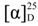Chromomycins A2 and A3 from Marine Actinomycetes with TRAIL Resistance-Overcoming and Wnt Signal Inhibitory Activities
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Identification of 1 and 2
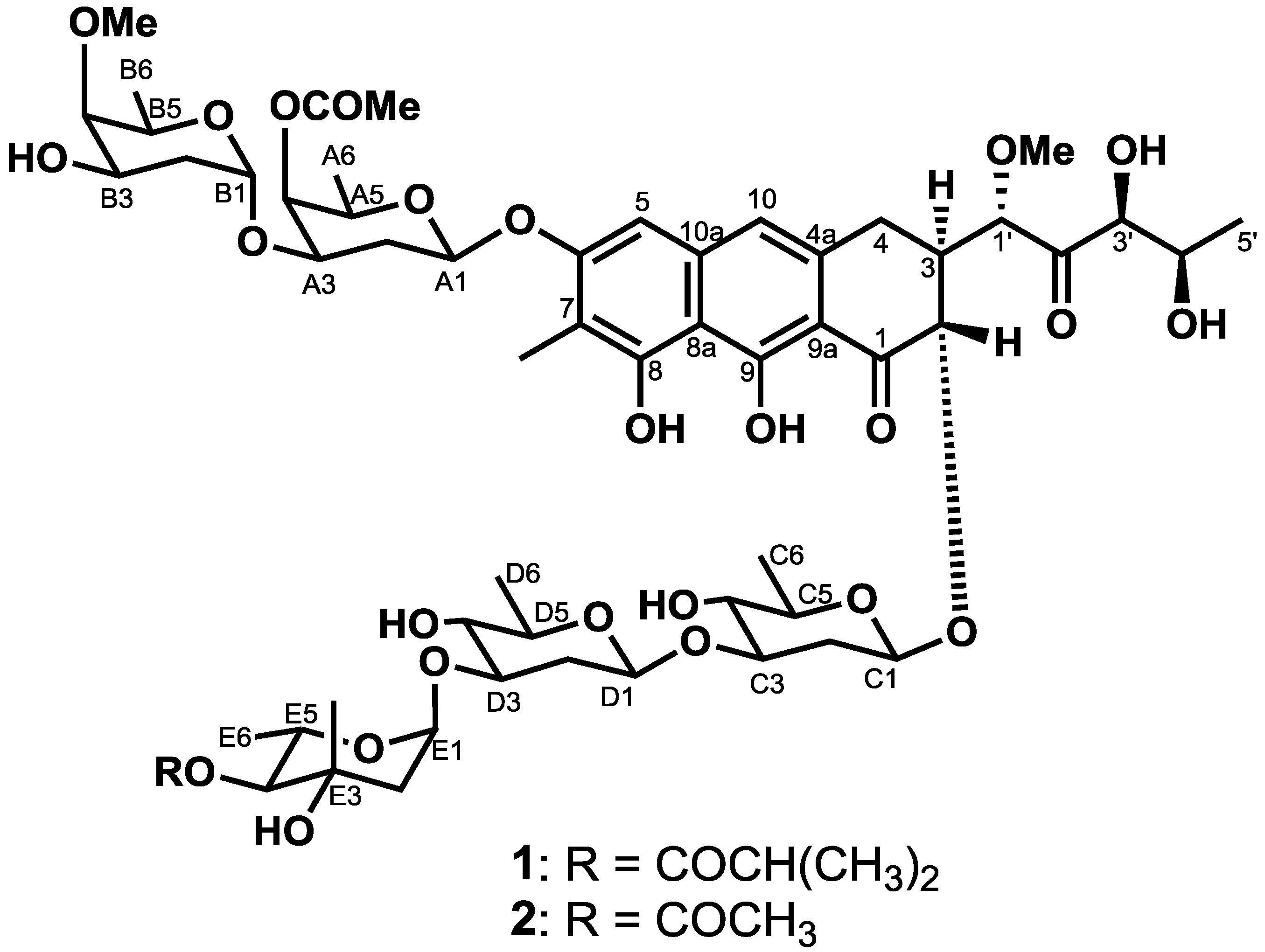
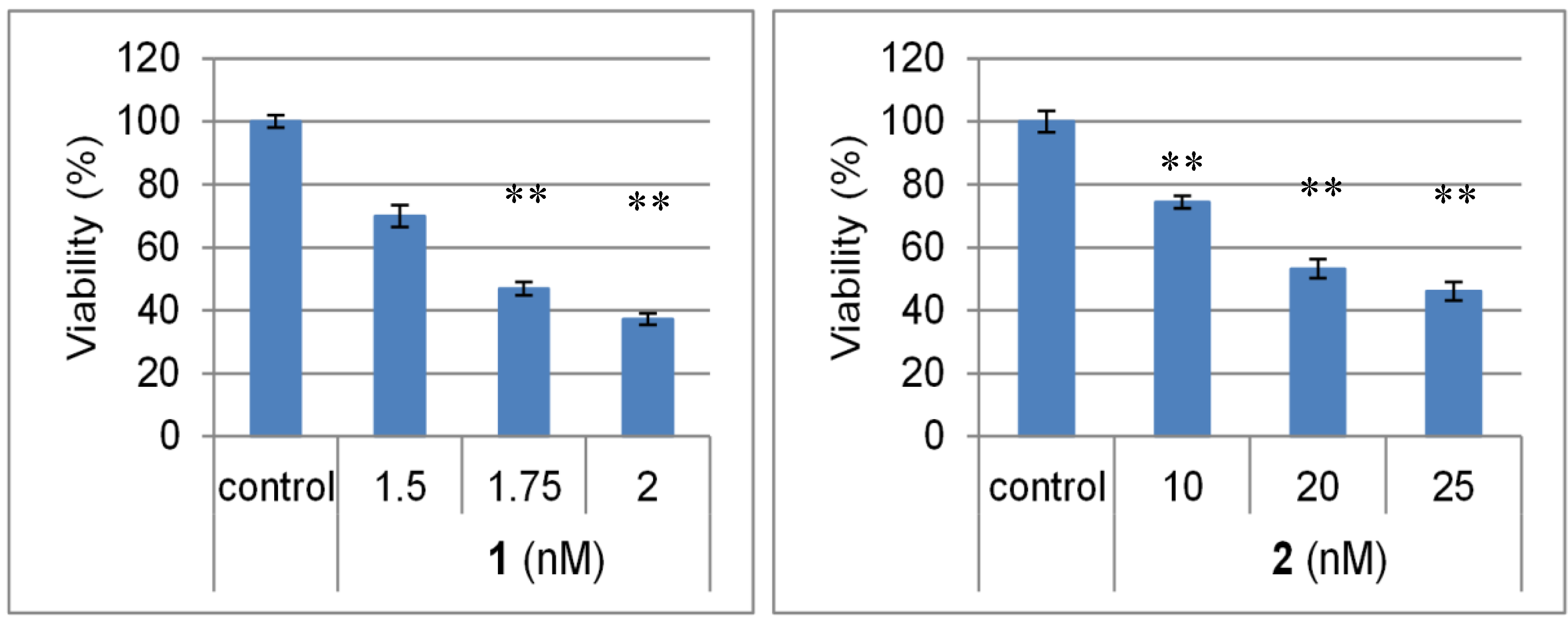
2.2. Biological Activities of 1 and 2

3. Experimental Section
3.1. General Experimental Procedures
3.2. Microbial Strain and Fermentation
3.3. Extraction and Isolation
| Position | 1 | 2 | |||
|---|---|---|---|---|---|
| δH (J in Hz) a | δH (J in Hz) b | δC c | δH (J in Hz) b | δC c | |
| 1 | 202.1 | 202.1 | |||
| 2 | 4.64, d (11.2) | 4.71, d (11.4) | 75.9 | 4.71, d (11.6) | 75.9 |
| 3 | 2.58, m | 2.58, m | 43.7 | 2.67, m | 43.7 |
| 4 | 3.00, d (15.2) | 3.06, d (13.1) | 26.9 | 3.13, m | 26.9 |
| 2.58, m | 2.58, m | 2.67, m | |||
| 5 | 6.58, s | 6.54, s | 100.8 | 6.60, s | 100.7 |
| 6 | 159.6 | 159.6 | |||
| 7 | 111.6 | 111.6 | |||
| 8 | 156.1 | 156.1 | |||
| 9 | 165.3 | 165.2 | |||
| 10 | 6.70, s | 6.66, s | 117.0 | 6.72, s | 117.0 |
| 4a | 134.6 | 134.5 | |||
| 8a | 108.1 | 108 | |||
| 9a | 108.1 | 108 | |||
| 10a | 138.4 | 138.3 | |||
| 7-CH3 | 2.10, s | 2.16, s | 8.2 | 2.16, s | 8.2 |
| 8-OH | 9.74 | 9.78, s | |||
| 9-OH | 15.67 | 15.68, s | |||
| 1′ | 4.67, s | 4.69, br s | 81.8 | 4.70, d (1.6) | 81.9 |
| 2′ | 211.2 | 211.2 | |||
| 3′ | 4.11, d (1.5) | 4.20, br s | 78.2 | 4.21, d (2.0) | 78.2 |
| 4′ | 4.21, qd (6.4, 1.5) | 4.36, q (6.0) | 67.9 | 4.37, qd (6.4, 2.0) | 67.8 |
| 5′ | 1.25 d (6.4) | 1.35, d (6.0) | 20.5 | 1.37, d (6.4) | 20.5 |
| 1′-OCH3 | 3.40, s | 3.49, s | 59.6 | 3.50, s | 59.6 |
| Sugar A | |||||
| A1 | 5.18, d (8.4) | 5.19, d (10.0, 2.0) | 97.3 | 5.21, dd (9.8, 2.1) | 97.3 |
| A2 | 2.16, m | 2.20, m | 32.9 | 2.26, m | 32.9 |
| 2.00, m | 2.02, m | 2.26, m | |||
| A3 | 3.96, m | 3.93, m | 69.9 | 4.00, m | 69.8 |
| A4 | 5.10, d (1.7) | 5.15, d (2.6) | 67.2 | 5.16, d (3.0) | 67.2 |
| A5 | 3.76, m | 3.80, q (6.4) | 69.7 | 3.82, q (6.4) | 69.7 |
| A6 | 1.19, d (6.0) | 1.27, d (6.4) | 16.8 | 1.28, d (6.4) | 16.8 |
