Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan
Abstract
:1. Introduction
2. Results and Discussion
2.1. Venom Fractionation and Purification
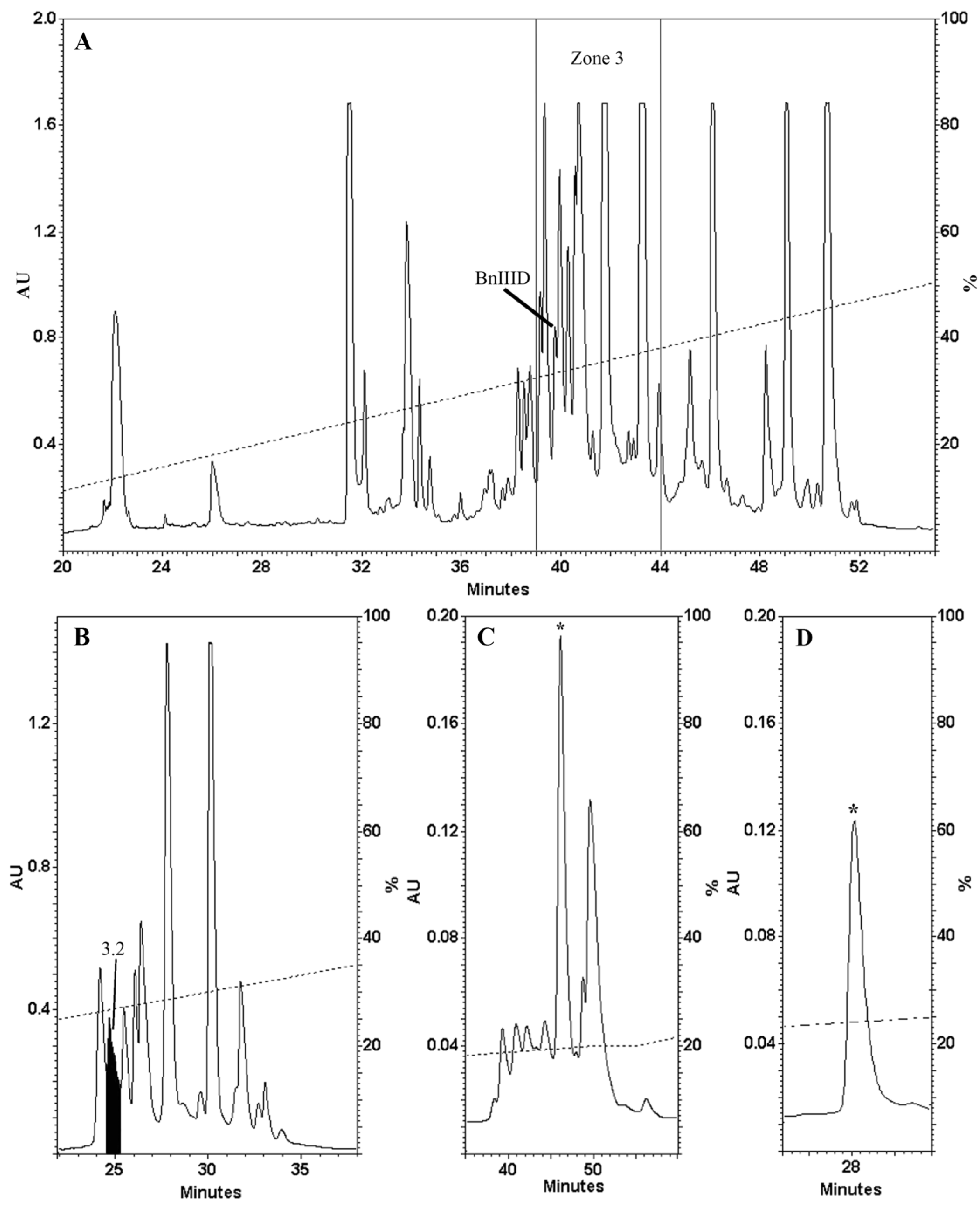
2.2. Determination of the Number of Disulphide Bonds

2.3. Presence of Gamma-Carboxylate Glutamate Residue
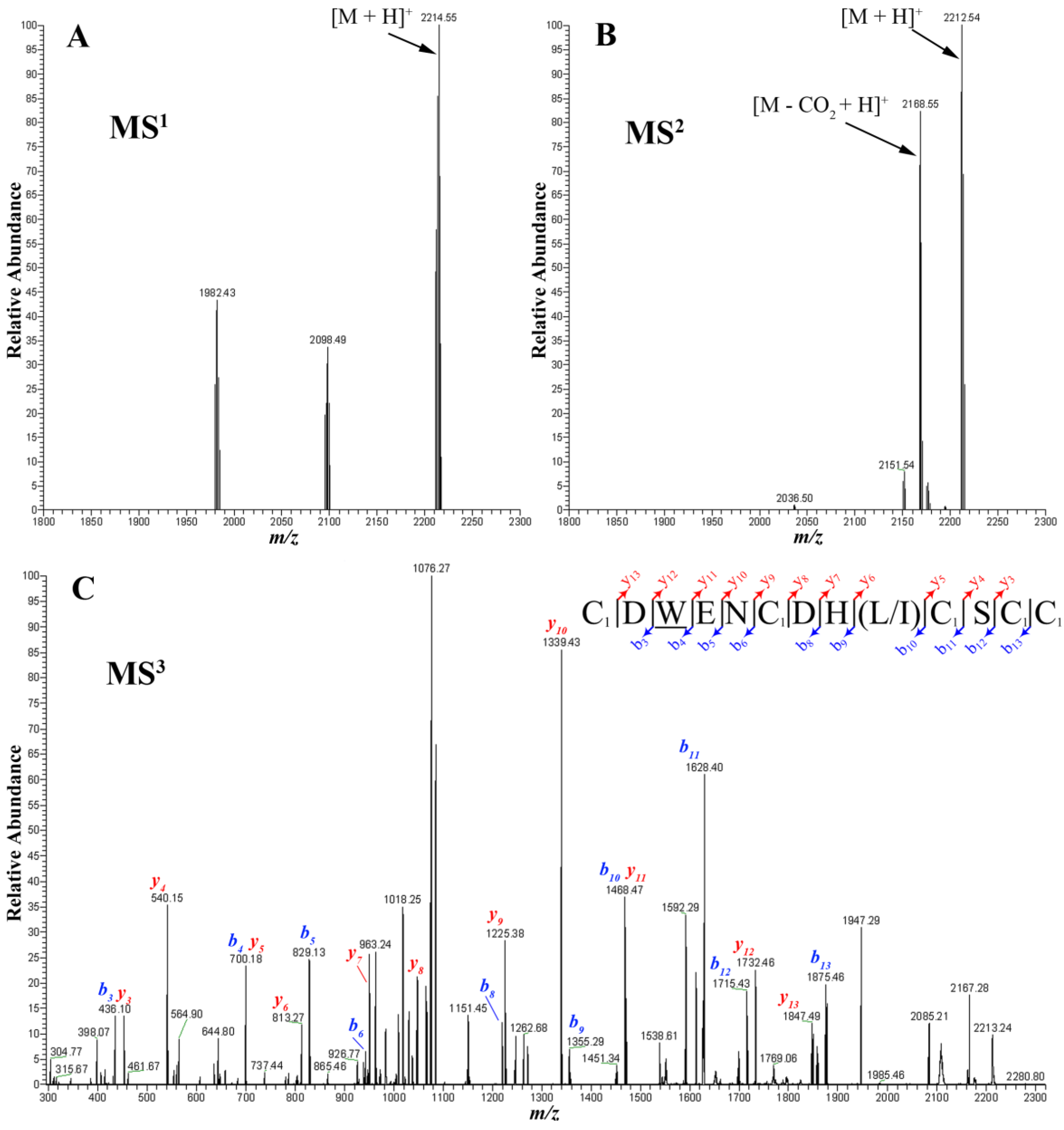
2.4. Peptide Sequencing
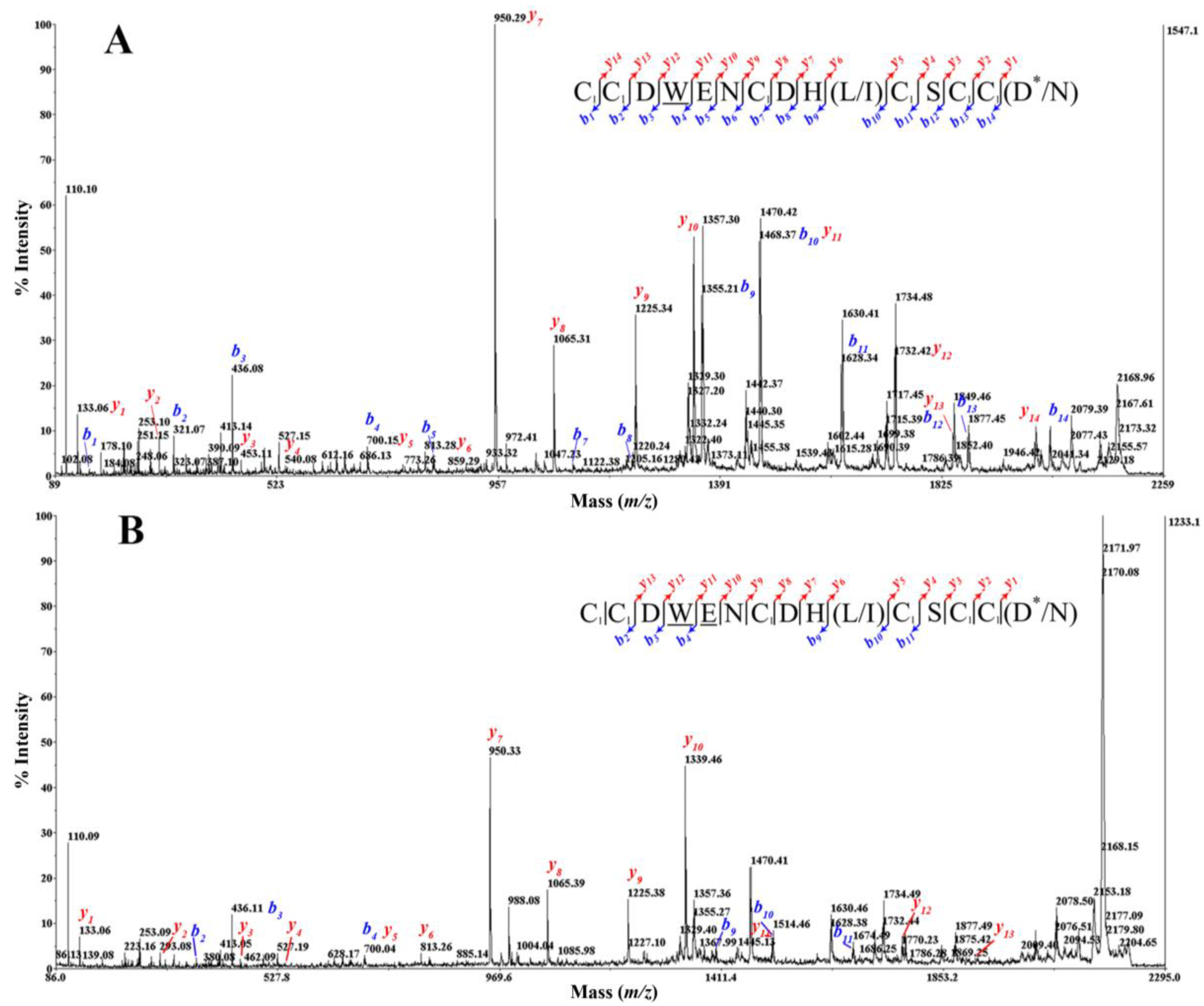
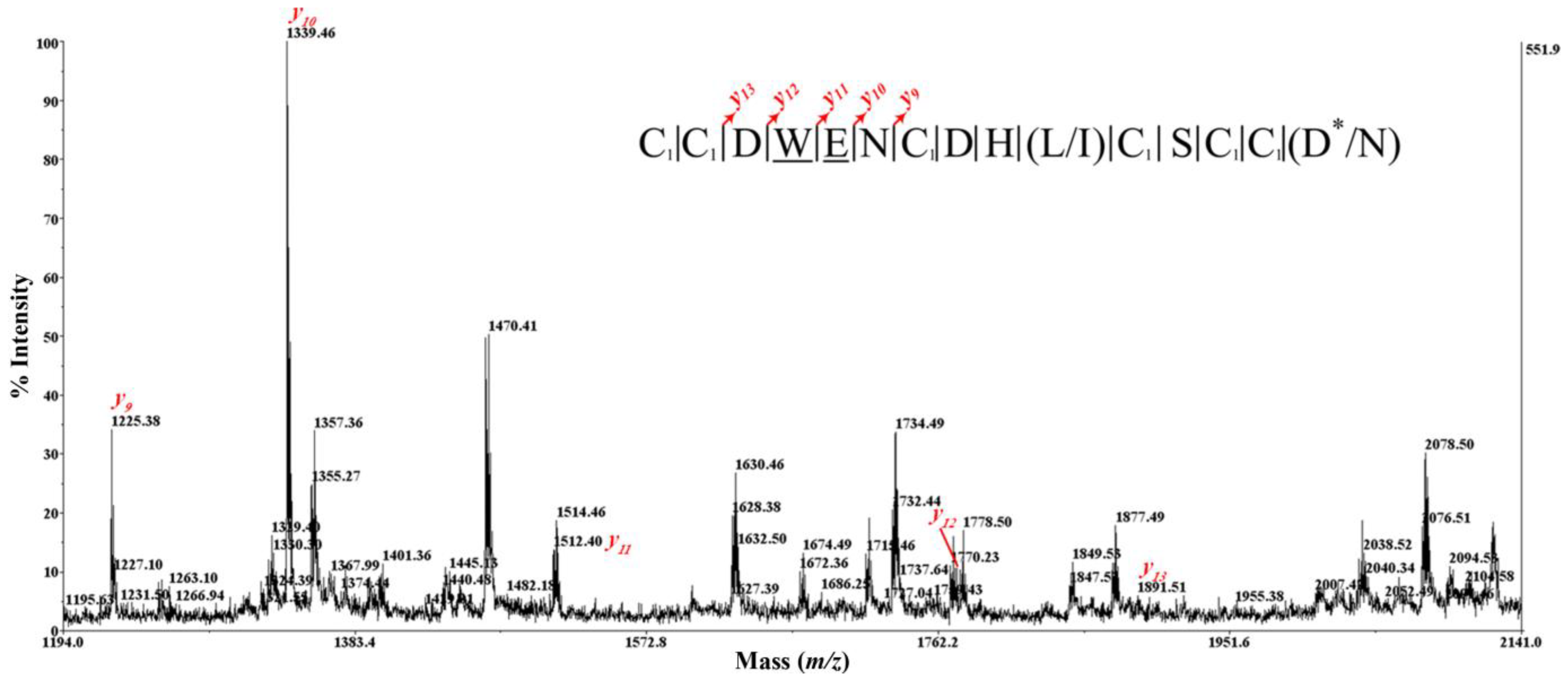
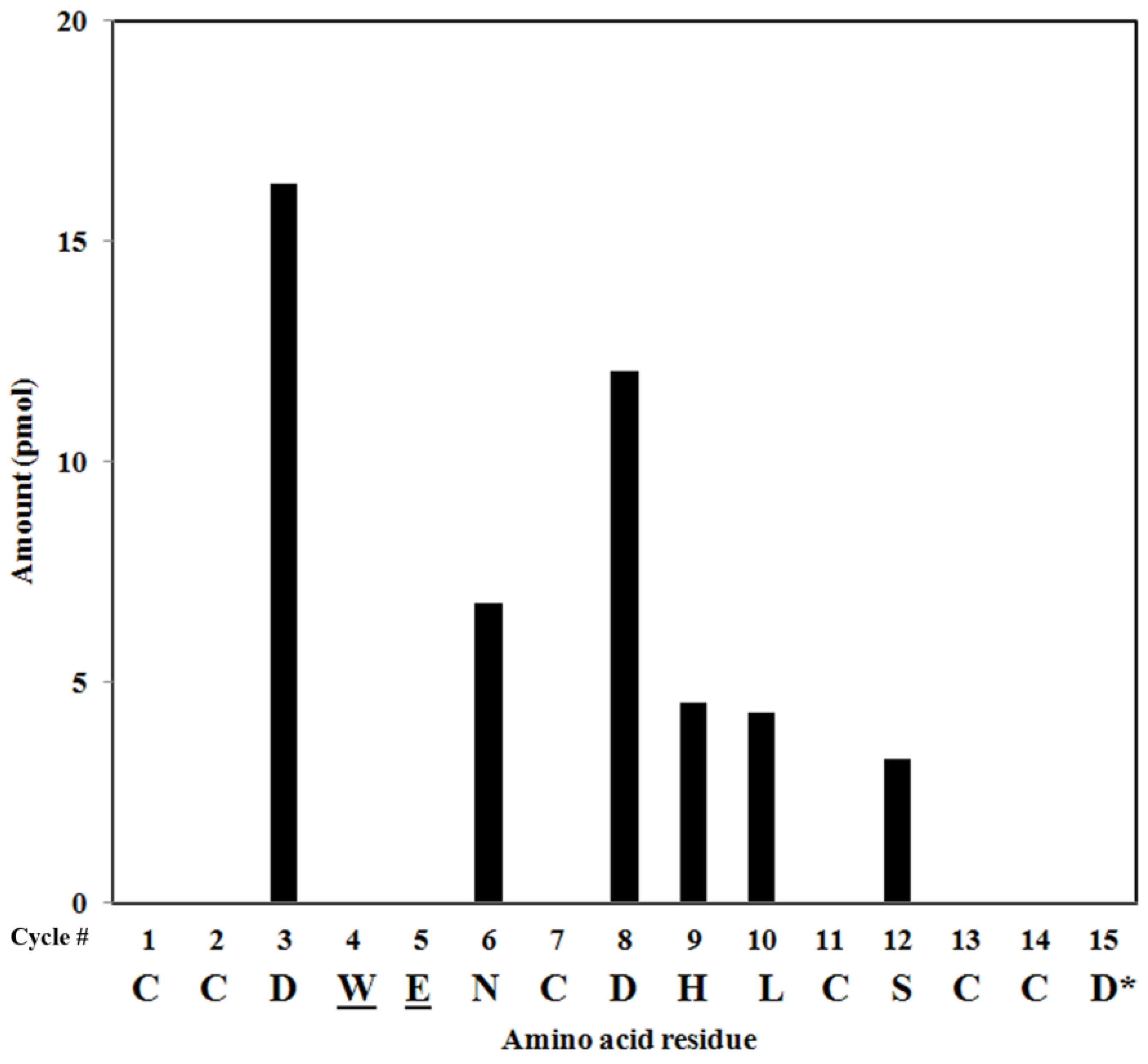
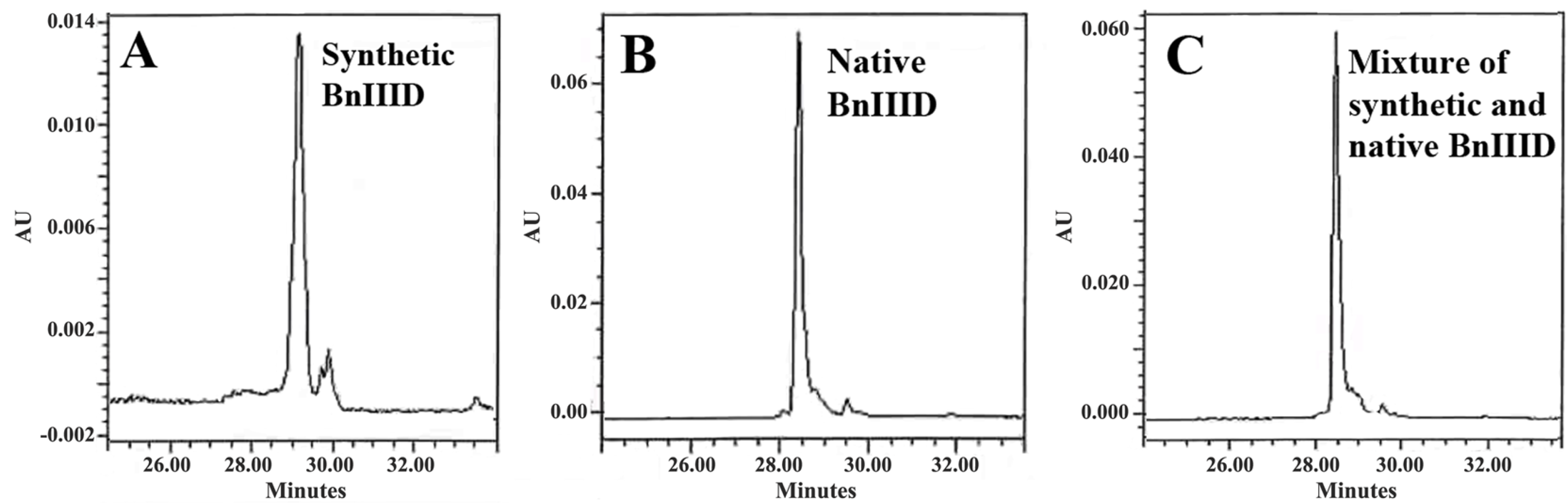
2.5. Peptide Synthesis
2.6. Sequence Similarity Analysis
| Name | Cone snail | Diet | Sequence | Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S3-S01 | C. striatus | p | - | C | C | P | K | E | W | C | N | R | D | C | S | C | C | T | - | [17,21] |
| S3-S02 | C. striatus | p | - | C | C | P | A | R | M | C | M | A | A | C | S | C | C | D | - | [17,21] |
| Ts3.6 | C. tessulatus | v | Q | C | C | D | W | Q | W | C | D | G | A | C | D | C | C | A | - | [22] |
| LtIIID | C. litteratus | v | - | C | C | D | W | E | W | C | D | E | L | C | S | C | C | W | - | [23] |
| Mr3.16 | C. marmoreus | m | V | C | C | S | F | G | S | C | D | S | L | C | Q | C | C | D * | - | [5] |
| MrIIIE | C. marmoreus | m | V | C | C | P | F | G | G | C | H | E | L | C | Y | C | C | D * | - | [18] |
| MrIIIF | C. marmoreus | m | V | C | C | P | F | G | G | C | H | E | L | C | L | C | C | D * | - | [18] |
| Mr3.18 | C. marmoreus | m | - | C | C | H | R | N | W | C | D | H | L | C | S | C | C | G | S | [5] |
| Mr3.8 | C. marmoreus | m | - | C | C | H | W | N | W | C | D | H | L | C | S | C | C | G | S | [18] |
| BnIIID | C. bandanus | m | - | C | C | D | W | E | N | C | D | H | L | C | S | C | C | D * | - | This work |
| Name | Organism | Diet | Sequence | Pharmacology | Reference | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BuIIIA | C. bullatus | p | V | T | D | R | C | C | K | G | K | R | E | - | C | G | R | W | - | - | C | R | D | H | S | R | C | C * | - | - | - | µ-conotoxin | [27] |
