Abstract
The diatom Phaeodactylum tricornutum can accumulate eicosapentaenoic acid (EPA) up to 30% of the total fatty acids. This species has been targeted for isolating gene encoding desaturases and elongases for long-chain polyunsaturated fatty acid (LC-PUFA) metabolic engineering. Here we first report the cloning and characterization of Δ5-elongase gene in P. tricornutum. A full-length cDNA sequence, designated PhtELO5, was shown to contain a 1110 bp open reading frame encoding a 369 amino acid polypeptide. The putative protein contains seven transmembrane regions and two elongase characteristic motifs of FLHXYHH and MYSYY, the latter being typical for microalgal Δ5-elongases. Phylogenetic analysis indicated that PhtELO5 belongs to the ELO5 group, tightly clustered with the counterpart of Thalassiosira pseudonana. Heterologous expression of PhtELO5 in Pichia pastoris confirmed that it encodes a specific Δ5-elongase capable of elongating arachidonic acid and eicosapentaenoic acid. Co-expression of PhtELO5 and IsFAD4 (a ∆4-desaturase from Isochrysis sphaerica) demonstrated that the high-efficiency biosynthetic pathway of docosahexaenoic acid was assembled in the transgenic yeast. Substrate competition revealed that PhtELO5 exhibited higher activity towards n-3 PUFA than n-6 PUFA. It is hypothesized that Phaeodactylum ELO5 may preferentially participate in biosynthesis of transgenic LC-PUFA via a n-3 pathway in the yeast host.
1. Introduction
Polyunsaturated fatty acids (PUFAs) are fatty acids of 18 carbons or more in length with two or more double bonds. They can be classified into n-6 and n-3 major groups, depending on the position of the first double bond proximate to the methyl end of the fatty acid. PUFAs are important structural components that confer membrane fluidity and selective permeability [1,2,3]. Deficiencies in long chain (LC) PUFAs such as DHA and ARA have been associated with disorders in cognitive function, early infancy and various other physiological processes. The n-3 PUFAs including DHA and EPA can be used as preventive drugs and supplements [3,4,5,6]. They can be obtained either through diet of marine fish or synthesized from dietary essential fatty acids. However, the marine fish industry is increasingly declining due to overfishing and environmental pollution. It is imperative to develop sustainable and alternative sources of LC-PUFAs [7,8]. One strategy is to make use of high-efficiency desaturase and elongase genes for transgenic production of LC-PUFAs [7].
The fatty acid desaturases and elongases play key roles in the processes of the conventional Δ6-pathway, alternative Δ8-pathway and microbial Δ4-pathaway [8,9]. The ω-3 (or n-3) LC-PUFA biosynthesis can start with ALA as substrate which can be produced by some yeasts and plants. The biosynthesis of DHA from ALA involves a series of desaturation and elongation reactions catalyzed by various desaturases and elongases. Δ5-elongase and Δ4-desaturase are rate-limiting enzymes for DHA biosynthesis [2,9]. Molecular characterizations of desaturase and elongase in transgenic organisms were explored with species of yeast, microalgae and plants [9,10,11]. Not a few desaturase genes have been characterized from species of mosses, algae, fungi and others [12,13,14,15,16,17,18]. There are few reports about the Δ5-elongases in Pavlova salina, Marchantia polymorpha and Thraustochytrium sp. [19,20,21,22,23]. Nevertheless, so far the Δ5-elongase of Phaeodactylum tricornutum has not been characterized. This diatom is rich in n-3 PUFAs with superior genetic resources revealed by genomic research [24,25]. We aimed to characterize the diatom elongase and desaturase which can be used for LC-PUFA metabolic engineering. In this study, we describe the isolation and characterization of Δ5-elongase gene from Phaeodactylum, and the functional stacking of Δ5-elongase and Δ4-desaturase in transgenic yeast realizing efficient formation of DHA.
2. Results
2.1. Isolation of Gene Encoding Δ5-elongase from P. tricornutum
NCBI search identified a sequence of P. tricornutum mRNA, XM_002176650.1 that predicted a hypothetical protein XP_002176686.1. NCBI conserved domain database (CDD) analysis indicated that this shortened protein belonged to the ELO-super family, sharing 53% identity with the closest homologue in T. pseudonana. In order to isolate the full length cDNA and the putative gene, primers (Table 1) were designed for amplification of target gene and coding sequence using the High Fidelity PCR system (Roche). The PCR fragments were fully sequenced and assembled. The contig1 of 1241 bp was identified from cDNA as a coding sequence containing an open reading frame (ORF) of 1110 bp in length; and the contig2 from genomic DNA is 1661 bp long containing a partial 5′-UTR (415 bp) and a putative structural gene (1212 bp) which includes a 102 bp long intron. Sequencing analysis indicated that two sequences of ORF of 1110 bp were identical and designated PhtELO5.

Table 1.
Microbial strains, plasmids and primers.
| Strain, Plasmid or Primer | Characteristic, Use and Source |
|---|---|
| Strains | |
| E.coli Top10 | E.coli host; for DNA manipulations, Transgene (Beijing, China) |
| P. pastoris GS115 | his4; Invitrogen (Invitrogen China Limited, Beijing, China) |
| PHC01 | GS115 transformed by empty pHBM906 vector, as control |
| PHE5.01 | GS115 carrying pHBM-PtELO5, for gene expression |
| PHE5d.01 | GS115 carrying pHBM-PtELO5-Δ1, for gene expression |
| PHE5d.02 | GS115 carrying pHBM-PtELO5-Δ2, for gene expression |
| PAC01 | GS115 carrying pAO815, as control |
| PAE5.01 | GS115 carrying PtELO5 cassette in pAO815, for expression |
| PDE01 | GS115 carrying FAD4-ELO5 cascade in pAO815, for co-expression |
| Plasmids | |
| pMD18-T | T-cloning vector, Apr, Takara; for gene cloning |
| pHBM906 | Apr, transformation vector for P. pastoris; stored in our lab |
| pHBM-ELO5 | Apr, PCR fragment containing PtELO5 coding sequence, generated with primers Ptelo5-U/ Ptelo5-D, cloned into pHBM906 |
| pAO815 | Apr, HIS4, P. pastoris expression vector with AOX1 promoter and terminator |
| pAO-FAD4 | Apr, PCR fragment containing IsFAD4 coding sequence, generated with primers ISFAD4E-F/ ISFAD4E-R, cloned into pAO815 |
| pAO-ELO5 | Apr, PCR fragment containing PtELO5 coding sequence, generated with primers PTELO5E-F/PTELO5E-R, cloned into pAO815 |
| pT-ELO5 | Apr, PCR fragment containing PtELO5 coding sequence, generated with primers ELO5BGL-F/ELO5BGL-R, cloned into pMD18-T |
| pAO-D4E5 | Apr, BglII digested fragment of PtELO5ORF subcloned into BamHI-digested and dephosphorylated pAO-FAD4 |
| * Primers | |
| Ptelo5-U1 | 5′- GGGAGACCAGATGGTCGACG-3′ |
| Ptelo5-U2 | 5′- TCGCGATACCCCGAATATAT-3′ |
| Ptelo5-U3 | 5′- CAGTTGTCCCTTCAGAACAGC-3′ |
| Ptelo5-U4 | 5′- TCGTGTAGAAGAGCGTGGCG-3′ |
| Ptelo5-D1 | 5′- GCTCTGTAATATAGTGCTCTG-3′ |
| Ptelo5-U | 5′- GTCATGTGTGGTCCCACAGATACAG-3′ |
| Ptelo5-D | 5′- GGCCACTACGARAGACCGGTCATC-3′ |
| Ptelo5-del1F | 5′- GTCgatccacccgtgccctctct-3′ |
| Ptelo5-del2F | 5′- GTCTTGCACAACTGGAAGGTTC-3′ |
| ISFAD4E-F | 5′- CCGCCGGAATTCGCCATGTGCAACGCGGCAGTCG-3′ |
| ISFAD4E-R | 5′- CCGCCGGAATTCTCAATCCGCCTTGAGCGTCTC-3′ |
| PTELO5E-F | 5′- CCGCCGGAATTCGCCATGTGTGGTCCCACAGATAC-3′ |
| PTELO5E-R | 5′- CCGCCGGAATTCCTACGAAGACCGGTCATCCC-3′ |
| ELO5BGL-F | 5′- GGAAGATCTAACATCCAAAGACGAAAGG-3′ |
| ELO5BGL-R | 5′- GGAAGATCTGCACAAACGAACGTCTCAC-3′ |
| Co-F | 5′- GCTCATGATCAACGGGCTCTACCA-3′ |
| Co-R | 5′- TCCCCACACTGCGAAGACACCTAC-3′ |
| 5′AOX1 | 5′- GACTGGTTCCAATTGACAAGC-3′ |
| 3′AOX1 | 5′- GCAAATGGCATTCTGACATCC-3′ |
* Note: Primers Ptelo5-U1 to U4 and -D1 were used for PCR amplifications against putative PhtELO5 structural gene and cDNA; Ptelo5-U, -del1F, -del2F, and -D for PCR-cloning of the full length or truncated PhtELO5 into pHBM906; ISFAD4E-F and ISFAD4E-R for making vector pAO-IsFAD4; PTELO5E-F and PTELO5E-R for making pAO-PhtELO5; ELO5BGL-F and ELO5BGL-R for making ELO5 expression cassette and the subsequent gene stacking cascade; Co-F,Co-R,5′AOX1 and 3′AOX1 in combination with others for PCR and sequencing confirmation of constructs.