| CH3-CO | 2.09, s | 2.15, s | 20.8 | 2.15, s | 20.8 |
| CH3-CO | 170.9 | 170.9 | |||
| Sugar B | |||||
| B1 | 5.05, d (2.8) | 5.10, br s | 95.2 | 5.10, d (2.4) | 95.1 |
| B2 | 1.78, m | 1.63–1.74, m | 33.5 | 1.63–1.75, m | 33.4 |
| 1.64, m | 1.63–1.74, m | 1.63–1.75, m | |||
| B3 | 3.89, dd (7.8, 2.8) | 3.93, m | 65.9 | 3.93, m | 65.8 |
| B4 | 3.14, d (2.8) | 3.21, d (3.2) | 81.5 | 3.21, d (3.2) | 81.5 |
| B5 | 3.82, q (6.5) | 3.93, m | 66.7 | 3.89, q (6.4) | 66.7 |
| B6 | 1.14, d (6.5) | 1.27, d (6.5) | 17.2 | 1.29, d (6.4) | 17.2 |
| B4-OCH3 | 3.51, s | 3.57, s | 62.3 | 3.58, s | 62.3 |
| Sugar C | |||||
| C1 | 5.01, d (9.2) | 5.07, br d (9.7) | 100.3 | 5.08, dd (9.7, 1.6) | 100.3 |
| C2 | 2.46, dd (12.2, 4.6) | 2.49, dd (11.4, 5.2) | 37.4 | 2.49, ddd (12.8, 5.1, 1.6) | 37.4 |
| 1.64, m | 1.63, m | 1.75, m | |||
| C3 | 3.59, m | 3.57, m | 82.3 | 3.59, m | 82.3 |
| C4 | 3.06, d (8.8) | 3.13, d (9.0) | 75.1 | 3.08, m | 75.1 |
| C5 | 3.27, m | 3.38, dq (9.0, 5.5) | 72.1 | 3.32, dq (9.1, 5.5) | 72.1 |
| C6 | 1.28, d (5.6) | 1.34, d (5.5) | 18.0 | 1.34, d (5.5) | 18.0 |
| Sugar D | |||||
| D1 | 4.55, d (9.6) | 4.59, d (9.0) | 99.7 | 4.60, d (9.6, 2.0) | 99.7 |
| D2 | 2.22, dd (11.9, 4.2) | 2.27, dd (13.1, 4.8) | 37.1 | 2.29, dd (5.9, 2.0) | 37.0 |
| 1.56, d (11.9) | 1.63, m | 1.75, m | |||
| D3 | 3.49, m | 3.49, m | 80.7 | 3.47, m | 80.5 |
| D4 | 3.03, d (8.0) | 3.10, d (8.5) | 75.2 | 3.08, m | 75.2 |
| D5 | 3.27, m | 3.30, qd (9.0, 6.0) | 72.3 | 3.40, dq (9.1, 6.0) | 72.2 |
| D6 | 1.31, d (6.0) | 1.37, d (6.0) | 17.8 | 1.38, d (6.0) | 17.8 |
| Sugar E | |||||
| E1 | 4.96 (t, 2.8) | 4.99, dd (3.9, 2.0) | 97.1 | 5.01, dd (4.0, 2.2) | 97.0 |
| E2 | 1.92, m | 2.00, m | 43.8 | 2.00, dd (13.8, 4.0) | 43.6 |
| E3 | 70.6 | 2.06, dd (13.8, 2.2) | 70.6 | ||
| E4 | 4.55, d (9.6) | 4.59, d (9.0) | 79.4 | 4.61, d (9.3) | 79.5 |
| E5 | 3.98, m | 3.88, q (6.7) | 67.0 | 4.00, m | 67.0 |
| E6 | 1.22, d (5.2) | 1.21, d (7.4) | 17.8 | 1.23, d (6.4) | 17.8 |
| E3-CH3 | 1.28, s | 1.33, s | 22.9 | ||