| BuIIIB | C. bullatus | p | V | G | E | R | C | C | K | N | G | K | R | G | C | G | R | W | - | - | C | R | D | H | S | R | C | C * | - | - | - | µ-conotoxin | [27] |
| CIIIA | C. catus | p | - | - | G | R | C | C | E | G | P | N | G | - | C | S | S | R | W | - | C | K | D | H | A | R | C | C * | - | - | - | µ-conotoxin | [28] |
| CnIIIA | C. consors | p | - | - | G | R | C | C | D | V | P | N | A | - | C | S | G | R | W | - | C | R | D | H | A | Q | C | C * | - | - | - | µ-conotoxin | [28] |
| CnIIIB | C. consors | p | - | - | Z | G | C | C | G | E | P | N | L | - | C | F | T | R | W | - | C | R | N | N | A | R | C | C | R | Q | Q | µ-conotoxin | [28] |
| GIIIA | C. geographus | p | - | - | R | D | C | C | T | O | O | K | K | - | C | K | D | R | Q | - | C | K | O | Q | R | - | C | C | A* | - | µ-conotoxin | [29] | |
| GIIIB | C. geographus | p | - | - | R | D | C | C | T | O | O | R | K | - | C | K | D | R | R | - | C | K | O | M | K | - | C | C | A* | - | µ-conotoxin | [30] | |
| GIIIC | C. geographus | p | - | - | R | D | C | C | T | O | O | K | K | - | C | K | D | R | R | - | C | K | O | L | K | - | C | C | A* | - | µ-conotoxin | [31] | |
| KIIIA | C. kinoshitai | p | - | - | - | - | C | C | N | - | - | - | - | - | C | S | S | K | W | - | C | R | D | H | S | R | C | C * | - | - | - | µ-conotoxin | [32] |
| LtIIIA | C. litteratus | v | - | - | D | E | C | C | E | O | Q | W | - | - | C | D | G | A | - | - | C | D | - | - | - | - | C | C | S | - | - | ι-conotoxin | [26] |
| MIIIA | C. magus | p | - | - | Z | G | C | C | N | V | P | N | G | - | C | S | G | R | W | - | C | R | D | H | A | Q | C | C * | - | - | - | µ-conotoxin | [33] |
| PIIIA | C. purpurascens | p | - | Z | R | L | C | C | G | F | O | K | S | - | C | R | S | R | Q | - | C | K | O | H | R | - | C | C * | - | - | - | µ-conotoxin | [34] |
| PIIIE | C. purpurascens | p | - | H | O | O | C | C | L | Y | G | K | - | - | C | R | R | Y | O | G | C | S | S | A | S | - | C | C | Q | R * | ψ-conotoxin | [35] | |
| PIIIF | C. purpurascens | p | - | G | O | O | C | C | L | Y | G | S | - | - | C | R | O | F | O | G | C | Y | N | A | L | - | C | C | R | K * | ψ-conotoxin | [36] | |