2.2. Properties of Putative Elongase PhtELO5
The translated peptide of PhtELO5 was analyzed by protein family (Pfam) search, Clustal W and TMHMM programs. The results indicated that elongase PhtELO5 is an ELO-family protein containing seven transmembrane regions and two characteristic motifs FLHXYHH and MYSYY. The motif MYSYY is common in all Δ5-elongases analyzed indicating that it may be unique for ELO5 elongases (Figure 1). These properties suggested that putative gene PhtELO5 is likely to encode fatty acid Δ5-elongase. Phylogenetic analysis indicated that Δ5-, Δ6- and Δ9-elongases included were generally grouped into respective ELO5-, ELO6- and ELO9 clusters in accordance with their functions. However, there are some exceptions, for example, Marchantia polymorpha elongase MapELO5 and Thrasutcohytrium elongase ELO1 were grouped into ELO6 cluster (Figure 2). Among microalgal Δ5-elongases, PhtELO5 was strongly clustered with T. pseudonana Δ5-elongase (ThpELO5) forming a tight clade but quite different from other ELO5s. Protein sequence distances analyzed using DNA-Star (Lasergene) indicated that PhtELO5 shares the highest identity of 51% with ThpELO5 and low identities of 32.6%–38.2% with other members within the ELO5 group. Interestingly, although Thrasutcohytrium elongase TsELO1 shares the lowest identity of 26.1% with PhtELO5, it reportedly possessed the activity of ELO5. This may be explained by the existence of motif MYSYY in the TsELO1 protein sequence. Together, it is likely that PhtELO5 encodes Δ5-elongase and its evolution history is largely different from those of other microalgae and protists [20,22,23].
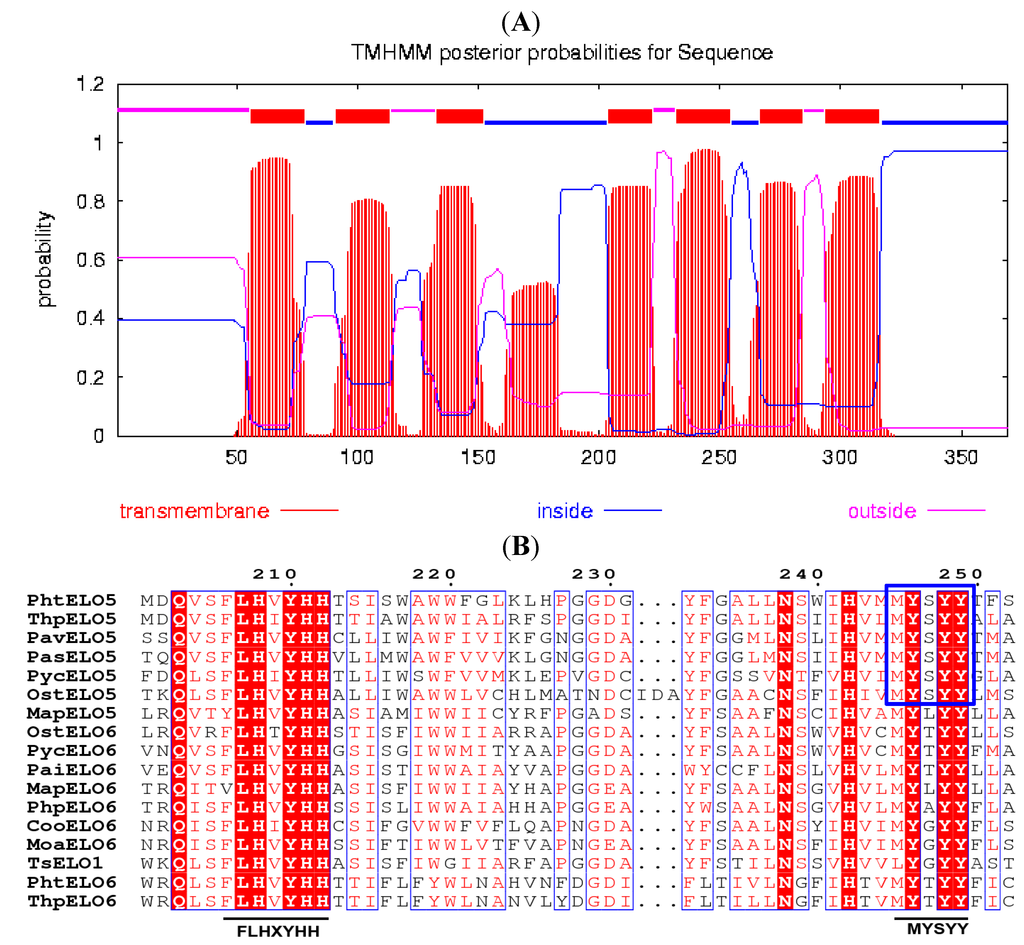
Figure 1.
Properties analysis of PhtELO5. (A) Prediction of seven transmembrane helices in PhtELO5 by online analysis of TMHMM program (Trans-Membrane prediction using Hidden Markov Models). The predicted regions of transmembrane helices are shown in red, other regions are predicted to be either inside (in blue) or outside (in pink) the membrane. (B) Multiple peptide sequence alignment (partly shown) was performed using the Clustal W and ESPript 3, highlighting the typical motifs of Δ5-elongase FLHXYHH and MYSYY which are underlined and boxed. Protein sequences used are the same as that in Figure 2.
