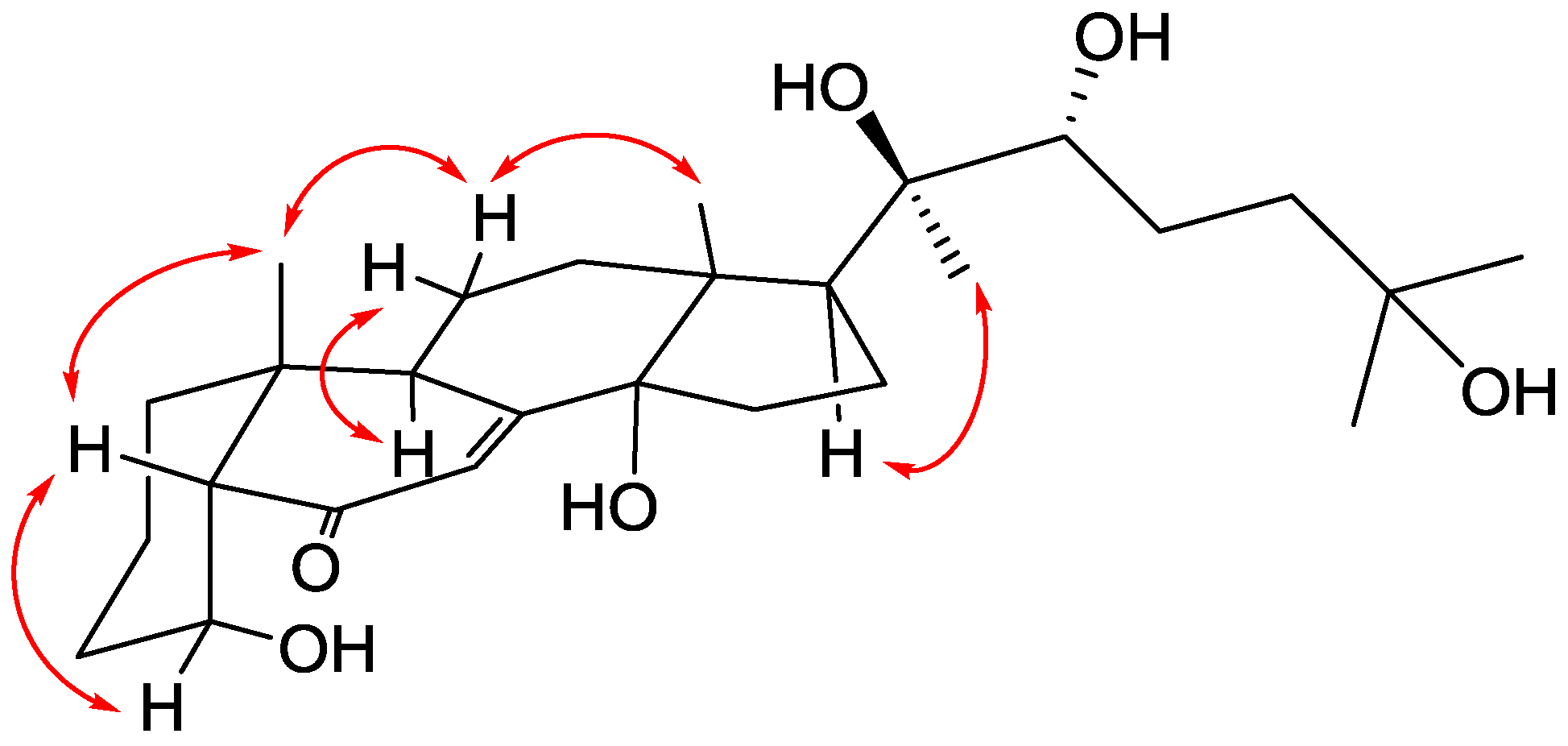
| Position | δH, Mult (J in Hz) | δC, Type | HMBC (1H–13C) |
|---|---|---|---|
| 1 | 1.88, m | 34.3, CH2 | |
| 1.11, m | |||
| 2 | 2.08, m | 35.7, CH2 | 4, 10 |
| 1.81, m | |||
| 3 | 2.17, m | 31.7, CH2 | |
| 1.92, dd (9.4, 2.7) | |||
| 4 | 3.85, m | 69.2, CH | 6 |
| 5 | 2.32, dd (13.3, 4.0) | 57.3, CH | 6 |
| 6 | 202.0, C | ||
| 7 | 6.25, d (2.8) | 121.4, CH | 5, 9, 14 |
| 8 | 166.3, C | ||
| 9 | 3.72, ddd (12.8, 6.0, 2.8) | 34.0, CH | 8, 11 |
| 10 | 36.8, C | ||
| 11α | 1.80, m | 20.9, CH2 | |
| 11β | 1.70, m | ||
| 12 | 2.60, td (12.8, 4.7) | 32.0, CH2 | 9, 11, 13, 14, 18 |
| 2.06, m | 11, 13, 18 | ||
| 13 | 48.2, C | ||
| 14 | 84.2, C | ||
| 15 | 2.00, m | 31.7, CH2 | 14 |
| 1.85, m | |||
| 16 | 2.48, m | 21.5, CH2 | |
| 2.10, m | |||
| 17 | 3.03, t (9.5) | 50.1, CH | |
| 18 | 1.25, s | 17.9, CH3 | 12, 13, 14, 17 |
| 19 | 1.01, s | 23.9, CH3 | 1, 5, 9, 10 |
| 20 | 76.9, C | ||
| 21 | 1.61, s | 21.7, CH3 | 17, 20, 22 |
| 22 | 3.90, dd (9.8, 3.3) | 77.6, CH | |
| 23 | 2.14, m | 27.5, CH2 | |
| 1.86, m | |||
| 24 | 2.28, dd (11.8, 3.4) | 42.7, CH2 | |
| 1.85, m | 25, 26, 27 | ||
| 25 | 69.6, C | ||
| 26 | 1.40, s | 30.1, CH3 | 24, 25, 27 |
| 27 | 1.40, s | 30.0, CH3 | 24, 25, 26 |
| Compound | EC50 (μM) a | CC50 (μM) b | SI c |
|---|---|---|---|