| PrIIIE | C. parius | p | - | A | A | R | C | C | T | Y | H | G | S | - | C | L | K | E | K | - | C | R | R | K | Y | - | C | C * | - | - | - | ψ-conotoxin | [37] |
| RIIIJ | C. radiatus | p | - | L | O | O | C | C | T | O | O | K | K | H | C | O | A | O | A | - | C | K | Y | K | O | - | C | C | K | S | - | κ-conotoxin | [38] |
| RIIIK | C. radiatus | p | - | L | O | S | C | C | S | L | N | L | R | L | C | O | V | O | A | - | C | K | R | N | O | - | C | C | T * | - | κ-conotoxin | [39] | |
| SIIIA | C. striatus | p | - | - | Z | N | C | C | N | G | G | - | - | - | C | S | S | K | W | - | C | R | D | H | A | R | C | C * | - | - | - | µ-conotoxin | [32] |
| SIIIB | C. striatus | p | - | - | Z | N | C | C | N | G | G | - | - | - | C | S | S | K | W | - | C | K | G | H | A | R | C | C * | - | - | - | µ-conotoxin | [40] |
| SmIIIA | C. stercusmuscarum | p | - | - | Z | R | C | C | N | G | R | R | G | - | C | S | S | R | W | - | C | R | D | H | S | R | C | C | - | - | - | µ-conotoxin | [41] |
| SxIIIA | C. striolatus | p | - | - | - | R | C | C | T | G | K | K | G | S | C | S | G | R | A | - | C | K | N | L | K | - | C | C | A * | - | µ-conotoxin | [42] | |
| SxIIIB | C. striolatus | p | - | - | Z | K | C | C | T | G | K | K | G | S | C | S | G | R | A | - | C | K | N | L | R | - | C | C | A * | - | µ-conotoxin | [42] | |
| TIIIA | C. tulipa | p | - | R | H | G | C | C | K | G | O | K | G | - | C | S | S | R | E | - | C | R | O | Q | H | - | C | C * | - | - | - | µ-conotoxin | [43] |
| BnIIID | C. bandanus | m | - | - | - | - | C | C | D | W | E | N | - | - | C | D | H | L | - | - | C | S | - | - | - | - | C | C | D * | - | unknown | This work | |
3. Experimental Section
3.1. Isolation and Purification of Native Conopeptides
3.2. Reduction-Alkylation Procedures
3.3. Mass Spectrometry Analysis
3.4. Automatic Amino Acid Sequencing
3.5. Chemical Synthesis
4. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Olivera, B.M. Conus peptides: Biodiversity-based discovery and exogenomics. J. Biol. Chem. 2006, 281, 31173–31177. [Google Scholar]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- Favreau, P.; Stocklin, R. Marine snail venoms: Use and trends in receptor and channel neuropharmacology. Curr. Opin. Pharmacol. 2009, 9, 594–601. [Google Scholar] [CrossRef]
- Olivera, B.M.E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol. Biol. Cell 1997, 8, 2101–2109. [Google Scholar]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteomics 2013, 12, 312–329. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Craig, A.G.; Bandyopadhyay, P.; Olivera, B.M. Post-translationally modified neuropeptides from Conus venoms. Eur. J. Biochem. 1999, 264, 271–275. [Google Scholar] [CrossRef]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V. Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: An important component of conotoxin discovery. Toxicon 2006, 47, 688–699. [Google Scholar] [CrossRef]
- Gerwig, G.J.; Hocking, H.G.; Stocklin, R.; Kamerling, J.P.; Boelens, R. Glycosylation of conotoxins. Mar. Drugs 2013, 11, 623–642. [Google Scholar] [CrossRef]
- Buczek, O.; Bulaj, G.; Olivera, B.M. Conotoxins and the posttranslational modification of secreted gene products. Cell. Mol. Life Sci. 2005, 62, 3067–3079. [Google Scholar] [CrossRef]
- Yates, J.R., III. Mass spectrometry. From genomics to proteomics. Trends Genet. 2000, 16, 5–8. [Google Scholar] [CrossRef]
- Nam, H.H.; Corneli, P.S.; Watkins, M.; Olivera, B.; Bandyopadhyay, P. Multiple genes elucidate the evolution of venomous snail-hunting Conus species. Mol. Phylogenet. Evol. 2009, 53, 645–652. [Google Scholar] [CrossRef]
- Nguyen, B.; Molgó, J.; Lamthanh, H.; Benoit, E.; Khuc, T.A.; Ngo, D.N.; Nguyen, N.T.; Millares, P.; le Caer, J.P. High accuracy mass spectrometry comparison of Conus bandanus and Conus marmoreus venoms from the South Central Coast of Vietnam. Toxicon 2013, 75, 148–159. [Google Scholar] [CrossRef]