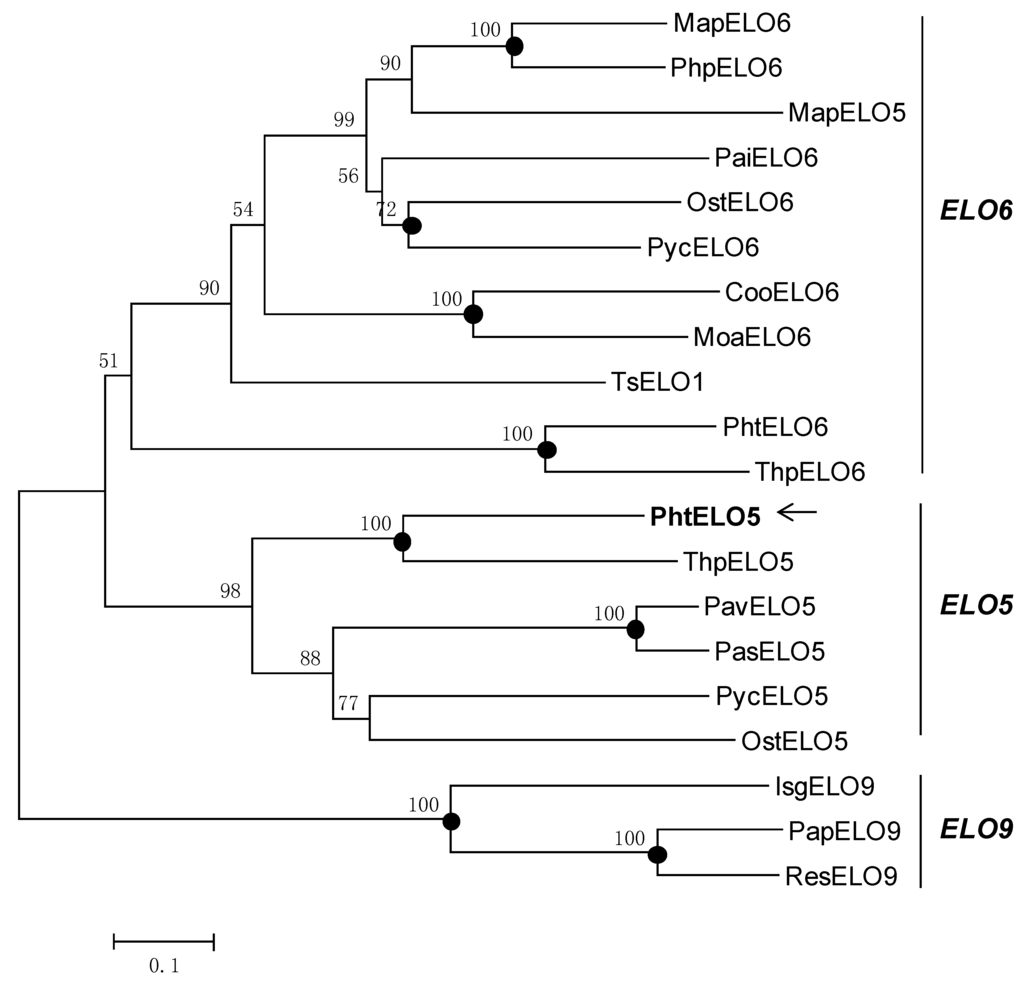
Figure 2.
Phylogenetic dendrogram of fatty acid elongase families. The evolutionary relationship was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown above the branches. Filled circles at nodes indicate phylogenetic branches that were also recovered by using maximum-parsimony algorithms. GenBank accession numbers of the sequences are ABR67690 (PavELO5; Pavlova viridis), AAV33630 (PasELO5; Pavlova sp. CCMP459), ACR53360 (PycELO5; Pyramimonas cordata), BAE71129 (MapELO5; Marchantia polymorpha), AAV67798 (OstELO5; Ostreococcus tauri), AAV67800 (ThpELO5; Thalassiosira pseudonana), AAT85662 (MapELO6; Marchantia polymorpha), AAW70157 (PtELO6; Phaeodactylum tricornutum), XP_003074750 (OstELO6; Ostreococcus tauri), AAV67799 (ThpELO6; Thalassiosira pseudonana), ACK99719 (PaiELO6; Parietochloris incisa), XP_001780388 (PhpELO6, Physcomitrella patens), AEA07666 (CooELO6; Conidiobolus obscurus), ACR53359 (PycELO6; Pyramimonas cordata), ADE06662 (MoaELO6; Mortierella alpina), ADN94475 (PapELO9; Pavlova pinguis), ADN94476 (ResELO9; Rebecca salina), AAL37626 (IsgELO9; Isochrysis galbana). TsELO1 (Thrasutcohytrium sp. ATCC26185, ref 22); PtELO5 (Phaeodactylum tricornutum, arrow shows this study).
2.3. Functional Analysis in Yeast: Confirmation of PhtELO5’s Activity as Δ5-Elongase
To verify whether the putative PhtELO5 has Δ5-elongase activity or not, we heterologously expressed this gene in P. pastoris. ELO5 expression vector was constructed by using plasmid pHBM906 which allows ELO5 ORF to be driven by the inducible AOX1 promoter [26]. Transgenic yeast cells expressing the PhtELO5 ORF were fed with precursor fatty acids (Δ5-) C20 PUFAs (referring to ARA and EPA [9,12]) and then were analyzed for fatty acid profiling. Clearly, control strain with empty vector pHBM906 saw no new fatty acids formed, whereas the ELO5-expressing cells fed with ARA or EPA were shown to have a detectable amount of DTA or DPA accordingly (Figure 3 and Figure 4, and Table 2). This result indicated that upon addition of (Δ5-) C20 PUFA substrate new fatty acids of DTA and DPA were formed as immediate products. However, it appeared that PhtELO5 displayed a higher rate of converting EPA into DPA(n-3) (93.1%) than that of converting ARA into DTA (79.4%). The difference in efficiency was confirmed by using another expression vector pAO815, which exhibited the rate of 90.8% and 83.2%, respectively (Table 3). It is likely that the difference in conversion efficiency may reflect PhtELO5’s substrate preference.
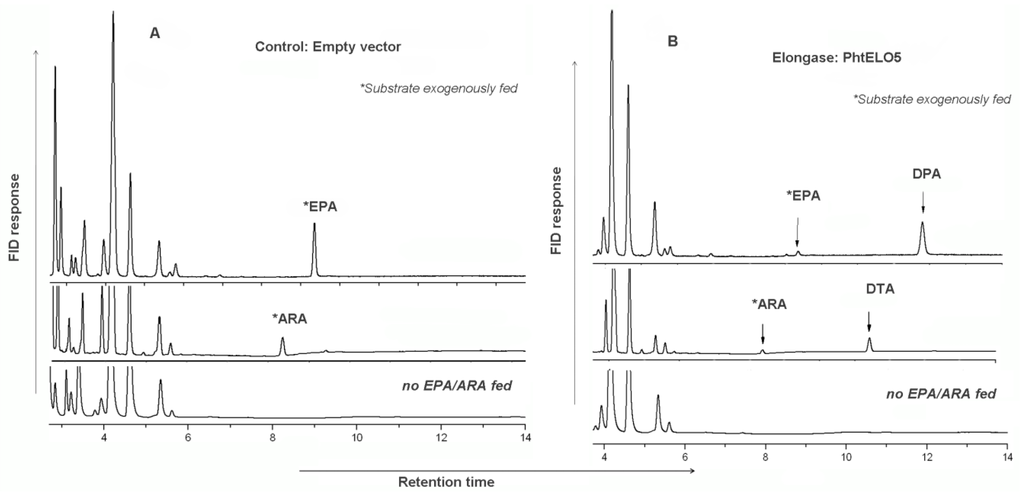
Figure 3.
Comparison of fatty acid profiles of the control and PhtELO5-expressing Pichia cells. The representative control strain PHC01 (transformed with empty vector pHBM906) and representative ELO5-expressing strain PHE5.01 were grown for 3 days with or without adding substrate and subjected to FA analysis. Samples of 100 µM of (Δ5-) C20 PUFAs were exogenously fed as substrates. (A): control strain PHC01; (B): PhtELO5-expressing strain PHE5.01. Fatty acid profiles of immediate product from substrate were clearly observed in gene expressed cells but not in controls. Stars indicate substrates which were initially fed and detected as left-over in samples.
