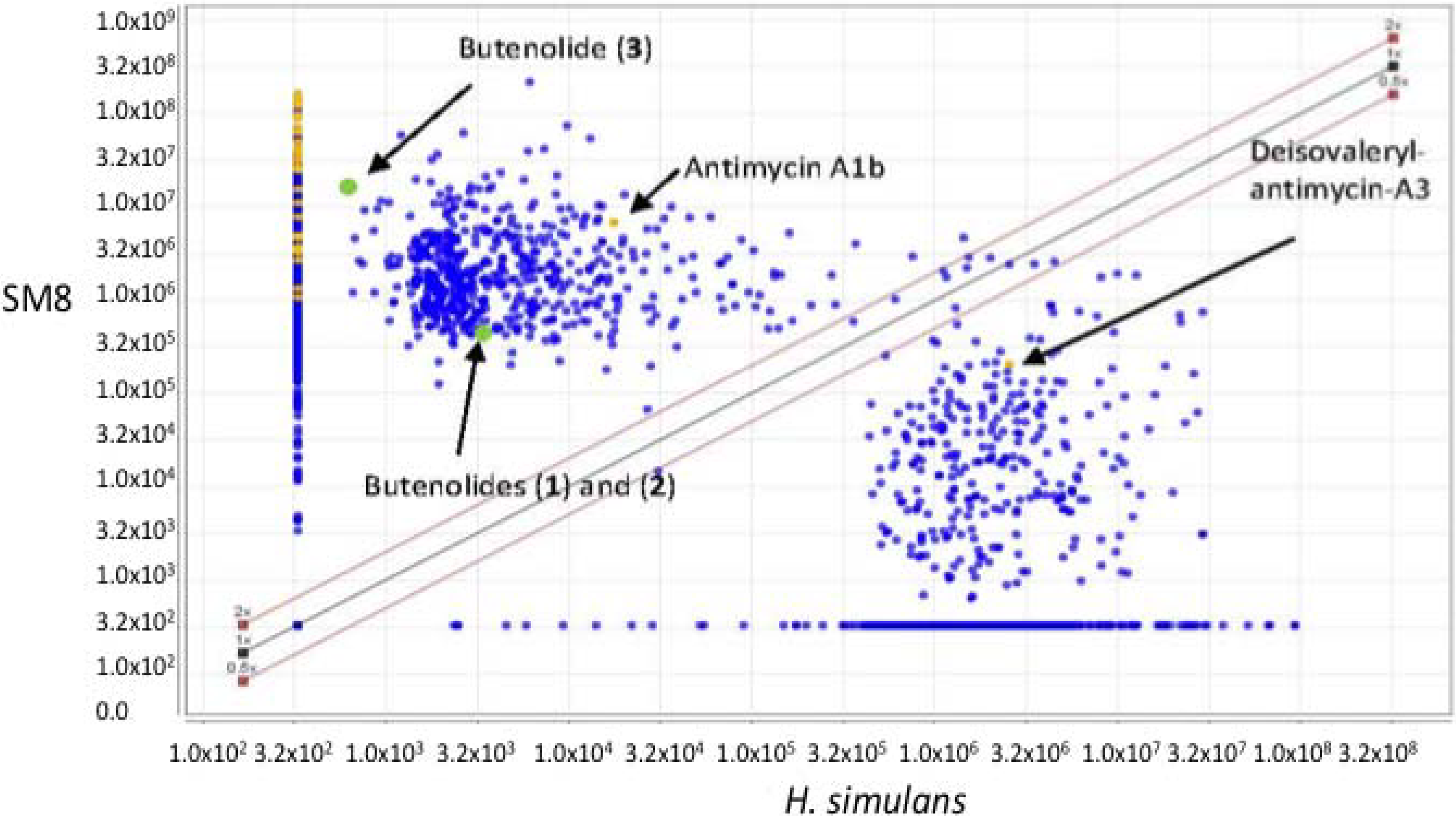
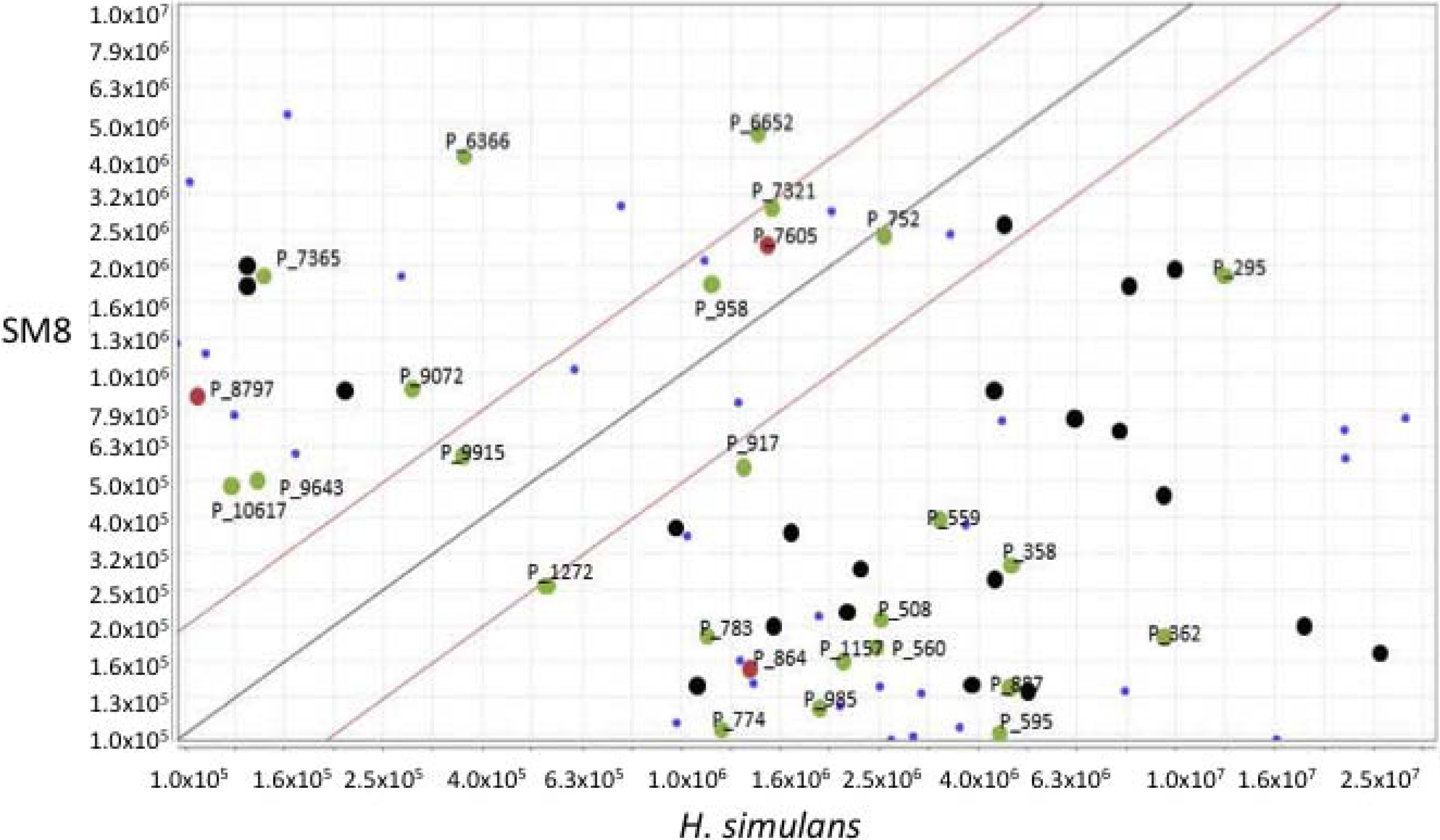
3.5.2. NMR and LC-HRESIMS of Active SM8 Fractions
Following activity assays, the active fractions were analyzed by NMR using a Jeol-LA400 FT-NMR spectrometer system (JEOL Ltd., Tokyo, Japan) with an AS400 magnet (Oxford Instruments, Inghilterra, UK) at 400 MHz for 1H and 100 MHz for 13C, using a Pulse Field Gradient “Autotune” 40TH5AT/FG broadband high sensitivity probe (JEOL Ltd., Tokyo, Japan) to accept 5 mm tubes. High-resolution mass spectrometry was carried out on a Dionex UltiMate-3000 (DIONEX, Sunnyvale, CA, USA) coupled to an Exactive-Orbitrap (Thermo Scientific, Bremen, Germany). The column was an ACE 5 C18 75 × 3.0 mm column from Hichrom Ltd, Reading, UK. Compounds were eluted with a flow rate of 300 μL·min−1 using water (A) and acetonitrile (B), both of which contained 0.1% formic acid, by a gradient starting with 10% B and increasing to 100% B in 30 min. The mobile phase was maintained at 100% B for 5 min, after which the column was equilibrated with 10% B. The injection volume was 10 μL and the tray temperature was maintained at 12 °C. High resolution mass spectrometry was carried out in both positive and negative ESI ionization modes with a spray voltage of 4.5 kV and capillary temperature of 268 °C. Spray voltage at 4.5 kV, spray current set 10.67 μA, and capillary voltage at 30 V. The mass range was acquired from m/z 150 to 1500.