- Craig, A.G.; Jimenez, E.C.; Dykert, J.; Nielsen, D.B.; Gulyas, J.; Abogadie, F.C.; Porter, J.; Rivier, J.E.; Cruz, L.J.; Olivera, B.M.; et al. A novel post-translational modification involving bromination of tryptophan. Identification of the residue, l-6-bromotryptophan, in peptides from Conus imperialis and Conus radiatus venom. J. Biol. Chem. 1997, 272, 4689–4698. [Google Scholar] [CrossRef]
- England, L.J.; Imperial, J.; Jacobsen, R.; Craig, A.G.; Gulyas, J.; Akhtar, M.; Rivier, J.; Julius, D.; Olivera, B.M. Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science 1998, 281, 575–578. [Google Scholar] [CrossRef]
- Nair, S.S.; Nilsson, C.L.; Emmett, M.R.; Schaub, T.M.; Gowd, K.H.; Thakur, S.S.; Krishnan, K.S.; Balaram, P.; Marshall, A.G. De novo sequencing and disulfide mapping of a bromotryptophan-containing conotoxin by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2006, 78, 8082–8088. [Google Scholar] [CrossRef]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef]
- Han, Y.H.; Wang, Q.; Jiang, H.; Liu, L.; Xiao, C.; Yuan, D.D.; Shao, X.X.; Dai, Q.Y.; Cheng, J.S.; Chi, C.W. Characterization of novel M-superfamily conotoxins with new disulfide linkage. FEBS J. 2006, 273, 4972–4982. [Google Scholar]
- Jimenez, E.C.; Craig, A.G.; Watkins, M.; Hillyard, D.R.; Gray, W.R.; Gulyas, J.; Rivier, J.E.; Cruz, L.J.; Olivera, B.M. Bromocontryphan: Post-translational bromination of tryptophan. Biochemistry 1997, 36, 989–994. [Google Scholar] [CrossRef]
- Jimenez, E.C.; Watkins, M.; Olivera, B.M. Multiple 6-bromotryptophan residues in a sleep-inducing peptide. Biochemistry 2004, 43, 12343–12348. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, L.; Wu, Y.; Zhu, X.; Feng, Y.; Chen, Z.; Li, Y.; Sun, D.; Ren, Z.; Xu, A. Characterizing the evolution and functions of the M-superfamily conotoxins. Toxicon 2013, 76, 150–159. [Google Scholar] [CrossRef]
- Conticello, S.G.; Gilad, Y.; Avidan, N.; Ben-Asher, E.; Levy, Z.; Fainzilber, M. Mechanisms for evolving hypervariability: The case of conopeptides. Mol. Biol. Evol. 2001, 18, 120–131. [Google Scholar] [CrossRef]
- Pi, C.; Liu, J.; Peng, C.; Liu, Y.; Jiang, X.; Zhao, Y.; Tang, S.; Wang, L.; Dong, M.; Chen, S.; Xu, A. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics 2006, 88, 809–819. [Google Scholar] [CrossRef]