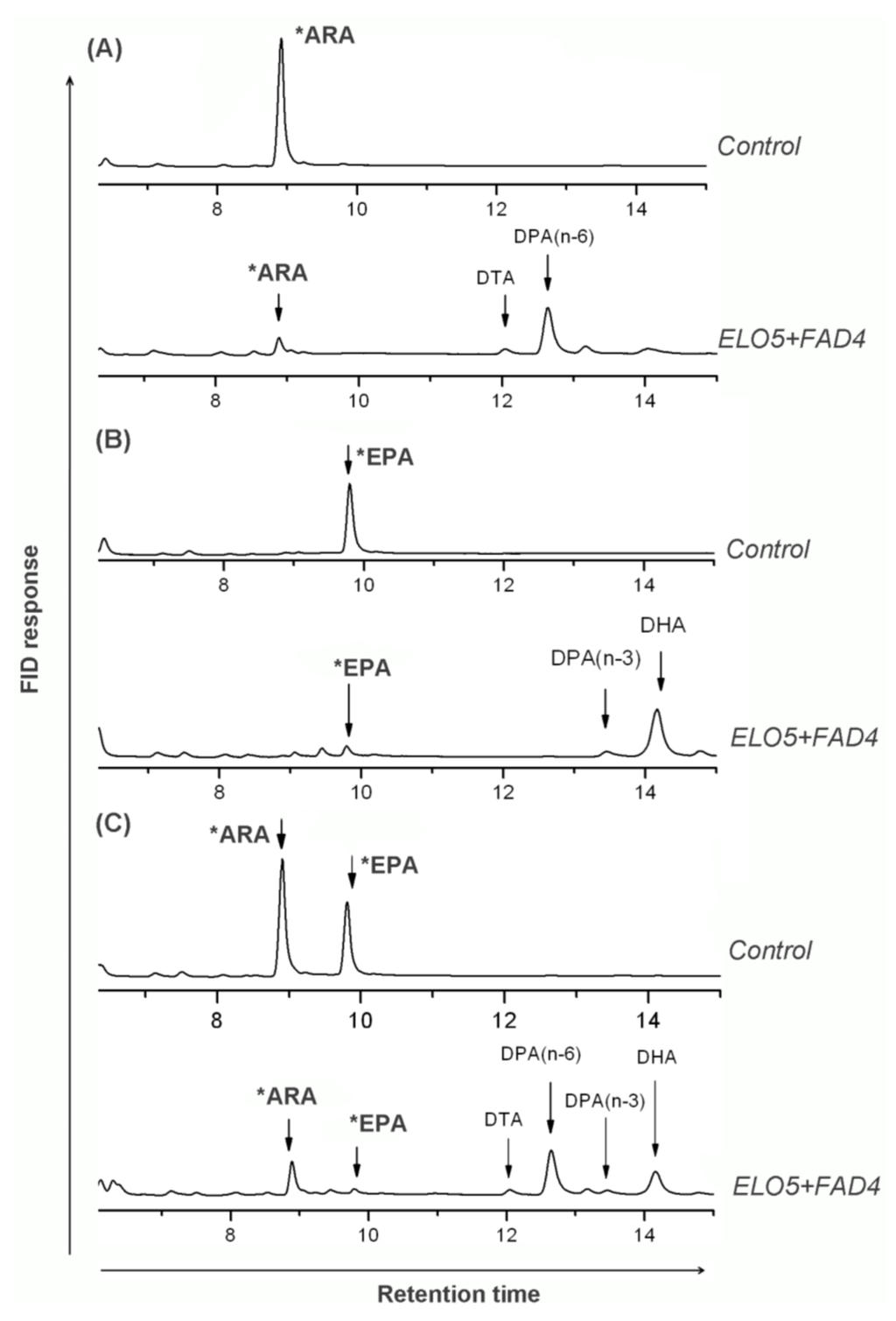
Figure 4.
Comparison of fatty acid profiles (partly shown) of the control and co-expressed Pichia cells fed with substrates. Control strain PAC01 (transformed with empty vector pAO815) and co-expressed strain PDE01 (transformed with pAO-D4E5) were fed with 100 µM of (Δ5-) C20 PUFAs and grown for 3 days, followed by FA analysis. Shown are fatty acid GC profiles of indicated strains fed with ARA (A), EPA (B) and ARA & EPA (C). Fatty acid profiles of immediate product from substrate were clearly observed in co-expressed cells but not in controls. Stars indicate substrates which were initially fed and detected as left-over in samples.

Table 2.
Fatty acid composition of transgenic P. pastoris GS115.
| Fatty Acid Composition
(% of Total Fatty Acids) | P. pastoris with Plasmids | |||||||
|---|---|---|---|---|---|---|---|---|
| PAC01(E) | PDE01(E) | PAC01(A) | PDE01 (A) | PAC01(EA) | PDE01(EA) | PAC01(−) | PDE01(−) | |
| C14:0 | 9.75 ± 0.13 | 15.56 ± 0.23 | 9.77 ± 0.11 | 16.2 ± 0.09 | 10.12 ± 0.24 | 17.12 ± 0.23 | 8.98 ± 0.19 | 13.79 ± 0.24 |
| C16:0 | 3.01 ± 0.13 | 10.08 ± 0.32 | 2.98 ± 0.15 | 9.90 ± 0.14 | 3.11 ± 0.18 | 9.29 ± 0.27 | 3.48 ± 0.12 | 9.25 ± 0.21 |
| C17:0 | 1.01 ± 0.07 | 1.24 ± 0.04 | 1.23 ± 0.05 | 1.13 ± 0.15 | 1.31 ± 0.13 | 1.47 ± 0.09 | 1.36 ± 0.09 | 1.09 ± 0.04 |
| C17:1 | 2.90 ± 0.18 | 1.13 ± 0.15 | 3.01 ± 0.17 | 1.13 ± 0.15 | 3.32 ± 0.14 | 1.59 ± 0.11 | 2.50 ± 0.10 | 0.81 ± 0.08 |
| C18:0 | 6.89 ± 0.33 | 6.03 ± 0.39 | 7.24 ± 0.32 | 7.44 ± 0.22 | 7.54 ± 0.32 | 6.69 ± 0.23 | 6.80 ± 0.22 | 6.43 ± 0.10 |
| C18:1 n-9 | 2.87 ± 0.14 | 2.31 ± 0.18 | 2.79 ± 0.12 | 2.00 ± 0.11 | 3.07 ± 0.16 | 2.90 ± 0.19 | 2.24 ± 0.16 | 1.77 ± 0.07 |
| C18:1 n-7 | 42.86 ± 2.68 | 34.04 ± 2.01 | 39.13 ± 2.05 | 32.95 ± 2.92 | 38.55 ± 1.63 | 29.10 ± 1.92 | 38.14 ± 2.398 | 33.46 ± 1.73 |
| C18:2 n-6 | 19.09 ± 0.75 | 21.31 ± 0.67 | 18.67 ± 0.33 | 20.28 ± 0.64 | 17.27 ± 0.47 | 17.32 ± 0.69 | 28.63 ± 1.18 | 27.22 ± 0.73 |
| C18:3 n-3 | 4.62 ± 0.13 | 4.39 ± 0.19 | 4.68 ± 0.19 | 4.07 ± 0.13 | 4.16 ± 0.23 | 4.14 ± 0.16 | 6.14 ± 0.43 | 4.69 ± 0.08 |
| C20:0 | 1.79 ± 0.19 | 1.32 ± 0.23 | 1.26 ± 0.23 | 1.67 ± 0.12 | 1.74 ± 0.14 | 1.66 ± 0.11 | 1.72 ± 0.08 | 1.51 ± 0.05 |
| C20:4 n-6 (ARA) | ND | ND | 7.82 ± 0.23 | 0.54 ± 0.04 | 3.32 ± 0.17 | 1.56 ± 0.05 | ND | ND |
| C20:5 n-3 (EPA) | 5.27 ± 0.02 | 0.26 ± 0.02 | ND | ND | 5.31 ± 0.02 | 0.30 ± 0.02 | ND | ND |
| C22:4 n-6 (DTA) | ND | ND | ND | 0.24 ± 0.01 | ND | 0.30 ± 0.02 | ND | ND |
| C22:5 n-6 (DPA) | ND | ND | ND | 2.44 ± 0.03 | ND | 3.00 ± 0.07 | ND | ND |
| C22:5 n-3 (DPA) | ND | 0.24 ± 0.03 | ND | ND | ND | 0.34 ± 0.01 | ND | ND |
| C22:6 n-3 (DHA) | ND | 2.35 ± 0.05 | ND | ND | ND | 1.82 ± 0.03 | ND | ND |
Note: (1) Cells transformed with empty vector pAO815 (representative strain PAC01) or recombinant vector pAO-D4E5 (representative strain PDE01) grown on different substrate containing media were tested. (2) PAC01(E): Strain PAC01 with EPA; PDE01(E): Strain PDE01 with EPA; PAC01(A): Strain PAC01 with ARA; PDE01(A): Strain PDE01 with ARA; PAC01(EA): Strain PAC01with EPA and ARA; PDE01(EA): Strain PDE01 with EPA and ARA; PAC01(−): Strain PAC01 without adding substrate; PDE01(−): Strain PDE01 without adding substrate. ND: not detected or not detectable.