| (CH3)2CH-CO | 1.13, d (6.8) | 1.19, d (7.4) | 19.0 | ||
| (CH3)2CH-CO | 2.58, m | 2.56, m | 34.2 | ||
| (CH3)2CH-CO | 177.5 | ||||
| CH3CO | 2.12, s | 20.9 | |||
| CH3CO | 171.4 | ||||
3.4. Viability Assay (FMCA Assay [25])
3.5. Luciferase Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takahashi, Y.; Omura, S. Isolation of new actinomycetes strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 2003, 49, 141–154. [Google Scholar]
- Abdelfattah, M.S.; Kazufumi, T.; Ishibashi, M. Izumiphenazines A–C: Isolation and structure elucidation of phenazine derivatives from Streptomyces sp. IFM 11204. J. Nat. Prod. 2010, 73, 1999–2002. [Google Scholar] [CrossRef]
- Abdelfattah, M.S.; Toume, K.; Ishibashi, M. Isolation and structure elucidation of izuminosides A–C: A rare phenazine glycosides from Streptomyces sp. IFM 11260. J. Antibiot. 2011, 64, 271–275. [Google Scholar] [CrossRef]
- Tamai, Y.; Toume, K.; Arai, M.A.; Hayashida, A.; Kato, H.; Shizuri, Y.; Tsukamoto, S.; Ishibashi, M. Nonactin and related compounds found in a screening program for Wnt signal inhibitory activity. Heterocycles 2012, 84, 1245–1250. [Google Scholar] [CrossRef]
- Tamai, Y.; Toume, K.; Arai, M.A.; Ishibashi, M. Griseoviridin and cyclic hydroxamates found in a screening program for Wnt signal inhibitor. Heterocycles 2012, 86, 1517–1524. [Google Scholar]
- Abdelfattah, M.S.; Toume, K.; Arai, M.A.; Masu, H.; Ishibashi, M. Katorazone, a new yellow pigment with a 2-azaquinone-phenylhydrazone structure produced by Streptomyces sp. IFM 11299. Tetrahedron Lett. 2012, 53, 3346–3348. [Google Scholar]
- Abdelfattah, M.S.; Toume, K.; Ishibashi, M. Yoropyrazone, a new naphthopyridazone alkaloid isolated from Streptomyces sp. IFM 11307 and evaluation of its TRAIL resistance-overcoming activity. J. Antibiot. 2012, 65, 245–248. [Google Scholar] [CrossRef]
- Abdelfattah, M.S.; Kazufumi, T.; Ishibashi, M. New pyranonaphthoquinones and a phenazine alkaloid isolated from Streptomyces sp. IFM 11307 with TRAIL resistance-overcoming activity. J. Antibiot. 2011, 64, 729–734. [Google Scholar] [CrossRef]