Multi-fragmentation (MSn) experiments were accomplished for the positive ionization mode on an Orbitrap analyser, CID (The collision-induced dissociation) was utilized with a normalized collision energy of 35%, activation Q of 0.250 ms, and activation time of 30.000 ms applied on ions of most intense, 2nd most intense, and 3rd most intense peaks for MS2, MS3, and MS4, respectively at an isolation width of 3 microns with 5 microscans. Resolution was at 15,000 m/Δm 50%, while the minimum ion signal threshold was set to 500. Fragment mass tolerance for molecular formula detection was set to ±5 ppm.
Data mining was performed using MZmine 2.10 [
49]. The following parameters were used:
The chromatograms were first cropped to 0.5–38.0 min using the crop filter under the dataset filtering function. The centroid mass detector was used for peak detection with the noise level set to 1.0 × 10
5 and the MS level set to 1. The chromatogram builder function was set to a minimum time span of 0.2 min, minimum height of 1.0 × 10
5 and
m/z tolerance of 0.001
m/z or 5.0 ppm. For chromatogram deconvolution the algorithm used was the local minimum search. The chromatographic threshold was set to 90.0%. The search minimum in RT range was 0.4 min, minimum relative height was 5.0%, minimum absolute height was 3.0 × 10
5, minimum ratio of peak top/edge was 2 and the peak duration range was 0.3–5.0 min. Isotopes were detected using the isotopic peaks grouper. The
m/z tolerance was 0.001
m/z or 5.0 ppm, RT tolerance was 0.2 absolute (min), the maximum charge was 2 and the representative isotope used was the most intense. Retention time normalization was performed using the RT normalizer. Again,
m/z tolerance was 0.001
m/z or 5.0 ppm while the RT tolerance and the minimum standard intensity were set to 5% (relative) and 5.0 × 10
3 respectively. The peak lists were all aligned using the join aligner (
m/z tolerance 0.001
m/z or 5.0 ppm, weight for
m/z: 20, RT tolerance: 5.0% relative, weight for RT: 20). The aligned peak list was gapfilled using the peak finder function (intensity tolerance: 1%,
m/z tolerance: 0.001
m/z or 5.0 ppm, RT tolerance: 0.5 min). An adduct search was performed with the RT tolerance set at 0.2 absolute (min), the
m/z tolerance at 0.001
m/z or 5.0 ppm and the maximum relative adduct peak height at 30%. The adducts searched for were Na
+, K
+, NH
4+ and ACN + H. A complex search was also performed using [M + H]
+ for the ESI positive mode and [M − H]
− for the ESI negative mode. The RT tolerance was set at 0.2 absolute (min),
m/z tolerance was kept at 0.001
m/z or 5.0 ppm, and the maximum complex peak height was set at 50%. A custom database search was then performed using the DNP 2012 database [
65].
3.5.3. Construction of SM8 antC Mutant
To inactivate the antimycin gene cluster, a two-step gene deletion procedure was used [
60]. Flanking regions from within the
antC gene of approximately 1.4 kb were amplified using the High-Fidelity PCR kit (Roche, Basel, Switzerland). Primer pairs (
Table 6) were designed to amplify upstream and downstream regions with restriction sites engineered suitable for cloning into the
Streptomyces/E. coli shuttle vector pKC1139 [
66]. PCR products from
antC1-F/
antC1-R and
antC2-F/
antC2-R were cloned into pJET1.2/blunt using the Fermentas PCR cloning kit. Following sequence verification the
antC flanking regions were purified as
HindIII-
XbaI and
XbaI-
EcoRI fragments, respectively and cloned into
HindIII and
EcoRI digested pKC1139 resulting in pKC1139A1A2.
Table 6.
Primers designed for the construction of the antC deletion strain. Engineered restriction sites are shown underlined.
Table 6.