- Corpuz, G.P.; Jacobsen, R.B.; Jimenez, E.C.; Watkins, M.; Walker, C.; Colledge, C.; Garrett, J.E.; McDougal, O.; Li, W.; Gray, W.R.; et al. Definition of the M-conotoxin superfamily: Characterization of novel peptides from molluscivorous Conus venoms. Biochemistry 2005, 44, 8176–8186. [Google Scholar] [CrossRef]
- Jacob, R.B.; McDougal, O.M. The M-superfamily of conotoxins: A review. Cell. Mol. Life Sci. 2010, 67, 17–27. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Pi, C.; Zeng, X.; Zhou, M.; Jiang, X.; Chen, S.; Ren, Z.; Xu, A. Identification of a novel M-superfamily conotoxin with the ability to enhance tetrodotoxin sensitive sodium currents. Arch. Toxicol. 2009, 83, 925–932. [Google Scholar] [CrossRef]
- Holford, M.; Zhang, M.M.; Gowd, K.H.; Azam, L.; Green, B.R.; Watkins, M.; Ownby, J.P.; Yoshikami, D.; Bulaj, G.; Olivera, B.M. Pruning nature: Biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon 2009, 53, 90–98. [Google Scholar] [CrossRef]
- Zhang, M.M.; Fiedler, B.; Green, B.R.; Catlin, P.; Watkins, M.; Garrett, J.E.; Smith, B.J.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Structural and functional diversities among mu-conotoxins targeting TTX-resistant sodium channels. Biochemistry 2006, 45, 3723–3732. [Google Scholar] [CrossRef]
- Wakamatsu, K.; Kohda, D.; Hatanaka, H.; Lancelin, J.M.; Ishida, Y.; Oya, M.; Nakamura, H.; Inagaki, F.; Sato, K. Structure-activity relationships of mu-conotoxin GIIIA: Structure determination of active and inactive sodium channel blocker peptides by NMR and simulated annealing calculations. Biochemistry 1992, 31, 12577–12584. [Google Scholar]
- Hill, J.M.; Alewood, P.F.; Craik, D.J. Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels. Biochemistry 1996, 35, 8824–8835. [Google Scholar] [CrossRef]
- Cruz, L.J.; Gray, W.R.; Olivera, B.M.; Zeikus, R.D.; Kerr, L.; Yoshikami, D.; Moczydlowski, E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J. Biol. Chem. 1985, 260, 9280–9288. [Google Scholar]