- Lombo, F.; Menendez, N.; Salas, J.A.; Mendez, C. The aureolic acid family of antitumor compounds: Structure, mode of action, biosynthesis, and novel derivatives. Appl. Microbiol. Biotechnol. 2006, 73, 1–14. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Koenuma, M.; Matsumoto, K.; Tori, K.; Terui, Y. NMR studies of chromomycins, olivomycins, and their derivatives. J. Antibiot. 1988, 41, 53–67. [Google Scholar]
- Hu, Y.; Espindola, A.P.D.M.; Stewart, N.A.; Wei, S.; Posner, B.A.; MacMillan, J.B. Chromomycin SA analogs from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 5183–5189. [Google Scholar] [CrossRef]
- Jin, C.Y.; Park, C.; Cheong, J.; Choi, B.T.; Lee, T.H.; Lee, J.D.; Lee, W.H.; Kim, G.Y.; Ryu, C.H.; Choi, Y.H. Genistein sensitizes TRAIL-resistant human gastric adenocarcinoma AGS cells through activation of caspase-3. Cancer Lett. 2007, 257, 56–64. [Google Scholar] [CrossRef]
- Mellier, G.; Huang, S.; Shenoy, K.; Pervaiz, S. TRAILing death in cancer. Mol. Aspects Med. 2010, 31, 93–112. [Google Scholar] [CrossRef]
- Minakawa, T.; Toume, K.; Arai, M.A.; Koyano, T.; Kowithayakorn, T.; Ishibashi, M. Prenylflavonoids isolated from Artocarpus champeden with TRAIL-resistance overcoming activity. Phytochemistry 2013, 96, 299–304. [Google Scholar] [CrossRef]
- Ahmed, F.; Toume, K.; Sadhu, S.K.; Ohtsuki, T.; Arai, M.A.; Ishibashi, M. Constituents of Amoora cucullata with TRAIL resistance-overcoming activity. Org. Biomol. Chem. 2010, 8, 3696–3703. [Google Scholar] [CrossRef]
- Horinaka, M.; Yoshida, T.; Shiraishi, T.; Nakata, S.; Wakada, M.; Nakanishi, R.; Nishino, H.; Matsui, H.; Sakai, T. Luteolin induces apoptosis via death receptor 5 upregulation in human malignant tumor cells. Oncogene 2005, 24, 7180–7189. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: From human mutations to treatments. Nat. Med. 2013, 19, 179–192. [Google Scholar] [CrossRef]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef]