Table 3.
Substrate conversion rates by FAD4-ELO5 co-expressed transgenic yeast.
| Conversation Rate (%) | ||
|---|---|---|
| Addition of Single Substrate | Addition of Double Substrates | |
| EPA→DPA(n-3) | 90.8 | 87.9 |
| DPA(n-3)→DHA | 90.8 | 84.4 |
| ARA→DTA | 83.2 | 67.9 |
| DTA→DPA(n-6) | 90.9 | 90.9 |
Based on domain analysis of PhtELO5, we carried out domain deletion experiments to verify the function of key ELO-conserved regions. Yeast cells expressing the truncated ELO5 deleted for 4VR region (4-vinyl reductase, ELO5-Δ1) or ELO5 deleted for 4VR region plus part of the first transmembrane helix (ELO5-Δ2) were shown to have no alteration in fatty acids profiling (Figure 5). The results indicated that at least in transgenic yeast, deletion of N-terminal 4VR domain and partial transmembrane regions had little influence on the activity of Δ5-elongase. This is in support of the prediction that PhtELO5’s core activity regions range from amino acid 90–328. Together, it is concluded that PhtELO5 indeed possesses the activity of Δ5-elongase as expected.
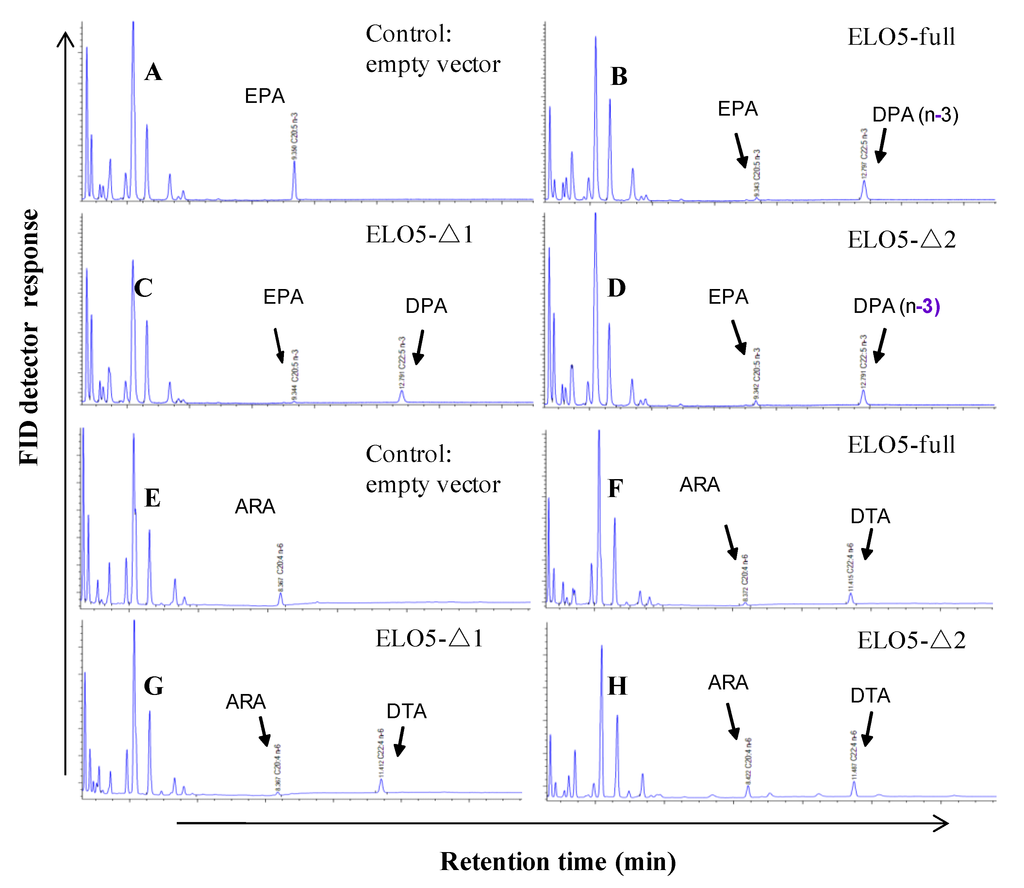
Figure 5.
Fatty acid profiling patterns of transgenic Pichia cells expressing various versions of PhtELO5. Yeast cells transformed with empty vector pHBM906, and expression vectors containing ELO5-full length, ELO5-Δ1 and ELO5-Δ2 were fed with (Δ5-) C20 PUFAs and analyzed for FA composition. (A) to (D): indicated strains fed with EPA; (E) to (H): indicated strains fed with ARA.
2.4. Co-expression of PhtELO5 and IsFAD4 Assembled Function of EPA and DHA Biosynthetic Pathway in Transgenic Yeast
Having confirmed the specificity of PhtELO5 in yeast by single gene expression, we wanted to know further what its performance would be in an assembled EPA and DHA biosynthetic pathway. We thus co-expressed microalgal Δ5-elongase and Δ4-desaturase in yeast to see if they were functional in a non-native system. The co-expression vector was successfully constructed using the stable vector pAO815 [26], which allowed PhtELO5 and IsFAD4 cassettes to be stacked in a cascade through linker sites of BglII and BamHI. The resultant target construct pAO-D4E5 contained ELO5 and FAD4 as a cascade driven by strong inducible promoter pAOX1 (Supplementary Information, Figure S1).
Yeast can only synthesize the shortest PUFAs like LA and ALA, it needs the addition of (Δ5-) C20-PUFAs to continue the DHA biosynthesis via microbial Δ4-pathway [9]. Control strain PAC01 and ELO5-FAD4 co-expressed strain (PDE01) were exogenously fed with ARA and/or EPA. Fatty acid analysis indicated that samples of control cells were unable to detect any new fatty acids formed. In contrast, samples of strain PDE01 expressing ELO5 and FAD4 were new fatty acids detected upon the addition of (Δ5-) C20-PUFAs (Figure 4 and Table 2). For instance, fed with ARA alone, strain PDE01 was shown to produce DTA and n-6 DPA in comparison with control samples where no new fatty acids identified; this indicated that co-expression indeed led to the conversion of single substrate ARA into intermediate product DTA and the latter subsequently into product n-6 DPA (Figure 4A). Fed with single substrate EPA, n-3 DPA and DHA were identified as new constituents besides the left-over EPA in FAME samples of strain PDE01. In this case, product n-3 DPA was formed from EPA and subsequently transformed into DHA (Figure 4B). Consistently, fed with ARA and EPA as dual substrates, four above mentioned products and two left-over substrates were all detected in PDE01 FAME samples. Taken together, this suggested that the co-expression successfully reconstituted the function of both Δ5 elongation and Δ4 desaturation orientating the n-3 pathway.
2.5. PhtELO5’s Converting Efficiencies Varied with Substrates
To test the effect of substrate competition, cells of double gene co-expressed strain PDE01 were fed with the same molar concentration of ARA and EPA concurrently. FA compositions and converting rates were determined to relatively estimate the enzymatic activity or preference upon type of (Δ5-) C20-PUFA substrates. As revealed in Table 2 and Table 3, in the presence of single substrate, the rates of conversion EPA→DPA (n-3), DPA (n-3) →DHA, ARA→DTA and DTA→DPA (n-6) were 90.8%, 90.8%, 83.2% and 90.9%, respectively. However in the presence of dual substrates, the rate of conversion (ARA→DTA) dropped drastically from 83.2% to 67.9%, while conversion rates of other substrates showed little change, ranging from 84.4% to 90.9%. The relatively lower conversion rate upon ARA was also observed in ELO5 single gene expression in strain PHD5.01 (79.4% versus 93.1%). Obviously, compared to EPA, fewer molecules of ARA were incorporated with elongase PhtELO5. It is interpreted that PhtELO5 is likely to have preference for EPA among (Δ5-) C20-PUFA substrates. This finding is consistent with the conclusion that Δ5-elongase is one of the rate-limiting enzymes involved in LC-PUFA biosynthesis [2,25].