Primers designed for the construction of the antC deletion strain. Engineered restriction sites are shown underlined.
| | Primer Sequences (5′–3′) |
|---|
| antC1-F | TATATAAAGCTTGGACGGCTACAGCTACAAGC |
| antC1-R | TATATATCTAGAATGAGGTATGCGGTGTCGTA |
| antC2-F | TATATATCTAGAGAGGTGGTTCGTGGAGGAG |
| antC2-R | TATATAGAATTCTGACGATGATGACGTCCTTG |
pKC1139A1A2 was conjugated to
Streptomyces sp. SM8 strain. Competent cells of the dam-/dcm-
E. coli C2925/pUZ8002 were transformed to apramycin resistance with pKC1139A1A2. The helper plasmid pUZ8002 can supply transfer functions to oriT-carrying plasmids, such as pKC1139. Cultures of
E. coli C2925/pUZ8002/pKC1139A1A2 were grown in 10 mL of LB containing 25 μg kanamycin/mL, 25 μg chloramphenicol/mL and 50 μg apramycin/mL overnight at 37 °C. The overnight culture was diluted (1:10) in LB containing the aforementioned antibiotics and grown at 37 °C to an OD600 of 0.5. The cells were washed twice with an equal volume of LB to remove antibiotics and resuspended in 0.1 vol. of LB. Approximately, 10
8 spores of the SM8 strain were added to 500 μL of 2× YT broth (1.6% peptone, 1% yeast extract, and 0.5% NaCl) and subjected to heat shock at 50 °C for 10 min, 0.5 mL of
E. coli cells were added and the spores were briefly mixed. The pellet was resuspended and plated onto MS-SW agar (mannitol 20 g, soya flour 20 g, agar 20 g, Instant Ocean
® 33 g made up to 1 L with dH
2O). Following incubation at 28 °C for 20 h the plates were overlayered with 1 mL antibiotic solution containing 0.5 mg nalidixic acid and 2 mg apramycin. These plates were further incubated at 28 °C until potential exconjugants were observed. The transconjugants were plated on to SYP-SW media containing nalidixic acid 25 μg·mL
−1 and apramycin 100 μg·mL
−1 [
60] and incubated at 28 °C until exconjugants were observed.
The shuttle vector pKC1139A1A2 has a (pSG5) temperature-sensitive origin of replication. In Streptomyces it can replicate only at temperatures between 28 and 30 °C. When potential exconjugants were plated on to SYP-SW containing apramycin 100 μg·mg−1 and incubated at 37 °C, the vector stops replicating and apramycin resistance is only maintained when the plasmid is integrated onto the chromosome by homologous recombination. Apramycin resistant recombinant clones were allowed to sporulate on SYP-SW agar at 37 °C. Spores were collected and plated out to single colonies at 28 °C without selection. Single colonies were then picked and screened for the loss of apramycin resistance, a phenotype indicating that the plasmid has been lost with deletion of the antC gene.
The wild-type and mutant SM8 extracts were compared to an antimycin A standard (Antimycin A, from
Streptomyces sp.) purchased from Sigma-Aldrich Co (St. Louis, MO, USA). The LC-HRMS analysis was performed on a Finnigan Surveyor system coupled to a Thermo-Finnigan LTQ Orbitrap system using the same column and method stated in
section 3.5.2.
3.5.4. Isolation of Compounds from Large Scale SM8 Culture
The large-scale SM8 extract, weighing 810.3 mg, was fractionated using a Sephadex® LH20 column (Pharmacia, Stockholm, Sweden) with methanol as the mobile phase at a flow rate of 1 mL/15 min (0.067 mL·min−1) and a collection volume of 1 mL/test tube. 169 fractions were collected. An aliquot of each tube was subjected to activity assays. The active fractions were subsequently pooled based on their activity and similarities in the TLC.