- Toume, K.; Kamiya, K.; Arai, M.A.; Mori, N.; Sadhu, S.K.; Ahmed, F.; Ishibashi, M. Xylogranin B: A potent Wnt signal inhibitory limonoid from Xylocarpus granatum. Org. Lett. 2013, 15, 6106–6109. [Google Scholar] [CrossRef]
- Li, X.; Ohtsuki, T.; Koyano, T.; Kowithayakorn, T.; Ishibashi, M. New Wnt/β-catenin signaling inhibitors isolated from Eleutherine palmifolia. Chem. Asian J. 2009, 4, 540–547. [Google Scholar] [CrossRef]
- Hayakawa, M.; Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 1987, 65, 501–509. [Google Scholar] [CrossRef]
- Hirsch, C.F.; Christensen, D.L. Novel method for selective isolation of actinomycetes. Appl. Environ. Microbiol. 1983, 46, 925–929. [Google Scholar]
- Lindhagen, E.; Nygren, P.; Larsson, R. The fluorometric microculture cytotoxicity assay. Nat. Protoc. 2008, 3, 1364–1369. [Google Scholar] [CrossRef]
- Simon, H.; Wittig, B.; Zimmer, C. Effect of netropsin, distamycin-A and chromomycin-A (3) on the binding and cleavage reaction of DNA gyrase. FEBS Lett. 1994, 353, 79–83. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Toume, K.; Tsukahara, K.; Ito, H.; Arai, M.A.; Ishibashi, M. Chromomycins A2 and A3 from Marine Actinomycetes with TRAIL Resistance-Overcoming and Wnt Signal Inhibitory Activities. Mar. Drugs 2014, 12, 3466-3476. https://doi.org/10.3390/md12063466
Toume K, Tsukahara K, Ito H, Arai MA, Ishibashi M. Chromomycins A2 and A3 from Marine Actinomycetes with TRAIL Resistance-Overcoming and Wnt Signal Inhibitory Activities. Marine Drugs. 2014; 12(6):3466-3476. https://doi.org/10.3390/md12063466
Chicago/Turabian StyleToume, Kazufumi, Kentaro Tsukahara, Hanako Ito, Midori A. Arai, and Masami Ishibashi. 2014. "Chromomycins A2 and A3 from Marine Actinomycetes with TRAIL Resistance-Overcoming and Wnt Signal Inhibitory Activities" Marine Drugs 12, no. 6: 3466-3476. https://doi.org/10.3390/md12063466
APA StyleToume, K., Tsukahara, K., Ito, H., Arai, M. A., & Ishibashi, M. (2014). Chromomycins A2 and A3 from Marine Actinomycetes with TRAIL Resistance-Overcoming and Wnt Signal Inhibitory Activities. Marine Drugs, 12(6), 3466-3476. https://doi.org/10.3390/md12063466