3. Discussion
Diatoms are successful groups of unicellular eukaryotic algae playing important roles in global carbon and silica pools. P. tricornutum is one of the most widely utilized model systems for studying the ecology, physiology, and molecular biology of diatoms. Although the genome of this model diatom has been sequenced, the current knowledge on fatty acid metabolic organization and biochemistry remains fragmentary [24,25]. Since the availability of the genome, the PUFA synthesis associated enzymes such as thioesterases, elongases, desaturases, acyl-CoA synthetases and acyltransferases have been increasingly explored [19,23,25]. However, P. tricornutum’s fatty acid Δ5-elongase has not been investigated. We therefore conducted the identification and function analysis of P. tricornutum Δ5-elongase.
We present herein three lines of evidence verifying that PhtELO5 functions as fatty acid Δ5-elongase. First, sequence analysis indicated that ELO5 is an ELO-family protein, having two typical motifs of FLHXYHH and MYSYY. The motif MYSYY is the typical characteristic for all microalgal Δ5-elongases reported; and it may be crucially required for its enzymatic activity. It is supported by the properties of Thrasutcohytrium TsELO1. Although the protein sequence of TsELO1 is largely distant from most Δ5-elongases, it possesses quite high activity of C20-Δ5 elongase [22]. This is likely due to the existence of motif MYSYY. Second, results of PhtELO5 single and double gene expression in yeast not only verified that PhtELO5 indeed possesses the activity of Δ5-elongase, but also demonstrated that gene stacking of microalgal Δ5-elongase and Δ4-desaturase can reconstitute the function of the EPA and DHA biosynthetic pathway in the yeast host. Upon addition of (Δ5-) C20-PUFA substrates, both ELO5 single- and double-expression strains were able to form new fatty acids of C22 PUFAs, strongly suggesting that PhtELO5 was a key player in orienting DHA synthesis viaΔ4-pathway [9]. Third, PhtELO5 was shown to have higher conversion efficiencies upon EPA than ARA, implying its preference for n-3 C20-PUFA. This observation is quite different from ELO5 of Pavlova and Thalassiosira which appeared to have little differences on (Δ5-) C20-PUFA substrates [20,21]. To check if PhtELO5 would have activity on saturated C20 fatty acid, we did the C20:0 feeding experiment and the results demonstrated that this saturated fatty acid is not the substrate of PhtELO5 (Supplementary Information, Figure S2).
In this study a high efficiency of converting (Δ5-) C20-PUFA substrates was repeatedly observed in ELO5 single and ELO-FAD4 double expressing cells. This is largely different from ELO5s of other microalgae where the conversion rates were generally less than 50% [12,20]. The high conversion rate may reflect the high enzymatic activity of PhtELO5 and IsFAD4. Initially we wanted to co-express Phaeodactylum Δ4-desaturase and Δ5-elongase but failed to get Δ4-desaturase functional; thus we then made use of previously described I. sphaerica Δ4-desaturase which had been proven to be efficient in converting substrate DPA [18]. Furthermore, PhtELO5 and IsFAD4 were constructed under a strong inducible promoter which may have facilitated the functional stacking of both enzymes in Pichia. Gene stacking through a combined construct was proven to have advantages such as fewer selective markers and higher expression level [27,28]. However, such very high conversion efficiencies were probably attributed to the continuous strong inducing condition we applied: cells were batch-fed with methanol every 24 h. Expression patterns of target genes may support the explanation. By using Quantitative real-time PCR, mRNA levels of ELO5 and FAD4 were shown to be steadily high over the period of cultivation, indicating that the target genes were stably expressed at a high level under current inductions (Supplementary Information, Figure S3). All these factors have contributed to maintaining the high activities of Δ5-elongase and Δ4-desaturase in the Pichia host.
In nature, yeast including P. postoris cannot synthesize PUFAs longer than LA and ALA [2,9]. Introduction of microalgal ELO5 and FAD4 into yeast cells can reconstruct the functional routes orienting EPA and DHA biosynthesis, as part of the microbial Δ4-pathway (Figure 6). Based on our finding, it is concluded that synthesis of C22 PUFAs can be realized via functional assembly of ELO5/FAD4 in the presence of (Δ5-) C20 PUFA substrates. ELO5 is likely to have a higher converting activity upon n-3 C20 PUFA substrate, whereas FAD4 activity remains comparable in both n-3 and n-6 pathways (Table 3, Figure 6). It is postulated that in transgenic yeast, Phaeodactylum ELO5 may determine the n-3 pathway as a dominant synthetic route for LC-PUFA production.
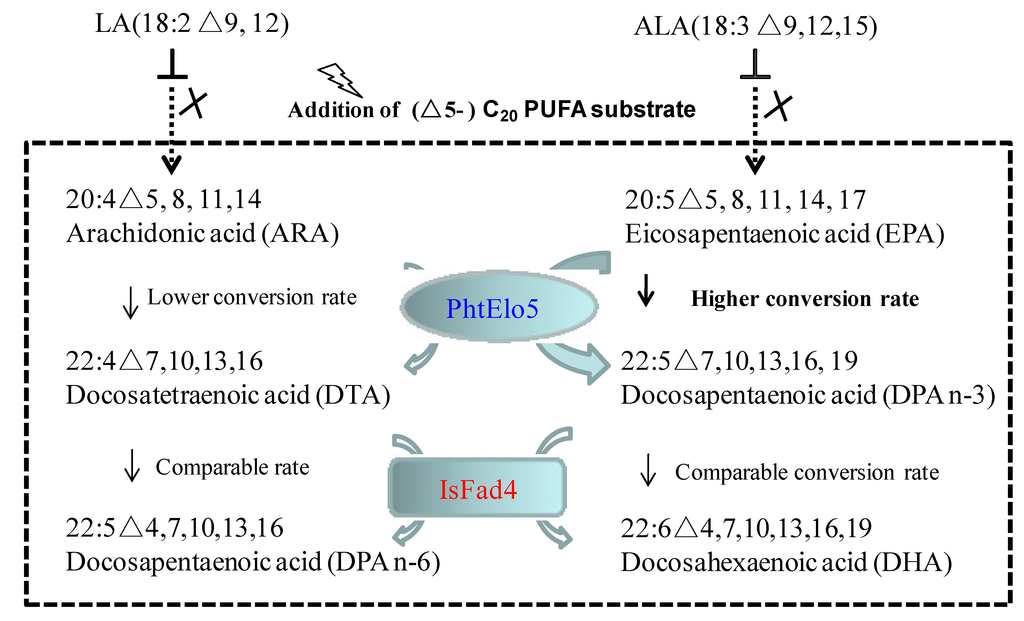
Figure 6.
Schematic illustration of the heterologous assembly of microalgal ELO5 and FAD4 implementing EPA and DHA syntheses in transgenic yeast.
4. Experimental Section
4.1. Strains and Culture Conditions
The diatom P. tricornutum Pt9 (CCMP633) was obtained from the Institute of Hydrobiology, Chinese Academy of Sciences (Wuhan, China). P. tricornutum was grown in f/2-enriched artificial sea water (f/2AW, pH8.5) medium at 25 °C with continuous aeration and illumination (50 mol photons m−2s−1, by cool daylight fluorescent tubes). P. pastoris strain GS115 was grown and maintained in YPD medium.
4.2. Sequence Analysis
DNA or protein sequence alignment and neighbor-joining tree construction were done with Clustal W and MEGA 5.2.2. Sequence distances were analyzed by DNA-star. Structural domain prediction was carried out using the online server program TMHMM and ESPript 3.