- Bulaj, G.; West, P.J.; Garrett, J.E.; Watkins, M.; Zhang, M.M.; Norton, R.S.; Smith, B.J.; Yoshikami, D.; Olivera, B.M. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry 2005, 44, 7259–7265. [Google Scholar] [CrossRef]
- Wilson, M.J.; Yoshikami, D.; Azam, L.; Gajewiak, J.; Olivera, B.M.; Bulaj, G.; Zhang, M.M. mu-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 10302–10307. [Google Scholar]
- Shon, K.J.; Olivera, B.M.; Watkins, M.; Jacobsen, R.B.; Gray, W.R.; Floresca, C.Z.; Cruz, L.J.; Hillyard, D.R.; Brink, A.; Terlau, H.; et al. mu-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J. Neurosci. 1998, 18, 4473–4481. [Google Scholar]
- Shon, K.J.; Grilley, M.; Jacobsen, R.; Cartier, G.E.; Hopkins, C.; Gray, W.R.; Watkins, M.; Hillyard, D.R.; Rivier, J.; Torres, J.; et al. A noncompetitive peptide inhibitor of the nicotinic acetylcholine receptor from Conus purpurascens venom. Biochemistry 1997, 36, 9581–9587. [Google Scholar] [CrossRef]
- Van Wagoner, R.M.; Jacobsen, R.B.; Olivera, B.M.; Ireland, C.M. Characterization and three-dimensional structure determination of psi-conotoxin Piiif, a novel noncompetitive antagonist of nicotinic acetylcholine receptors. Biochemistry 2003, 42, 6353–6362. [Google Scholar] [CrossRef]
- Lluisma, A.O.; Lopez-Vera, E.; Bulaj, G.; Watkins, M.; Olivera, B.M. Characterization of a novel psi-conotoxin from Conus parius Reeve. Toxicon 2008, 51, 174–180. [Google Scholar] [CrossRef]
- Chen, P.; Dendorfer, A.; Finol-Urdaneta, R.K.; Terlau, H.; Olivera, B.M. Biochemical characterization of kappaM-RIIIJ, a Kv1.2 channel blocker: evaluation of cardioprotective effects of kappaM-conotoxins. J. Biol. Chem. 2010, 285, 14882–14889. [Google Scholar]
- Ferber, M.; Sporning, A.; Jeserich, G.; DeLaCruz, R.; Watkins, M.; Olivera, B.M.; Terlau, H. A novel Conus peptide ligand for K+ channels. J. Biol. Chem. 2003, 278, 2177–2183. [Google Scholar]