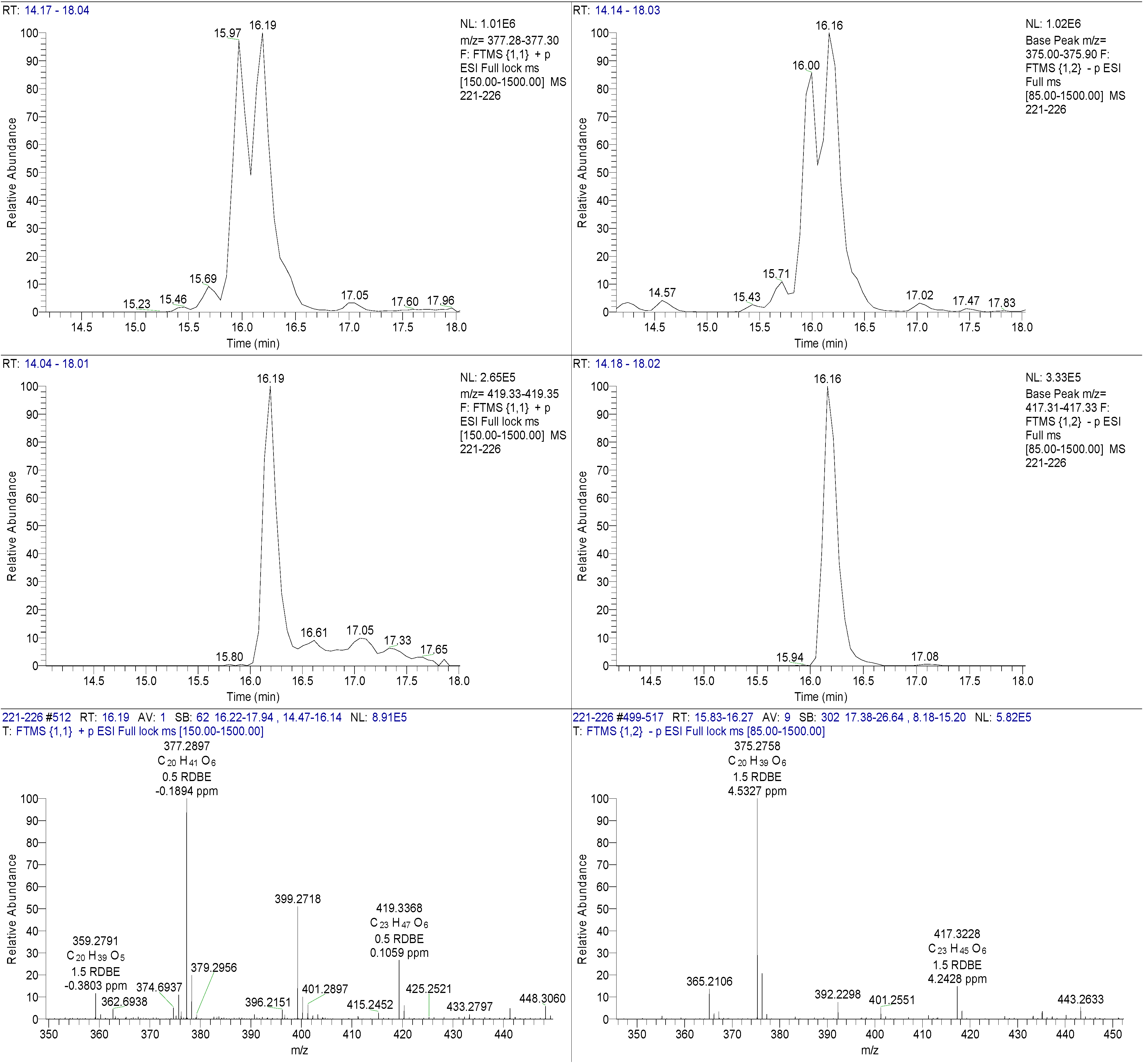
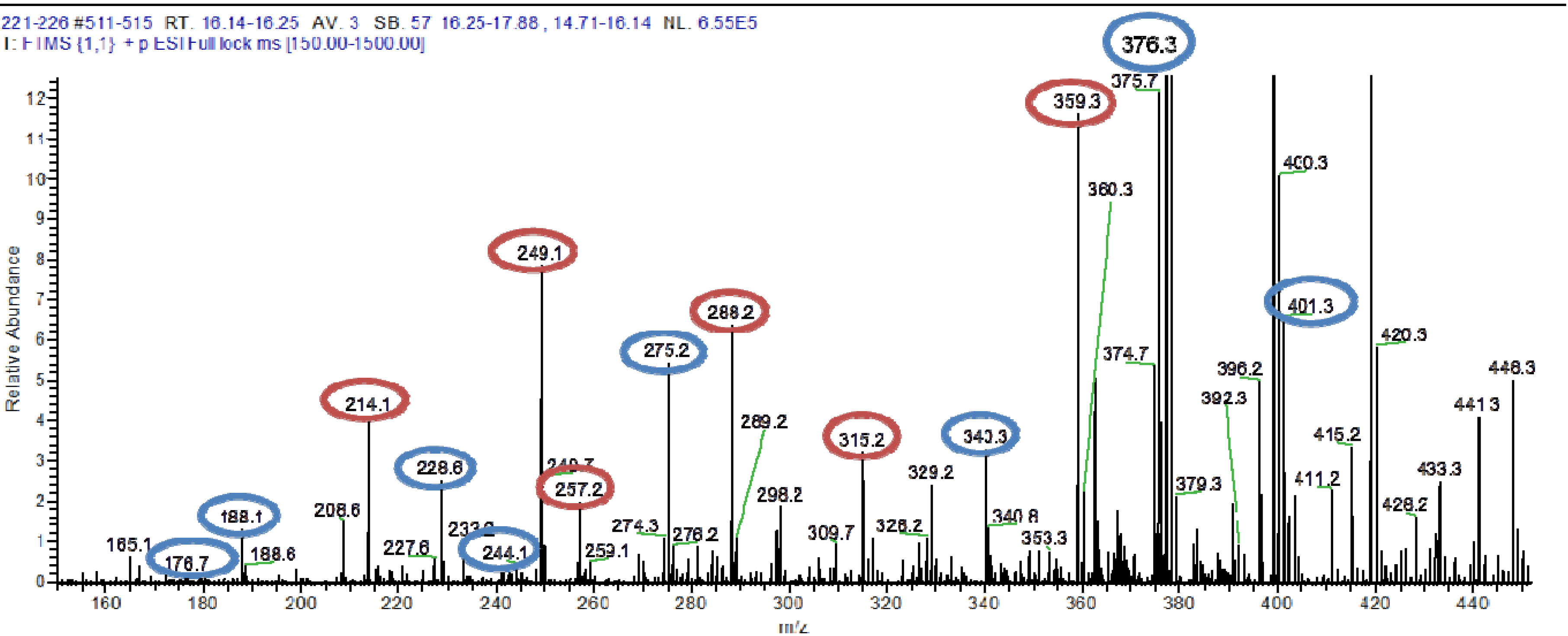
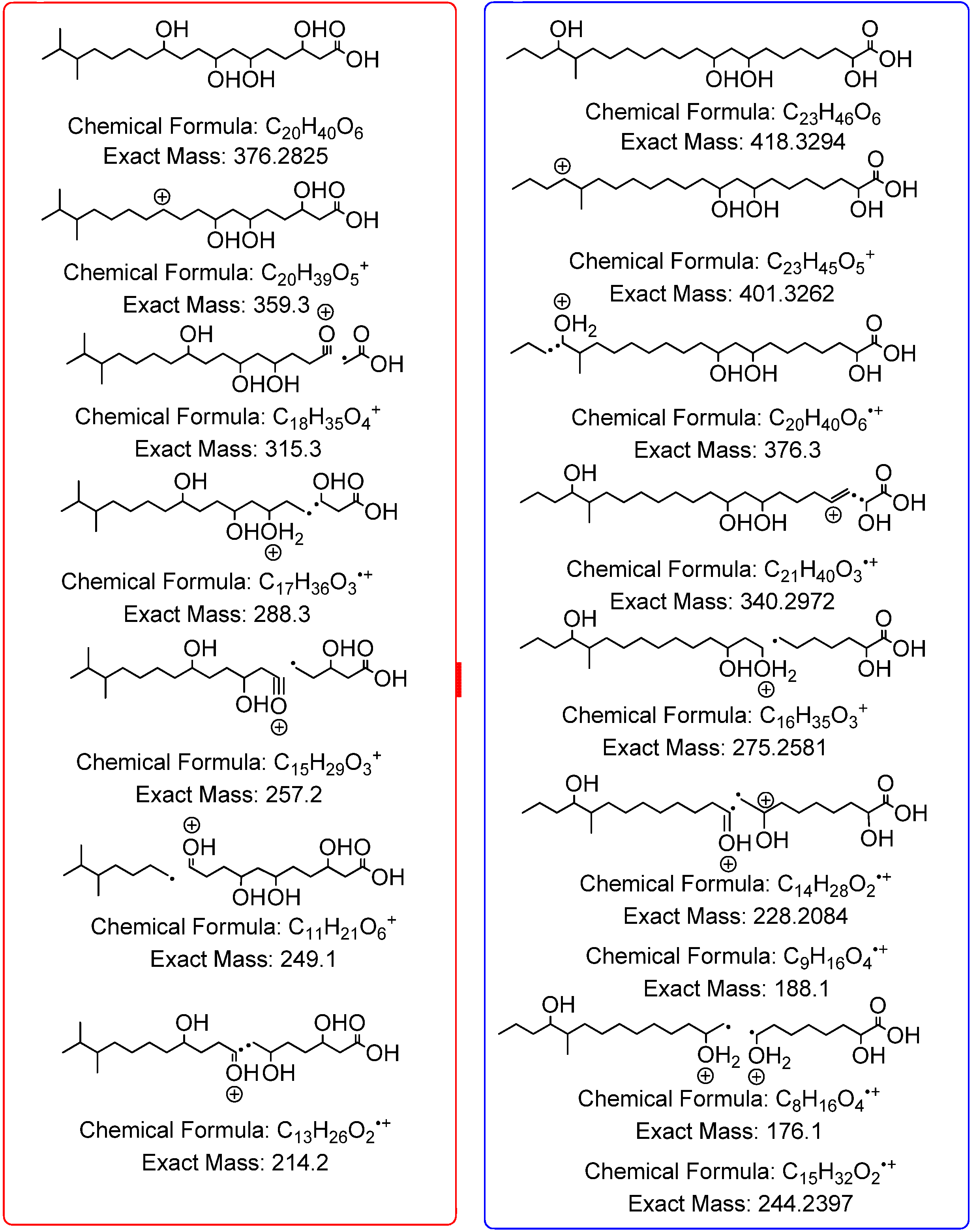
The pooled fraction 127–156 displayed prominent antifungal activity. This fraction was further purified using a silica column with the following solvent systems: 95:5 hexane:ethyl acetate, 90:10 hexane:ethyl acetate, 80:20 hexane:ethyl acetate, and 50:50 hexane:ethyl acetate, followed by washing of the column with 70:30 dichloromethane:methanol, 50:50 acetone:methanol and 100% methanol. The fractions and purified compounds were subsequently subjected to activity assays and analyzed using NMR and LC-HRESIMS.