4.3. Identification and Isolation of the Δ5-Elongase cDNA from P. tricornutum
DNA, RNA and cDNA preparation were performed by standard protocols. Molecular tool enzymes and reagent kits were used according to instructions provided by the manufacturers. At the time of study there was only a partial sequence of P. tricornutumt mRNA available (NCBI Reference Sequence: XM_002176650.1). This partial start and partial stop fragment was linked to a sequence in chromosome 1. In order to isolate its full length cDNA and putative gene, three forward primers Phtelo5-U1, -U2 and -U3 and one reverse primer Phtelo5-D1 (Table 1) were designed for DNA amplification against genomic DNA and the first strand cDNA. For this purpose, the High Fidelity PCR system (Roche) was employed to minimize the error rate. The specific PCR products were gel purified and cloned into pMD18-T vector (TaKaRa, Dalian, China) after the fragments were prepared by the A-tailing procedure. Inserts were then fully sequenced with complete coverage. Assembly of the sequences allowed the identification of a full length of ORF from cDNA and the coding sequences. It was designated PhtELO5 and subjected to cloning into yeast expression vectors.
4.4. Plasmid Construction and Transformation of Yeast Cells
Plasmids and primers used in this study are featured in Table 1. PCR fragments were cloned into pMD18-T (Takara) and confirmed by sequencing. To characterize the function of ELO5, the PhtELO5 was cloned into pHBM906 and expressed in P. pastoris. Briefly, PhtELO5 was cloned into pMD18-T through cloning sites of NotI and CpoI to generate pMD-ELO5. The full length of PhtELO5 was released from pMD-ELO5 by NotI and CpoI digestions, and subcloned into NotI and CpoI-cut pHBM906 (P. pastoris expression vector). In the resultant vector pHBM-ELO5, ELO5 was placed in between the promoter and transcription terminator regions of the AOX1 gene. Linearized by SalI, pHBM-ELO5 was introduced into P. pastoris strain GS115 by electroporation (Bio-Rad). Concurrently, the empty pHBM906 vector was introduced into GS115 as control. For co-expression of ELO5 and FAD4 in P. pastoris, the expressing vector pAO815 was used to stack the two genes in a cascade. First, IsFAD4 and PtELO5 were individually cloned into pAO815 via EcoRI cloning site to form vector pAO-FAD4 and pAO-ELO5, respectively. Then, pAO-FAD4 was digested with BamHI followed by dephosphorylation with calf intestine alkaline phosphatase according to the manufacturer’s instructions; and PhtELO5 expression cassette (5′AOX1-PhtELO5-TT) was amplified from vector pAO-ELO5 with primer ELO5BGL-F and ELO5BGL-R using a high fidelity PCR. Finally, after digestion with BglII, the PhtELO5 cassette was ligated to BamHI-digested and dephosphorylated vector pAO-FAD4, resulting in the target vector designated pAO-D4E5. Various PCR and sequencing were carried out to confirm the accuracy of sequence and structure of the stacked cassettes prior to the transformation.
4.5. Heterologous Expression of PtELO5 and IsFAD4 in P. pastoris
To verify the function of PhtELO5, the recombinant vectors pHBM-ELO5 and/or pAO-D4E5 were introduced in Pichia pastoris. The representative strains of three independent positive transformants were selected and grown on 50 mL MGY liquid medium (1.34% YNB with amino acids and ammonium sulfate, 10−5% biotin and 1% glycerol) on a shaker (220 rpm) at 28 °C. Cultures of the stationary phase were harvested and washed with sterile deionized water twice and inoculated (at final rate of OD600 = 1) into the 500 mL glass Erlenmeyer flasks containing 50 mL MM medium (1.34% YNB with amino acids and ammonium sulfate, 10−5% biotin and 0.5% methanol). Methanol, 0.5% (v/v), was supplemented every 24 h to keep inducing gene expression. All yeast cultures were grown for 72 or 96 h at 20 °C and used for fatty acid analysis.
Heterologous gene expressions were estimated by Quantitative real-time PCR. Total RNA was isolated from cells using the TRIzol reagent (Invitrogen China Limited, Beijing, China). First-strand cDNA was synthesized using a PrimeScript RT reagent kit with gDNA eraser (TaKaRa Biotechnology (Dalian), Dalian, China). qRT-PCR was performed using iCycler iQ5 real-time PCR system (Bio-Rad, Hercules, CA, USA) and SYBR Premix Ex Taq II (Tli RNaseH plus kit) (TaKaRa Biotechnology (Dalian), Dalian, China), according to the manufacturer’s protocols. Full-length PhtELO5 or IgFAD4 cDNA was amplified with primer pairs listed in Table 1. The yeast actin gene ACT1 was used as an internal standard. The relative mRNA level of the target gene was normalized against that of standard.
4.6. PUFA Substrate Feeding
P. pastoris transgenic strains carrying various expressing vectors were grown in BMGY medium individually. Each overnight culture was diluted to an OD600 of 0.5 and refreshed by growing for further hours till the OD600 value reached approximately 1.0. The cultures were harvested by centrifugation and resuspended in the same volume of fresh BMGY medium containing 1% NP40, 0.5% methanol and exogenously fed with 100 µM each of (Δ5-) C20 FUFAs. The cultures were grown for 72–96 h with addition of 0.5% methanol every 24 h. Cell were harvested, washed once with three volumes of 0.5% Triton X-100, and once with three volumes of distilled water. The pellets were subjected to analysis of fatty acid composition (% of total fatty acids).
4.7. Fatty Acid Analysis
Microalgae or yeast cells were dried by the Vacuum Freeze-drying System. Fatty acid methyl esters (FAMEs) were extracted with petroleum ether and trans-methylated with 0.4 M NaOH in methanol [29]. All samples were analyzed using a 7890A gas chromatography (Agilent technologies, Santa Clara, CA, USA) equipped with a flame ionization detector (FID) and an HP-FFAP capillary column (30m × 250 μm × 0.25 μm). High purity nitrogen was used as carrier gas. Standards of fatty acid mixtures were purchased from Sigma-Aldrich (St. Louis, MO, USA). FAMEs were identified by comparison of retention times with those of the authentic standards. The relative amount of FAME was quantified by comparing each peak area with that of the internal standard. 1 μg/μL of behenic acid (22:0) was added into samples as internal standard. The conversion efficiency was calculated as percentage of the relative amount of product divided by the sum of product and substrate (which is left-over), namely conversion rate (%) = [product]/([substrate] + [product]) × 100% [18,30].
5. Conclusions
We herein report the characterization and functional analysis of P. tricornutum Δ5-elongase gene PhtELO5. Heterologous expression in Pichia confirmed that ELO5 possesses strong activity of Δ5-elongase capable of elongating (Δ5-) C20 PUFA substrates. Substrate competition revealed that PhtELO5 had a higher converting activity towards 20:5n-3 than 20:4n-6 fatty acids. Functional stacking of Phaeodactylum ELO5 and Isochrysis FAD4 in Pichia reconstituted a high-efficiency biosynthetic pathway leading to transgenic production of DHA.
Abbreviations
| ARA | arachidonic acid |
| ALA | α-linolenic acid |
| DHA | docosahexaenoic acid |
| DTA | Docosatetraenoic acid |
| DPA | docosapentaenoic acid |
| EPA | eicosapentaenoic acid |
| FA | fatty acid |
| FAMEs | Fatty acid methyl esters |
| FID | flame ionization detector |
| GC-MS | Gas Chromatography-Mass Spectrometry |
| LA | linoleic Acid |
| LC | long chain |
| ORF | open reading frame |
| PUFA | polyunsaturated fatty acid |
Supplementary Files
Acknowledgments
This study was supported by National 863 High Tech Project of China (2011AA100904) and the National Natural Science Foundation of China (31270346). This work was partially funded by Special Grant of Oil Crops Research Institute (2013HT005). We thank Yu Longjiang of Huazhong University of Science and Technology, and Hu Hanhua of Institute of Hydrobiology, Chinese Academy of Sciences for the gifts of strains, plasmid and reagents.