- Schroeder, C.I.; Ekberg, J.; Nielsen, K.J.; Adams, D.; Loughnan, M.L.; Thomas, L.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Neuronally micro-conotoxins from Conus striatus utilize an alpha-helical motif to target mammalian sodium channels. J. Biol. Chem. 2008, 283, 21621–21628. [Google Scholar] [CrossRef]
- West, P.J.; Bulaj, G.; Garrett, J.E.; Olivera, B.M.; Yoshikami, D. Mu-conotoxin SmIIIA, a potent inhibitor of tetrodotoxin-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry 2002, 41, 15388–15393. [Google Scholar] [CrossRef]
- Walewska, A.; Skalicky, J.J.; Davis, D.R.; Zhang, M.M.; Lopez-Vera, E.; Watkins, M.; Han, T.S.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. NMR-based mapping of disulfide bridges in cysteine-rich peptides: Application to the mu-conotoxin SxIIIA. J. Am. Chem. Soc. 2008, 130, 14280–14286. [Google Scholar] [CrossRef]
- Lewis, R.J.; Schroeder, C.I.; Ekberg, J.; Nielsen, K.J.; Loughnan, M.; Thomas, L.; Adams, D.A.; Drinkwater, R.; Adams, D.J.; Alewood, P.F. Isolation and structure-activity of mu-conotoxin TIIIA, a potent inhibitor of tetrodotoxin-sensitive voltage-gated sodium channels. Mol. Pharmacol. 2007, 71, 676–685. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nguyen, B.; Caer, J.-P.L.; Mourier, G.; Thai, R.; Lamthanh, H.; Servent, D.; Benoit, E.; Molgó, J. Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan. Mar. Drugs 2014, 12, 3449-3465. https://doi.org/10.3390/md12063449
Nguyen B, Caer J-PL, Mourier G, Thai R, Lamthanh H, Servent D, Benoit E, Molgó J. Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan. Marine Drugs. 2014; 12(6):3449-3465. https://doi.org/10.3390/md12063449
Chicago/Turabian StyleNguyen, Bao, Jean-Pierre Le Caer, Gilles Mourier, Robert Thai, Hung Lamthanh, Denis Servent, Evelyne Benoit, and Jordi Molgó. 2014. "Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan" Marine Drugs 12, no. 6: 3449-3465. https://doi.org/10.3390/md12063449
APA StyleNguyen, B., Caer, J.-P. L., Mourier, G., Thai, R., Lamthanh, H., Servent, D., Benoit, E., & Molgó, J. (2014). Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan. Marine Drugs, 12(6), 3449-3465. https://doi.org/10.3390/md12063449