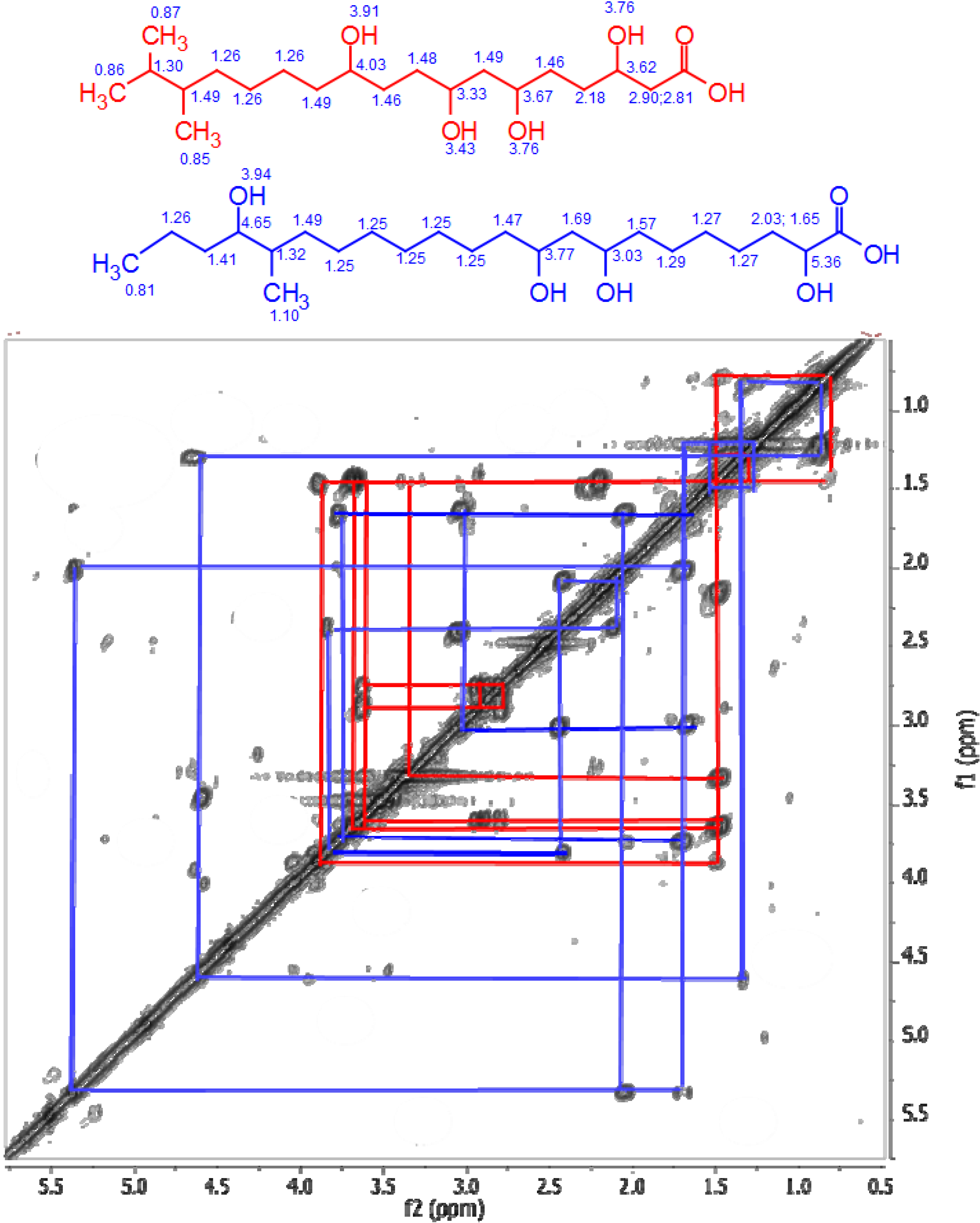
4,10-Dihydroxy-10-methyl-dodec-2-en-1,4-olide (1): Rf 0.56 (80:20 MeOH:H2O, C-18 silica gel), blue-violet with anisaldehyde-sulfuric acid; UVmax 219.0 (MeOH); 1H NMR (400 MHz, DMSO) δ 7.84 (1 H, dd, J = 5.7, 1.4 Hz, H-3), 6.20 (1 H, dd, J = 5.7, 1.8 Hz, H-2), 5.15 (1 H, m, H-4), 1.71 (1 H, m, H-5a), 1.55 (1 H, dd, J = 14.3, 6.9 Hz, H-5b), 1.33 (2 H, q, J = 7.5 Hz, H2-11), 1.33 (2 H, H2-6), 1.28 (brd. s), 0.98 (3 H, s, CH3-13), 0.79 (3 H, t, J = 7.5 Hz, CH3-12); 13C NMR (100 MHz, DMSO) δ 173.6 (1-C=O), 159.3 (3-CH), 120.8 (2-CH), 83.7 (4-CH), 71.2 (10-C), 41.5 (9-CH2), 34.4 (5-CH2), 34.4 (11-CH2), 30.1 (7-CH2), 26.9 (13-CH3), 