Author Contributions
Conceived and designed the experiments: MLJ, BG, CJH. Performed the experiments: MLJ, BG. Analyzed the data: BG, MLJ, CJH. Contributed reagents/materials/analysis tools: XW, YMG, YBZ. Wrote and discussed on the final form of the manuscript: CJH, MLJ, YMG, XW, YBZ, BG.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bistrian, B.R. Clinical aspects of essential fatty acid metabolism: Jonathan Rhoads Lecture. J. Parenter. Enteral. Nutr. 2003, 27, 168–175. [Google Scholar] [CrossRef]
- Leonard, A.E.; Pereira, S.L.; Sprecher, H.; Huang, Y.S. Elongation of long-chain fatty acids. Prog. Lipid Res. 2004, 43, 36–54. [Google Scholar] [CrossRef]
- Lands, B. Consequences of essential fatty acids. Nutrients 2012, 4, 1338–1357. [Google Scholar] [CrossRef]
- Agbaga, M.P.; Mandal, M.N.; Anderson, R.E. Retinal very long-chain PUFAs: New insights from studies on ELOVL4 protein. J. Lipid Res. 2010, 51, 1624–1642. [Google Scholar] [CrossRef]
- Kihara, A. Very long-chain fatty acids: Elongation, physiology and related disorders. J. Biochem. 2012, 152, 387–395. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Wu, J.H. (n-3) fatty acids and cardiovascular health: Are effects of EPA and DHA shared or complementary? J. Nutr. 2012, 142, 614S–625S. [Google Scholar] [CrossRef]
- Nevejan, N.; Saez, I.; Gajardo, G.; Sorgeloos, P. Supplementation of EPA and DHA emulsions to a Dunaliella tertiolecta diet: Effect on growth and lipid composition of scallop larvae, Argopecten purpuratus (Lamarck, 1819). Aquaculture 2003, 217, 613–632. [Google Scholar] [CrossRef]
- Williams, C.M.; Burdge, G. Long-chain n-3 PUFA: Plant v. marine sources. Proc. Nutr. Soc. 2006, 65, 42–50. [Google Scholar] [CrossRef]
- Sayanova, O.; Napier, J.A. Transgenic oilseed crops as an alternative to fish oils. Prostaglandins Leukot. Essent. Fatty Acids 2011, 85, 253–260. [Google Scholar] [CrossRef]
- Sandager, L. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002, 277, 6478–6482. [Google Scholar] [CrossRef]
- Tonon, T.; Harvey, D.; Larson, T.R.; Graham, I.A. Identification of a very long chain polyunsaturated fatty acid Delta4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 2003, 553, 440–444. [Google Scholar] [CrossRef]
- Meyer, A.; Kirsch, H.; Domergue, F.; Abbadi, A.; Sperling, P.; Bauer, J.; Cirpus, P.; Zank, T.K.; Moreau, H.; Roscoe, T.J.; et al. Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J. Lipid Res. 2004, 45, 1899–1909. [Google Scholar] [CrossRef]
- Hsiao, T.Y.; Holmes, B.; Blanch, H.W. Identification and functional analysis of a delta-6 desaturase from the marine microalga Glossomastix chrysoplasta. Mar. Biotechnol. 2007, 9, 154–165. [Google Scholar] [CrossRef]
- Lu, Y.; Chi, X.; Yang, Q.; Li, Z.; Liu, S.; Gan, Q.; Qin, S. Molecular cloning and stress-dependent expression of a gene encoding Delta(12)-fatty acid desaturase in the Antarctic microalga Chlorella vulgaris NJ-7. Extremophiles 2009, 13, 875–884. [Google Scholar] [CrossRef]
- Domergue, F.; Lerchl, J.; Zähringer, U.; Heinz, E. Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur. J. Biochem. 2002, 269, 4105–4113. [Google Scholar] [CrossRef]
- Wan, X.; Zhang, Y.; Wang, P.; Jiang, M. Molecular cloning and expression analysis of a delta 6-fatty acid desaturase gene from Rhizopus stolonifer strain YF6 which can accumulate high levels of gamma-linolenic acid. J. Microbiol. 2011, 49, 151–154. [Google Scholar] [CrossRef]
- Magny, S.; Thomassen, D.R.; Berge, G.M.; Ostbye, T.K.; Ruyter, B. High dietary EPA does not inhibit Δ5 and Δ6 desaturases in Atlantic salmon (Salmo salar L.) fed rapeseed oil diets. Aquaculture 2012, 360, 78–85. [Google Scholar]
- Guo, B.; Jiang, M.; Wan, X.; Gong, Y.; Liang, Z.; Hu, C. Identification and Heterologous Expression of a Δ4-Fatty Acid Desaturase Gene from Isochrysis sphaerica. J. Microbiol. Biotechnol. 2013, 23, 1413–1421. [Google Scholar] [CrossRef]
- Yamato, K.T.; Sakai, Y.; Fukuzawa, H.; Ohyama, K.; Kohchi, T. Isolation and functional characterization of fatty acid delta5-elongase gene from the liverwort Marchantia polymorpha L. FEBS Lett. 2006, 580, 149–154. [Google Scholar] [CrossRef]
- Robert, S.S.; Petrie, J.R.; Zhou, X.R.; Mansour, M.P.; Blackburn, S.I.; Green, A.G.; Singh, S.P.; Nichols, P.D. Isolation and characterisation of a delta5-fatty acid elongase from the marine microalga Pavlova salina. Mar. Biotechnol. 2009, 11, 410–418. [Google Scholar] [CrossRef]
- Petrie, J.R.; Liu, Q.; Mackenzie, A.M.; Shrestha, P.; Mansour, M.P.; Robert, S.S.; Frampton, D.F.; Blackburn, S.I.; Nichols, P.D.; Singh, S.P. Isolation and characterisation of a high-efficiency desaturase and elongases from microalgae for transgenic LC-PUFA production. Mar. Biotechnol. 2010, 12, 430–438. [Google Scholar] [CrossRef]
- Ohara, J.; Sakaguchi, K.; Okita, Y.; Okino, N.; Ito, M. Two fatty acid elongases possessing C18-Δ6/C18-Δ9/C20-Δ5 or C16-Δ9 elongase activity in Thraustochytrium sp. ATCC 26185. Mar. Biotechnol. 2013, 15, 476–486. [Google Scholar] [CrossRef]
- Tavares, S.; Grotkjær, T.; Obsen, T.; Haslam, R.P.; Napier, J.A.; Gunnarsson, N. Metabolic engineering of Saccharomyces cerevisiae for production of Eicosapentaenoic Acid, using a novel Δ5-Desaturase from Paramecium tetraurelia. Appl. Environ. Microbiol. 2011, 77, 1854–1861. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Mühlroth, A.; Li, K.; Røkke, G.; Winge, P.; Olsen, Y.; Hohmann-Marriott, M.F.; Vadstein, O.; Bones, A.M. Pathways of Lipid Metabolism in Marine Algae, Co-Expression Network, Bottlenecks and Candidate Genes for Enhanced Production of EPA and DHA in Species of Chromista. Mar. Drugs 2013, 11, 4662–4697. [Google Scholar] [CrossRef]
- Li, Y.T.; Li, M.T.; Fu, C.H.; Zhou, P.P.; Liu, J.M.; Yu, L.J. Improvement of arachidonic acid and eicosapentaenoic acid production by increasing the copy number of the genes encoding fatty acid desaturase and elongase into Pichia pastoris. Biotechnol. Lett. 2009, 31, 1011–1017. [Google Scholar] [CrossRef]
- Halpin, C. Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotechnol. J. 2005, 3, 141–155. [Google Scholar] [CrossRef]
- Zhu, S.; Li, Y.; Vossen, J.H.; Visser, R.G.; Jacobsen, E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012, 21, 89–99. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Huang, Y.S.; Chaudhary, S.; Thurmond, J.M.; Bobik, E.G.; Yuan, L.; Chan, G.M. Cloning of Δ12- and Δ6-Desaturases from Mortierella alpina and recombinant production of γ-linolenic acid in Saccharomyces cerevisiae. Lipids 1999, 34, 649–659. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).