| 1 | 54.01 ± 2.17 | NT d | – |
| 2 | >100 | >200 | – |
| 3 | >100 | >200 | – |
| 4 | 46.45 ± 0.59 | NT d | – |
| 9 | 4.50 ± 0.46 | 132.55 ± 3.21 | >29.5 |
| 2′CMC e | 13.23 ± 0.07 | 94.8 ± 4.15 | >7.2 |
| EC50 (μM) a | |||
|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 |
| 7.62 ± 0.17 | 4.50 ± 0.46 | 5.84 ± 0.19 | 6.51 ± 0.30 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-C.; Chang, F.-R.; Chen, S.-R.; Wu, Y.-H.; Hu, H.-C.; Wu, Y.-C.; Backlund, A.; Cheng, Y.-B. Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki. Mar. Drugs 2016, 14, 151. https://doi.org/10.3390/md14080151
Lee J-C, Chang F-R, Chen S-R, Wu Y-H, Hu H-C, Wu Y-C, Backlund A, Cheng Y-B. Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki. Marine Drugs. 2016; 14(8):151. https://doi.org/10.3390/md14080151
Chicago/Turabian StyleLee, Jin-Ching, Fang-Rong Chang, Shu-Rong Chen, Yu-Hsuan Wu, Hao-Chun Hu, Yang-Chang Wu, Anders Backlund, and Yuan-Bin Cheng. 2016. "Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki" Marine Drugs 14, no. 8: 151. https://doi.org/10.3390/md14080151
APA StyleLee, J.-C., Chang, F.-R., Chen, S.-R., Wu, Y.-H., Hu, H.-C., Wu, Y.-C., Backlund, A., & Cheng, Y.-B. (2016). Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki. Marine Drugs, 14(8), 151. https://doi.org/10.3390/md14080151