25.0 (6-CH2), 23.8 (8-CH2), 8.8 (12-CH3); HRESIMS: Calculated: m/z 227.1602 [M + H]+; Experimental: m/z 227.1637 [M + H]+.
4,11-Dihydroxy-10-methyl-dodec-2-en-1,4-olide (2): Rf 0.56 (80:20 MeOH:H2O, C-18 silica gel), blue-violet with anisaldehyde-sulfuric acid; UVmax 219.0 (MeOH); 1H NMR (400 MHz, DMSO) δ 7.84 (1 H, dd, J = 5.7, 1.4 Hz, H-3), 6.20 (1 H, dd, J = 5.7, 1.8 Hz, H-2), 5.15 (1 H, m, H-4), 3.42 (2 H, m, H2-11), 1.71 (1 H, m, H-5a), 1.55 (1 H, dd, J = 14.3, 6.9 Hz, H-5b), 1.33 (2 H, H2-6), 1.28 (brd. s), 0.96 (3 H, d, J = 6.3 Hz, CH3-12), 0.77 (3 H, td J = 6.6 Hz, CH3-13); 13C NMR (100 MHz, DMSO) δ 173.6 (1-C=O), 159.3 (3-CH), 120.8 (2-CH), 83.7 (4-CH), 70.0 (11-CH2), 41.0 (10-CH), 34.4 (5-CH2), 32.8 (9-CH2), 30.1 (7-CH2), 27.2 (8-CH2), 25.0 (6-CH2), 19.9 (12-CH3), 15.1 (13-CH3); HRESIMS: Calculated: m/z 227.1602 [M + H]+; Experimental: m/z 227.1637 [M + H]+.
4-Hydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide (3): UVmax 223.0 (MeOH); 1H NMR (400 MHz, DMSO) δ 7.84 (1 H, d, J = 5.9 Hz, H-3), 6.20 (1 H, dd, J = 5.6, 1.9 Hz, H-2), 5.14 (1 H, dd, J = 7.2, 5.1 Hz, H-4), 2.09 (3 H, s, CH3-12), 1.70 (1 H, m, H-5a), 1.55 (1 H, dd, J = 14.9, 7.4 Hz, H-5b), 1.27 (m), 0.98 (3 H, d, J = 6.9 Hz, CH3-13); HRESIMS: Calculated: m/z 225.1446 [M + H]+; Experimental: m/z 225.1486 [M + H]+.